Abstract
Herpes simplex virus type 1 (HSV-1) virions, like those of all herpesviruses, contain a proteinaceous layer termed the tegument that lies between the nucleocapsid and viral envelope. The HSV-1 tegument is composed of at least 20 different viral proteins of various stoichiometries. VP22, the product of the UL49 gene, is one of the most abundant tegument proteins and is conserved among the alphaherpesviruses. Although a number of interesting biological properties have been attributed to VP22, its role in HSV-1 infection is not well understood. In the present study we have generated both a UL49-null virus and its genetic repair and characterized their growth in both cultured cells and the mouse cornea. While single-step growth analyses indicated that VP22 is dispensable for virus replication at high multiplicities of infection (MOIs), analyses of plaque morphology and intra- and extracellular multistep growth identified a role for VP22 in viral spread during HSV-1 infection at low MOIs. Specifically, VP22 was not required for either virion infectivity or cell-cell spread but was required for accumulation of extracellular virus to wild-type levels. We found that the absence of VP22 also affected virion composition. Intracellular virions generated by the UL49-null virus contained reduced amounts of ICP0 and glycoproteins E and D compared to those generated by the wild-type and UL49-repaired viruses. In addition, viral spread in the mouse cornea was significantly reduced upon infection with the UL49-null virus compared to infection with the wild-type and UL49-repaired viruses, identifying a role for VP22 in viral spread in vivo as well as in vitro.
Herpes simplex virus type 1 (HSV-1) virions, like those of all herpesviruses, are composed of a nucleocapsid harboring the double-stranded linear DNA genome, a proteinaceous layer surrounding the nucleocapsid termed the tegument, and a host-derived lipid membrane envelope that contains viral glycoproteins. The tegument layer is a unique feature among herpesviruses and is composed of at least 20 different viral proteins of various stoichiometries. Tegument proteins have been shown to play a variety of roles in infection including the regulation of viral and host gene expression and the promotion of virus assembly and egress (6, 19, 34, 39). Because tegument proteins enter the cell upon fusion of the viral envelope with the host cell membrane, they can potentially exert their activities prior to viral gene expression to provide herpesviruses an advantage early in infection.
VP22, encoded by the UL49 gene, is one of the most abundant HSV-1 tegument proteins, with an average of 2,000 copies present in each virion (18, 22). VP22 is conserved among the alphaherpesviruses and has been studied in HSV-1, herpes simplex virus type 2 (HSV-2), bovine herpesvirus, and pseudorabies virus (PRV). Localization studies suggest that this protein is dynamically trafficked, as VP22 is present within both the cytoplasm and nucleus at different times in infection (16, 36). Although VP22 is present in infected cells in both phosphorylated and nonphosphorylated isoforms, only hypophosphorylated VP22 is incorporated into virions (4, 17, 21, 24, 30, 35, 36). In addition, VP22 has been shown to induce the stabilization and hyperacetylation of microtubules (15) and to interact with a number of proteins including histones H1 and H4 (40), the viral glycoproteins gE, gM, and gD (7, 20), and the viral transactivator of immediate-early gene expression, VP16 (14).
Despite these interesting observations, the role of VP22 in HSV-1 infection remains unclear. An HSV-1 recombinant virus that expresses a truncated form of VP22 (HSV-1 RF177) expresses low levels of the amino-terminal 212 residues of the 301-residue VP22 (37). The truncated form of VP22 expressed by RF177 showed normal cellular localization and was incorporated into extracellular virions. In addition, single-step growth kinetics, capsid assembly, and viral egress of RF177 were nearly indistinguishable from those of wild-type HSV-1. However, RF177 viral plaques were reduced in size by nearly 40% at 48 h postinfection compared to plaques produced by the wild-type virus, suggesting a role for VP22 in HSV-1 cell-cell spread. Recently, an HSV-1 UL49-null mutant was constructed and described (13). Although VP22 was found to alter the expression, localization, and virion incorporation of the viral transactivator ICP0, no difference in plaque size was reported. Thus, the role(s) of VP22 in HSV-1 infection, and specifically in viral spread, requires further study.
To determine the contribution of VP22 to HSV-1 infection, we generated HSV-1 UL49-null and UL49-repaired viruses and characterized their growth relative to the wild-type parental virus in both cultured cells and the mouse cornea. Single-step growth analyses indicated that VP22 was not required for efficient virus assembly while multistep growth analyses, plaque measurements, and mouse corneal spread assays showed that VP22 was required for efficient viral spread. Interestingly, the absence of VP22 did not appear to affect either intracellular virus production or cell-cell spread but decreased extracellular virus at least 90%. Thus, the defect in viral spread associated with the UL49-null virus is due to decreased secondary infection by an extracellular route.
MATERIALS AND METHODS
Viruses and cells.
Viral stocks of wild-type HSV-1F, the UL49 deletion virus, and the UL49 repair virus were propagated exclusively on V49 cells, a Vero-derived cell line that constitutively expresses VP22 (37). Vero and V49 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 4.0 mM l-glutamine, 4.5 g/liter glucose, 125 U/ml penicillin, 0.125 mg/ml streptomycin, and either 10% newborn calf serum or 10% fetal bovine serum, respectively.
Plasmid construction.
The recombinant plasmid used to generate the UL49R virus by shuttle mutagenesis was constructed as follows. First, the ApoI-BamHI segment (HSV-1 bp 104768 to 107022) of the BamHI F-fragment of HSV-1F was subcloned from pRB128 (a gift from Bernard Roizman) into pUC19. Next, the ApoI site was replaced with a BamHI site using the ApoI/BamHI linker 5′-AATTGGATCC-3′ and standard cloning methods. This fragment was then released via BamHI digest and cloned into the BamHI site of pST76K_SR. The shuttle mutagenesis plasmid pST76K_SR contains a Kanr gene for positive selection upon transformation, a recA gene for recombination in Escherichia coli, a temperature-sensitive origin of replication for selection of cointegrates, and an sacB sucrose-sensitivity gene for selection of cointegrate resolution (1, 5). The resulting pST76K_SR-derived shuttle mutagenesis plasmid carrying the UL49 gene with flanking arms for recombination was named pJB386.
Construction of WT, UL49 deletion, and UL49 repair viruses.
HSV-1(F) (wild type [WT]), the UL49 deletion virus (UL49−), the UL49 repair virus (UL49R), and the UL49.5 deletion virus (UL49.5−) were constructed using the HSV-1F bacterial artificial chromosome (BAC) pYEbac102 (42) (Fig. 1; see also Fig. 3). The UL49− BAC, in which the entire UL49 open reading frame (ORF; HSV-1 bp 105480 to 106396) (29) was replaced with an FRT-BamHI site, was generated according to the method of Lee et al. (25) as follows. E. coli EL250 cells, which harbor (i) a defective λ prophage in which the λ exo, bet, and gam recombination and nuclease-inhibition genes are under the control of the λ cI857 temperature-sensitive repressor and (ii) an arabinose-inducible flpe recombinase gene, were transformed with pYEbac102. A chloramphenicol-resistant (Cmr) clone that maintained the full-length HSV-1(F) genome as a BAC was identified and used for the mutagenesis. A linear recombination fragment containing a Kanr gene flanked by Flp recognition target (FRT) sites, one BamHI site, and short sequences homologous to regions up- and downstream of the UL49 ORF was generated by PCR and gel purified. pKD13, a plasmid containing a FRT-Kanr-FRT-BamHI cassette (9) was used as a template for the PCR. The primers used for the PCR were 5′-ACGCAACGCCAACACCGAATGAACCCCTGTTGGTGCTTTATTGTCTGGGTACGGAATTCCGGGGATCCGTCGAC-3′ and 5′-ACCCAGGCCTAATTGTCCGCGCATCCGACCCTAGCGTGTTCGTGCGTGTAGGCTGGAGCTGCTTC-3′ (Integrated DNA Technologies, Coralville, IA); nucleotides in italics are homologous to the BAC target sequences up- and downstream, respectively, of UL49; nucleotides in Roman type are homologous to the FRT-Kanr-FRT-BamHI template cassette of pKD13; and nucleotides in boldface indicate the BamHI site. To prepare E. coli EL250 cells carrying pYEbac102 for electroporation, liquid cultures were grown at 30°C in Luria-Bertani (LB) medium containing 30 μg/ml chloramphenicol (CHL) to an optical density at 600 nm of 0.6; cultures were temperature shifted to 42°C for 15 min to induce Exo, Beta, and Gam expression and then chilled on ice, washed three times with 10% ice-cold glycerol, and concentrated 500-fold. A total of 75 μl of competent cells was transformed via electroporation (2.3 kV, 200 Ω, and 25μF) with ∼500 ng of the gel-purified recombination fragment. Following growth at 30°C on LB agar plates containing 30 μg/ml CHL and 50 μg/ml kanamycin (KAN), Cmr Kanr recombinants were genotypically verified by restriction fragment length polymorphism (RFLP) of alkaline lysis-extracted BAC DNA. Next, the Kanr gene was excised through Flp-mediated recombination between the flanking FRT sites after induction of Flp recombinase by growth in 0.1% arabinose overnight at 30°C. Kanamycin-sensitive clones were identified through replica plating on LB agar plates containing either CHL (30 μg/ml) or KAN (50 μg/ml), and the BACs were again genotypically verified by RFLP of alkaline lysis-extracted BAC DNA. The resulting BAC was named pYEbac102-UL49−.
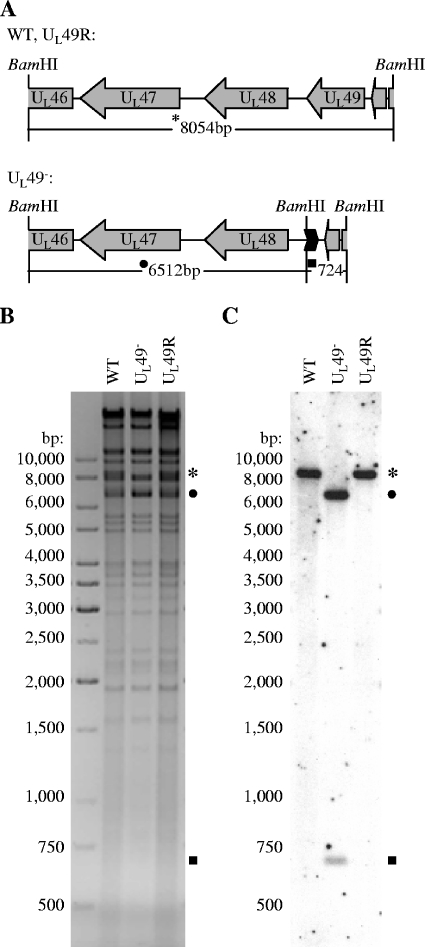
FIG. 1.
Genetic analysis of WT, UL49−, and UL49R viral DNAs. (A) Schematic representation of the BamHI F fragment in WT and UL49R (upper) and UL49− (lower) viral genomes. Replacement of the UL49 gene with a FRT-BamHI cassette in the UL49− viral genome yielded 6,512-bp and 724-bp BamHI fragments in place of the 8,054-bp BamHI F-fragment present in the WT and UL49R viral genomes. (B) Negative scanned image of an agarose gel containing BamHI-digested WT, UL49−, and UL49R viral DNAs visualized with ethidium bromide staining. (C) Scanned autoradiograph of Southern DNA hybridization of the same gel in which [α-32P]dCTP-labeled HSV-1 BamHI F-fragments were used as probes.
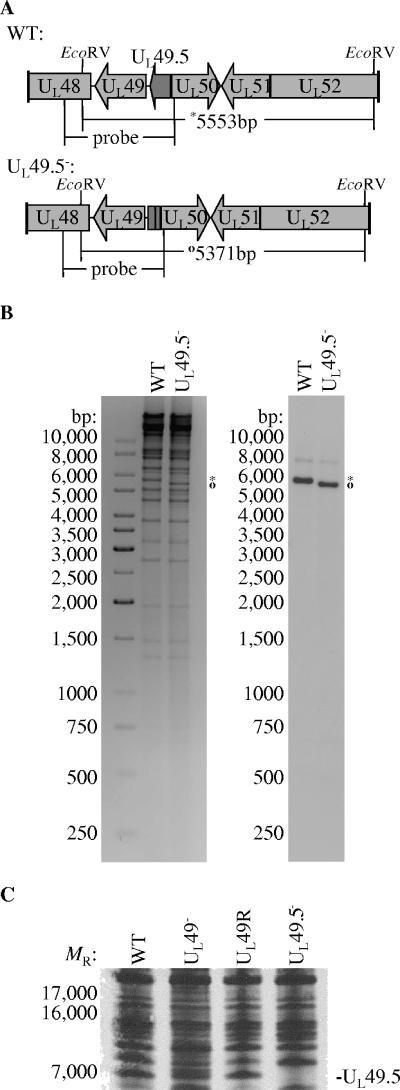
FIG. 3.
Analysis of UL49.5 expression upon infection with the WT, UL49−, and UL49R viruses. (A and B) Genetic analysis of UL49.5− viral DNA. (A) Deletion of nucleotides encoding UL49.5 residues 9 to 22 in the UL49.5− viral genome yielded a 5,371-bp EcoRV fragment (○) in place of the 5,553-bp EcoRV fragment (*) present in the WT viral genome. (B) One-percent agarose gel electrophoresis of EcoRV-digested WT and UL49.5− viral DNAs visualized with ethidium bromide staining (negative image, left) and Southern hybridization (right) using the ApoI-BamHI segment of the HSV-1 BamHI F-fragment labeled with [α-32P]dCTP as a probe. (C) Scanned autoradiographic image of a 16% Tricine-SDS-polyacrylamide gel containing electrophoretically separated [35S]methionine-cysteine-labeled proteins from lysates of cells infected with WT, UL49−, UL49R, and UL49.5− viruses. The location of the UL49.5 gene product in the WT, UL49−, and UL49R lanes is indicated.
The UL49R BAC was constructed by reintroducing the UL49 ORF into pYEbac102-UL49− in its native position using a RecA-based method of shuttle mutagenesis (5, 31). First, pYEbac102-UL49− DNA was electroporated into E. coli DH10B cells as above, and a Cmr clone was verified genotypically by RFLP. Next, E. coli DH10B cells harboring pYEbac102-UL49− were transformed with 200 ng of the shuttle plasmid pJB386 (see above) and incubated at 30°C on a bacterial shaker in LB medium. Cells were plated on LB agar containing CHL (30 μg/ml) and KAN (50 μg/ml) and incubated overnight at 42°C to select for integration of the shuttle plasmid into pYEbac102-UL49− via one of the UL49 flanking regions. Cointegrates were genotypically verified via RFLP of BAC DNA extracted from liquid cultures grown at 42°C in LB containing 30 μg/ml CHL and 50 μg/ml KAN. To resolve the cointegrates, positive clones were grown in LB plus CHL (30 μg/ml) for 4 h at 37°C, sucrose was then added to a final concentration of 10% (wt/vol), and the cultures were shaken for another 4 h at 37°C. Cells were plated on LB agar containing 30 μg/ml CHL and 10% sucrose and incubated overnight at 30°C to select for loss of the shuttle plasmid. Kanamycin-sensitive, chloramphenicol-resistant clones identified through replica plating on LB agar plates containing either 50 μg/ml KAN or 30 μg/ml CHL were genotypically verified through RFLP of alkaline lysis-extracted BAC DNA. The resulting BAC was named pYEbac102-UL49R.
The UL49.5− BAC, in which HSV-1 bp 106746 to 106928 were deleted, was generated according to the method of Tischer et al. (43) as follows. A linear recombination fragment containing a Kanr gene, an I-SceI endonuclease site, and short sequences homologous to regions up- and downstream of HSV-1F bp 106746 and 106928 was generated by PCR and gel purified. The primers used for the PCR were 5′-ACACAGGGCGGGTTCAGGCGTGCCCGGCAGCCAGTAGCCTGCCGCTAAGGCGACGAGCAATAGGGATAACAGGGTAATCGATTT-3′ and 5′-GGGCGGGCCTGTTGTTTGTCTTGCTCGTCGCCTTAGCGGCAGGCTACTGGCTGCCGGGCAGCCAGTGTTACAACCAATTAACC-3′ (Integrated DNA Technologies, Coralville, IA); nucleotides in italics are homologous to the BAC target sequences upstream of HSV-1(F) bp 106746, nucleotides in Roman type are homologous to the BAC target sequences downstream of HSV-1F bp 106928, and nucleotides in boldface are homologous to the Kanr-I-SceI template cassette. E. coli EL250 cells carrying pYEbac102 were transformed via electroporation (2.3 kV, 200 Ω, 25μF) with ∼200 ng of the gel-purified recombination fragment. Following growth at 30°C on LB agar plates containing 30 μg/ml CHL and 50 μg/ml KAN, Cmr Kanr recombinants were genotypically verified by RFLP of alkaline lysis-extracted BAC DNA. A genotypically correct clone was transformed with pBAD (43) which encodes an arabinose-inducible I-SceI restriction endonuclease, and transformants were selected via growth at 30°C on LB agar plates containing 30 μg/ml CHL, 50 μg/ml KAN, and 100 μg/ml ampicillin. Next, the Kanr gene was excised through Red-mediated recombination following induction of I-SceI with 0.5% arabinose. Kanamycin-sensitive clones were identified through replica plating on LB agar plates containing either CHL (30 μg/ml) or KAN (50 μg/ml), and the BACs were again genotypically verified by RFLP of alkaline lysis-extracted BAC DNA. The resulting BAC was named pYEbac102-UL49.5−.
WT, UL49−, UL49R, and UL49.5− viruses were generated from pYEbac102, pYEbac102-UL49−, pYEbac102-UL49R, and pYEbac102-UL49.5−, respectively, as follows. Flasks (25 cm2) of V49 cells (for the WT, UL49−, and UL49R viruses) or Vero cells (for the UL49.5− virus) at ∼80% confluence were cotransfected with ∼300 ng of alkaline lysis-extracted BAC DNA and ∼2.5 μg of a Cre recombinase expression vector (pCAGGS-nlsCre; a gift from Michael Kotlikoff, Cornell University) using Superfect transfection reagent (QIAGEN, Valencia, CA). Cre recombinase expressed in BAC-transfected cells mediated excision of the BAC vector sequences via recombination between the flanking loxP sites. Viral plaques were purified and propagated on either V49 cells for the WT, UL49−, and UL49R viruses or Vero cells for the UL49.5− virus.
Viral DNA analyses.
Viral DNAs were purified from filled capsids as follows. Vero cells in a 175-cm2 flask were infected with WT, UL49−, or the UL49R virus at a multiplicity of infection (MOI) of 5 and held at 34°C until a cytopathic effect was visible in ∼80% of the cells. Cells were washed with phosphate-buffered saline (PBS), pelleted, resuspended in 2.5 ml of Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and allowed to swell on ice. Cells were lysed by the addition of NP-40 to 1% and spun to pellet cellular debris. The supernatant containing DNA-filled capsids was transferred to a new tube, and viral DNA was released by incubation at 37°C for 30 min in the presence of 0.5 mg/ml proteinase K and 1% sodium dodecyl sulfate (SDS). Viral DNA was then extracted four times with phenol-chloroform-isoamyl alcohol (48:48:4), precipitated with sodium acetate and ethanol, and resuspended in water.
The genetically manipulated regions of the UL49− and UL49R genomes were amplified by PCR from purified viral DNA, and nucleotide sequencing was performed to confirm that the desired mutations had been introduced.
Purified viral DNAs were digested with BamHI or EcoRV, separated by electrophoresis through a 1% agarose gel, visualized with ethidium bromide, and transferred to Hybond-N+ nylon membrane (Amersham Biosciences, Piscataway, NJ). Probes used in Southern hybridization were generated from the HSV-1 BamHI F-fragment (HSV-1 bp 98967 to 107022) or the ApoI-BamHI segment of the HSV-1 BamHI F-fragment (HSV-1 bp 104768 to 107022) by [α-32P]dCTP nick translation labeling. Southern hybridizations were performed according to standard methods.
35S-labeled protein analysis.
35S-labeled cell lysates were prepared by infection of 25-cm2 flasks of Vero cells with WT, UL49−, UL49R, or UL49.5− viruses at an MOI of 5 at 37°C. From 8 to 12 h postinfection (hpi), the medium overlying each infected flask was replaced with Dulbecco's modified Eagle's medium (high glucose, plus pyridoxine hydrochloride, no l-glutamine, no l-methionine, and no l-cysteine) supplemented with 200 μCi of Trans-35S label ([35S]methionine-cysteine). At 12 hpi cells were collected, washed with PBS, resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (50 mM Tris-Cl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol), heated at 56°C for 10 min, and sonicated briefly. Labeled lysates were separated by electrophoresis through a Tricine-SDS-16% polyacrylamide gel and transferred to a 0.2-μm-pore-size polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA). Labeled proteins were visualized by autoradiography on Pierce CL-X Posure film (Pierce Biotechnology, Rockford, IL).
Virion preparations.
Extracellular virions were prepared from clarified overlying medium of Vero cells infected with WT, UL49−, or UL49R viruses at an MOI of 0.1 at 37°C for 48 h. Virions were pelleted through a 30% sucrose cushion, resuspended in 0.5 ml of 10 mM Tris-Cl (pH 7.4), and separated through a 14% to 26% Dextran T10 (Amersham Pharmacia Biotech, Uppsala, Sweden) gradient. Gradients were fractionated and titrated on Vero cells to determine the PFU/ml of each fraction and probed with an anti-VP5 antibody via immunoblotting (see below) to determine the relative amount of VP5 present in each fraction.
Intracellular virions were prepared from Vero cells infected with WT, UL49−, or UL49R viruses at an MOI of 0.1 at 37°C for 36 h. Cells were pelleted, washed with PBS, and frozen at −80°C. Cells were later thawed, resuspended in 1.0 mM PO4(Na) (pH 7.4), and lysed by dounce homogenization. Sucrose [1.25 M in 1.0 mM PO4(Na), pH 7.4] was added to stabilize nuclei, which were then pelleted by centrifugation. Virions present in the supernatant were pelleted through a sucrose cushion and separated through a Dextran T10 gradient as for extracellular virions. Virions observed as a light-scattering band were removed by needle and syringe, resuspended in 10 mM Tris-Cl (pH 7.4), and pelleted by centrifugation. The virion pellet was resuspended in SDS-PAGE sample buffer and boiled for 5 min.
Immunoblot analyses.
Cell lysates were prepared by infection of 25-cm2 flasks of Vero cells with WT, UL49−, or UL49R viruses at an MOI of 5 at 37°C. At 18 hpi, cells were collected, washed with PBS, boiled in SDS-PAGE sample buffer, and sonicated briefly. Cell lysates and virion preparations were separated by SDS-12% PAGE, and the proteins were transferred to nitrocellulose or polyvinylidene difluoride membranes. Free binding sites on the membranes were blocked by 10% skim milk in PBS before the addition of either anti-VP22 (4) (diluted 1:5,000), anti-VP16 (sc-17547; Santa Cruz Biotechnology, Santa Cruz, CA) (diluted 1:500), anti-pUL28 (3) (diluted 1:1,000), anti-VP5 (8) (diluted 1:3,000), anti-ICP0 (Abcam, Cambridge, MA) (diluted 1:5,000), anti-gE (23) (diluted 1:2,500), anti-gD (Rumbaugh-Goodwin Institute for Cancer Research, Plantation, FL) (diluted 1:1,000), or anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) (diluted to 0.5 μg/ml) antibodies. After extensive washing to remove unbound antibody, bound antibodies were detected with anti-rabbit (Amersham Biosciences, Piscataway, NJ) or anti-goat (Jackson Immunoresearch Laboratories, West Grove, PA) immunoglobulin G conjugated to horseradish peroxidase and visualized by ECL (Amersham Biosciences, Piscataway, NJ) chemiluminescence on Pierce CL-X Posure film (Pierce Biotechnology, Rockford, IL).
Plaque area determination in cultured cells.
Vero cells were grown in 35-cm2 dishes, infected with WT, UL49−, or UL49R viruses at 10 PFU/dish, overlaid with medium 199 containing 1% newborn calf serum (medium 199V), and held at 37°C for 44 h. Cells were fixed with 90% acetone and analyzed by indirect immunofluorescence using a rabbit polyclonal anti-gM primary antibody (2) and a fluorescein isothiocyanate-conjugated anti-rabbit secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA). Plaques were visualized under a fluorescence microscope (Zeiss Axiovert 25), and 70 plaques from each virus were photographed with a digital camera (Zeiss AxioCam HRc). Plaque areas were determined using ImageJ software available from the National Institutes of Health (http://rsb.info.nih.gov/ij/index.html). Statistical analyses (two-tailed t tests) were performed using Microsoft Excel software.
Single-step and multistep growth analyses.
To examine replication kinetics, Vero cells were infected with WT, UL49−, or UL49R viruses at an MOI of 5 (single-step growth analyses) or 0.001 (multistep growth analyses) and incubated at 37°C for 1 h to allow for virus adsorption. Cells were then washed twice with citrate buffer (135 mM NaCl, 10 mM KCl, 40 mM citric acid [pH 3.0]) and once with an excess of medium 199V to neutralize and remove unbound virus. Fresh medium 199V was added to the cells, and the infections were held at 37°C. At the indicated times postinfection, the medium containing extracellular virus was removed, clarified, and titrated on Vero cells. Infected cells were frozen immediately at −80°C. At a later date, the cells were thawed, scraped into 0.5-ml thrice-autoclaved milk, and briefly sonicated; virus was titrated on Vero cells to determine intracellular growth kinetics.
To study the kinetics of intracellular multistep growth in the absence of secondary infections initiated by virus released to the medium, 199V medium containing 0.3% human immunoglobulin (Sigma-Aldrich, St. Louis, MO) was added to cells following infection and citrate buffer-199V medium washes. At the indicated times postinfection, cells were scraped, pelleted, and washed three times with PBS to remove the neutralizing human immunoglobulins. Cell pellets were resuspended in 0.5 ml of thrice-autoclaved skim milk, briefly sonicated, and frozen at −80°C. At a later date, virus in the resuspended pellets was titrated on Vero cells.
Particle-PFU analyses.
Extracellular virions were prepared from clarified overlying medium of Vero cells infected with WT, UL49−, or UL49R viruses at an MOI of 0.1 at 37°C for 48 h. Virions were pelleted through a 30% sucrose cushion, resuspended in TBSal (10 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2.6 mM KCl, 20 mM MgCl, 1.8 mM CaCl), and briefly sonicated. Part of each virion preparation was titrated on Vero cells to determine the PFU/ml. Part of each virion preparation was diluted in ultrapure H2O, mixed with a known quantity of 0.3-μm polystyrene latex beads (Ladd Research, Williston, VT), and spotted and dried on Formvar and carbon-coated nickel grids. After drying, the samples were stained with 2% aqueous uranyl-acetate and viewed with a Philips 201 transmission electron microscope. Viral particles and latex beads were counted, with the beads serving as an internal control, and the number of viral particles/ml of preparation was calculated. The number of viral particles/PFU was calculated from the number of viral particles/ml and number of PFU/ml of each virion preparation.
Corneal viral spread analyses.
All procedures involving animals adhered to the Society for Neuroscience Guidelines for the Use of Animals in Research and the guidelines of the University of California San Francisco Committee on Animal Research. Male BALB/c mice 6 to 8 weeks old were anesthetized with an intraperitoneal injection of Avertin (28), followed by topical corneal administration of 0.05% proparacaine and 1% atropine in the eye. The eyes were blotted with sterile cotton, and immediately the corneas were scarified in a grid pattern of 10 horizontal and 10 vertical strokes with a 27-gauge needle. A 5-μl drop of virus in minimal essential medium containing 106 PFU of virus was applied to the cornea. Anesthesia was maintained for 20 to 30 min following inoculation to allow for viral adsorption. The animals were monitored daily and showed no behavioral signs of infection.
The mice were allowed to survive 24 (n = 3), 48 (n = 3), or 72 (n = 3) h after infection. They were reanesthetized with Avertin and killed by intracardiac perfusion with normal saline. The corneas were dissected and prepared for immunohistochemistry according to standard procedure (33). In brief, the corneas were first incubated overnight in blocking solution composed of 3% normal goat serum, 0.1% Triton-100 in PBS (pH 7.2). They were incubated overnight in primary antiserum (horseradish peroxidase-conjugated polyclonal antiserum raised in rabbits against human HSV-1; 1:100 dilution) (Accurate Chemical and Scientific Corp., Westbury, N.Y.), and the presence of HSV antigens was determined with nickel-enhanced diaminobenzine. The corneas were flat mounted on glass slides, and viral spread was estimated using digitized photographs of corneal whole mounts at ×25 magnification and ImageJ software. For each measurement the distance from the edge of the scratch to the farthest boundary of contiguous infected cells was traced. Measurements were collected from at least five fields for each eye.
RESULTS
Generation of UL49 deletion and repair viruses.
To identify the role(s) of VP22 during HSV-1 infection, we generated both a UL49-null virus (UL49−) that lacks the entire UL49 ORF and a repair virus (UL49R) in which the UL49 ORF is restored using an HSV-1F BAC system (42) as described in Materials and Methods. The deletion in the UL49-null virus spanned bp 105480 to 106396, a region from 5 bp before the start codon to 5 bp after the stop codon of the UL49 ORF. This entire sequence was restored to its original location in the UL49R virus.
When propagated on V49 cells and analyzed on Vero cells, the UL49− plaque phenotype (see below) remained consistent. However, when the UL49− virus was propagated on Vero or rabbit skin cells, some of the plaques became dramatically larger within the first two to three passages (http://www.vet.cornell.edu/labs/baines/). Therefore, all of the present studies were performed with virus stocks propagated on the complementing V49 cell line.
To verify the expected genotypes of the recombinant viruses, viral DNA was purified, digested with BamHI, and electrophoretically separated through a 1% agarose gel. The digested DNAs were transferred to a nitrocellulose membrane and probed with [α-32P]dCTP-labeled BamHI F-fragment derived from WT HSV-1(F). Figure 1A shows the expected size of the UL49-containing BamHI F-fragment of the WT and UL49R viruses compared to that of the UL49− virus in which the UL49 ORF was replaced by an FRT-BamHI site. Figures 1B and 1C show the ethidium bromide-stained agarose gel and Southern hybridization, respectively, of BamHI-digested viral DNA. As expected, the BamHI F-fragment (predicted size of 8,054 bp) was present in the digested WT and UL49R viral DNAs and hybridized with the BamHI F-fragment probe (Fig. 1B and C, asterisks). In contrast, BamHI fragments of approximately 6,500 bp (filled circle) and 700 bp (filled square) present in the digested UL49− viral DNA hybridized with the BamHI F-fragment probe. These results were consistent with the expected BamHI fragment sizes (6,512 bp and 724 bp) of the UL49 deletion as designed. To ensure that the nucleotide sequences of the UL49− and UL49R viruses were correct, the genetically manipulated regions of the genomes were amplified from purified viral DNA by PCR and sequenced. All nucleotide sequences were found to be correct as designed.
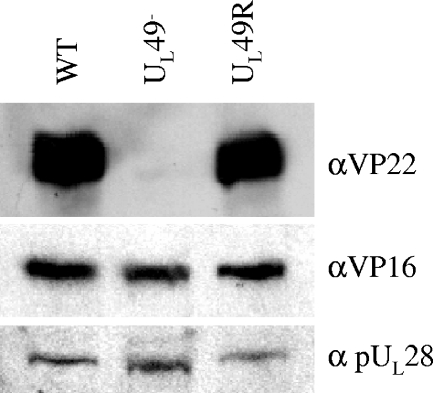
To confirm the deletion and repair of the UL49 ORF, the expression of VP22 in cells infected with the recombinant viruses was examined. Vero cells were infected at an MOI of 5 with WT, UL49−, or UL49R virus and then collected at 18 hpi and lysed. Proteins within the lysates were analyzed by immunoblotting with an anti-VP22 polyclonal antibody (4). As expected, VP22 was present in lysates of WT- and UL49R-infected cells but absent from lysates of UL49−-infected cells (Fig. 2, top).
FIG. 2.
Immunoblot analysis of VP16 expression upon infection with the WT, UL49−, and UL49R viruses. Lysates of Vero cells infected with the WT, UL49−, or UL49R viruses for 18 h were separated by SDS-PAGE and probed for the presence of VP22 (top) and VP16 (middle). As a loading control, the immunoblot was also probed with an antibody directed against pUL28 (bottom).
To ensure that the deletion of UL49 did not affect expression of the UL48 gene product VP16, immunoblots of the above lysates were probed with an anti-VP16 polyclonal antibody (Fig. 2, middle). As a loading control, these immunoblots were also probed with a polyclonal antibody directed against another late protein encoded by the UL28 gene (Fig. 2, bottom). VP16 was expressed in approximately equimolar amounts in cells infected with the WT, UL49−, and UL49R viruses, confirming that UL48 expression was unaffected by the deletion of UL49.
Due to the lack of a UL49.5 antibody, we used Tricine-SDS-PAGE separation of 35S-labeled infected cell lysates to ensure that the deletion of UL49 did not affect expression of the UL49.5 gene product. To determine the position of the UL49.5 gene product on the gel, we constructed a UL49.5 deletion mutant and compared 35S-labeled proteins present in lysates derived from cells infected with this mutant to those infected with the WT, UL49−, and UL49R viruses. Construction of the UL49.5 deletion mutant is described in Materials and Methods and Fig. 3. Figure 3C shows that the UL49.5 gene product was present in approximately equimolar amounts in cells infected with the WT, UL49−, and UL49R viruses, confirming that UL49.5 expression was unaffected by the deletion of UL49.
The UL49− virus produces reduced-sized plaques in cultured cells.
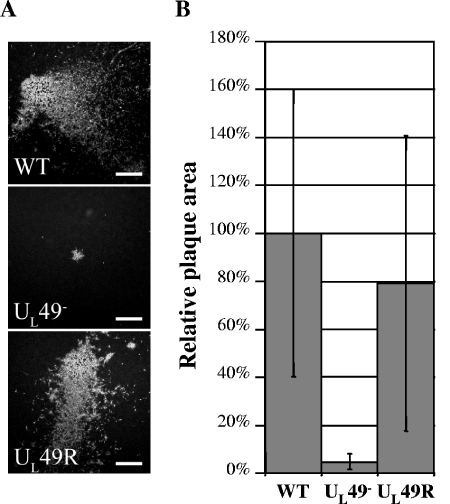
As a first step to examine the growth phenotype of the UL49-null virus, Vero cell monolayers were infected with the WT, UL49−, and UL49R viruses, and plaques were observed 44 h after infection (Fig. 4A). Quantitative analysis of these viral plaques showed that the UL49− virus produced plaques that were, on average, 5% the area of those formed by the WT virus (Fig. 4B). Even with the high inherent variability in plaque size among the plaques examined, this difference was statistically significant (P < 0.000001). By contrast, the UL49R virus produced plaques that were approximately 78% the size of those formed by the WT virus, but this difference was not statistically significant. To determine whether the reduced plaque size observed with the UL49− virus was due to a defect in virus assembly, egress, and/or cell-cell spread, further experiments were performed.
FIG. 4.
Analysis of plaques produced by the WT, UL49−, and UL49R viruses in cultured cells. Vero cell monolayers were infected with the WT, UL49−, or UL49R viruses at an MOI of ∼1 × 10−5 PFU/cell for 44 h. Cells were then fixed and prepared for immunofluorescence microscopy using an antibody directed against the viral protein gM. Plaques were visualized under a fluorescence microscope and photographed with a digital camera. (A) Representative photographs of WT, UL49−, and UL49R viral plaques. Scale bar, 500 μm. (B) Mean areas of 70 UL49− and 70 UL49R plaques relative to the mean area of 70 WT plaques. Error bars represent 1 standard deviation.
Characterization of growth properties of the UL49− virus in cultured cells.
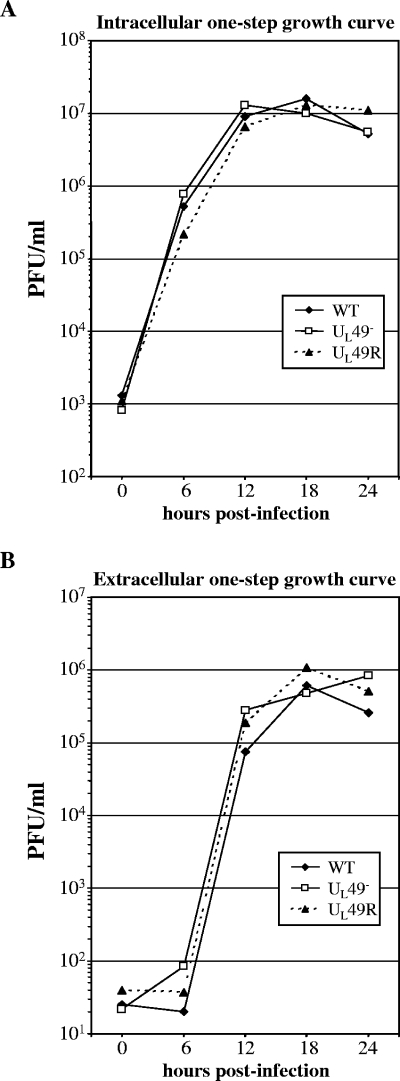
To determine whether the small plaques produced by the UL49− virus are due to a defect in virus assembly, single-step growth analyses were performed. Vero cells infected at an MOI of 5 with the WT, UL49−, and UL49R viruses were collected at various times postinfection and lysed, and intracellular virus was quantified by plaque assay (Fig. 5A). Medium overlaying the infected cell monolayers was clarified and assayed separately (Fig. 5B). Single-step growth kinetics of both intra- and extracellular UL49− virus did not differ significantly from those of either the WT or UL49R viruses. Together, the above data indicate that the reduced plaque size observed with the UL49− virus is not due to a defect in virus assembly.
FIG. 5.
Single-step growth analyses of the WT, UL49−, and UL49R viruses. Vero cell monolayers were infected with the WT, UL49−, and UL49R viruses at an MOI of 5 PFU/cell for 1 h to allow virus adsorption. The cells were then washed extensively with citrate buffer to neutralize and remove unbound virus. The cells were overlaid with medium and held at 37°C. At the indicated times postinfection, the infected cells (A) and the overlaying medium (B) were analyzed separately by plaque assay to determine intracellular and extracellular viral yields, respectively.
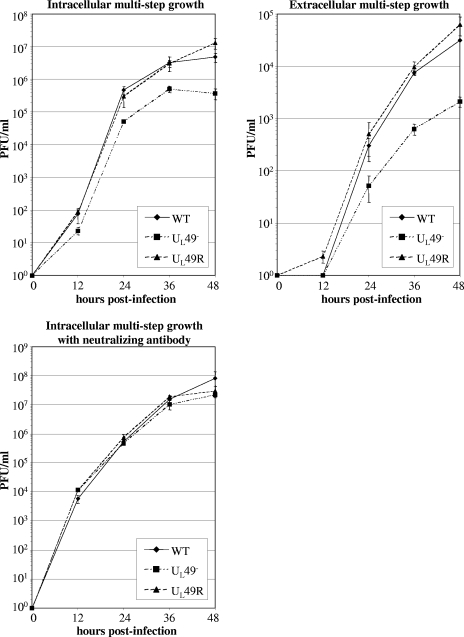
Many viral growth defects are subtle enough to only be observed after multiple rounds of infection. Also, viral growth initiated by high MOIs, such as in the above experiment, is measured in the presence of some amount of VP22 delivered from the infecting virions that were propagated on a complementing cell line. To minimize such effects and further study the role of VP22 in viral growth, we performed multistep growth analyses in cells infected with 0.001 PFU/cell. To study extracellular virus as a component of viral spread, we titrated the overlying medium and infected cell monolayers separately. Figure 6A shows that intracellular UL49− virus titers were decreased, up to ∼11-fold at 48 hpi, compared to both the WT and UL49R viruses. Extracellular UL49− virus titers were also decreased, again up to ∼11-fold at 48 hpi, compared to the WT and UL49R viruses as shown in Fig. 6B.
FIG. 6.
Multistep growth analyses of the WT, UL49−, and UL49R viruses in the presence and absence of neutralizing antibody. Vero cell monolayers were infected with the WT, UL49−, and UL49R viruses at an MOI of 0.001 PFU/cell for 1 h to allow virus adsorption. The cells were then washed extensively with citrate buffer to neutralize and remove unbound virus, overlaid with medium, and held at 37°C. Cells infected for intracellular multistep growth analysis in the presence of neutralizing antibody were overlaid with medium containing 0.3% human gamma globulins. At the indicated times postinfection, these cells were pelleted, washed three times with PBS to remove the neutralizing antibody, and lysed. Also at the indicated times postinfection, medium was removed from cells grown in the absence of neutralizing antibody and clarified. Intracellular virus was titrated from the cell lysates and extracellular virus was titrated from the clarified medium. The growth curves shown represent the means and standard deviations (error bars) of three independent experiments. Growth curves in the upper panels were performed at different times using different passages of Vero cells than those of the lower panel. Therefore, titers between upper and lower panels are not comparable.
Virion release is decreased in the absence of VP22.
The decrease in UL49− intra- and extracellular multistep growth could be due to one or more of the following mechanisms: (i) decreased release of UL49− virions into the medium, resulting in decreased secondary infection and spread through the cell monolayer; (ii) decreased infectivity of individual extracellular UL49− virions, resulting in decreased secondary infection; (iii) decreased or defective virus assembly within UL49−-infected cells, resulting in fewer intra- and extracellular infectious particles; and (iv) decreased spread from infected cells to adjacent cells (cell-cell infection), resulting in decreased spread through the monolayer. To differentiate between these possibilities, we performed multistep intracellular growth analyses in the presence of neutralizing antibody in the overlaying medium. This treatment dramatically reduces extracellular virus and therefore focuses the analysis onto viral spread in the absence of secondary infections from extracellular particles. As shown in Fig. 6C, when extracellular virus was eliminated from multistep growth, the UL49− virus generated intracellular titers approximately equivalent to those of the WT and UL49R viruses. Thus, the above data show the viral spread defect observed upon UL49− virus infection was primarily due to decreased viral release and/or decreased infectivity of extracellular UL49− particles rather than to decreased virus assembly or cell-cell spread.
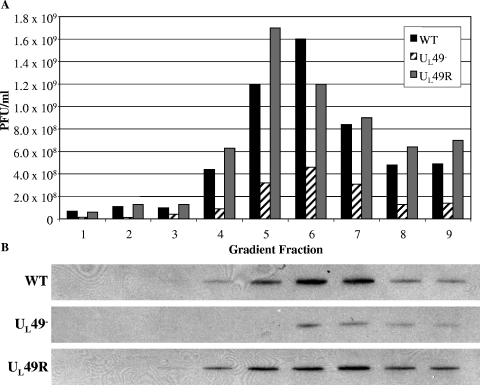
To determine whether the viral spread defect associated with the UL49− virus was due to decreased viral release, decreased infectivity of extracellular UL49− particles, or both, we compared the infectivity of purified WT, UL49−, and UL49R extracellular particles. As shown in Fig. 7A, fractions from gradients containing virions purified from the clarified overlying medium of infected cells contained substantially fewer UL49− PFU than either WT or UL49R PFU. Immunoblot analyses of these gradient fractions using an antibody against the major capsid protein, VP5, showed that the UL49− gradient fractions also contained less VP5 (Fig. 7B), indicating that UL49− viral infections produce fewer extracellular particles as well as fewer extracellular PFU compared to WT or UL49R infections. To further compare the infectivity of extracellular UL49− virions to extracellular wild-type and UL49R virions, we performed particle-PFU analyses. The particle-to-PFU ratios of purified WT, UL49−, and UL49R extracellular virions were 234:1, 550:1, and 418:1, respectively; these ratios were not readily distinguishable within the high inherent variability of this technique. These findings indicate that extracellular UL49− particles are not measurably less infectious than WT or UL49R extracellular particles and, together with the above data on multistep viral growth in the presence and absence of neutralizing antibody, show that the spread defect associated with the UL49− virus is largely attributable to decreased viral release.
FIG. 7.
Analysis of extracellular WT, UL49−, and UL49R virions. Extracellular virions were purified from the clarified medium overlying cells infected with either the WT, UL49−, or UL49R viruses by gradient centrifugation. (A) The number of PFU/ml in gradient fractions was determined by plaque assay on Vero cells. Fraction 1 represents the bottom of the gradient, and fraction 9 represents the top of the gradient. (B) Immunoblots of the same gradient fractions probed with an antibody against the major capsid protein VP5. Each lane is aligned with the corresponding fraction in panel A.
To correlate the decreased viral release of the UL49-null virus with the effect of extracellular virus on WT HSV-1 spread, we examined the effect of neutralizing antibody on WT plaque size. We found that in the presence of 1% human immunoglobulin, WT HSV-1 produces plaques that are 15% the size of those produced in the absence of neutralizing antibody (data not shown), showing that extracellular virus contributes greatly to HSV-1 plaque size and viral spread.
Protein composition of UL49− virions.
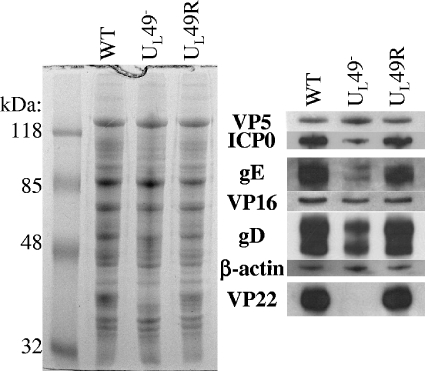
The defective viral release observed upon infection with the UL49− virus could be a direct or indirect consequence of the absence of VP22. Differences in intracellular virion composition could contribute to differences in viral release. Therefore, we investigated whether VP22 contributes indirectly to virion release, through a possible role in virion assembly, by examining the protein composition of WT, UL49−, and UL49R intracellular virions. The left panel of Fig. 8 shows a Coomassie-stained SDS-polyacrylamide gel separation of purified intracellular virions, whereas the right panels show immunoblot analyses of the same samples. The locations of the probed proteins are aligned with their counterparts in the Coomassie-stained gel. The major capsid protein, VP5, was probed as a loading control. Recently, Elliott and others showed that VP22 is absolutely required for the incorporation of ICP0 into virions (13, 38). We found that although ICP0 was decreased in UL49− virions, VP22 was not absolutely required for its incorporation. In HSV-1, VP22 has been shown to interact with both VP16 and gD (7, 14). As previously reported (13), we found that the absence of VP22 had no effect on incorporation of VP16, whereas levels of gD were decreased in UL49− virions compared to WT and UL49R virions. Although PRV VP22 was found to interact with PRV gE, virion incorporation of gE was not affected in a PRV UL49 deletion virus (20). In contrast, we found that levels of gE were dramatically decreased in HSV-1 UL49− virions compared to WT and UL49R HSV-1 virions. Michael et al. (32) recently reported extensive variability in levels of gE present in different PRV virion preparations. However, we found both the levels of gE present in WT and UL49R virions and the relative decrease in gE levels present in UL49− virions to be fairly consistent from virion preparation to preparation (data not shown). Therefore, the relative decrease of gE incorporation into UL49− virions shown in Fig. 8 is a consistent and reproducible aspect of the HSV-1 UL49-null virus phenotype. Finally, virion incorporation of actin was increased in PRV UL49− virions (10, 32) but was not affected in our HSV-1 UL49− virus. The above data indicate that VP22 is involved in, but not required for, incorporation of ICP0, gD, and gE into HSV-1 virions. In addition, the protein composition of VP22− virions varies between HSV-1 and PRV, suggesting that the role(s) VP22 plays in virion assembly varies between the two systems.
FIG. 8.
Analysis of the protein composition of WT, UL49−, and UL49R virions. Intracellular virions were purified from cells infected with either the WT, UL49−, or UL49R viruses. Virions proteins were electrophoretically separated on a 12% SDS-polyacrylamide gel and visualized by either Coomassie staining (left panel) or immunoblot analyses using antibodies against VP5, ICP0, gE, VP16, gD, β-actin, and VP22 (right panels). The immunoblots are aligned with the position of each probed protein in the Coomassie-stained gel.
VP22 is required for efficient viral spread in the mouse cornea.
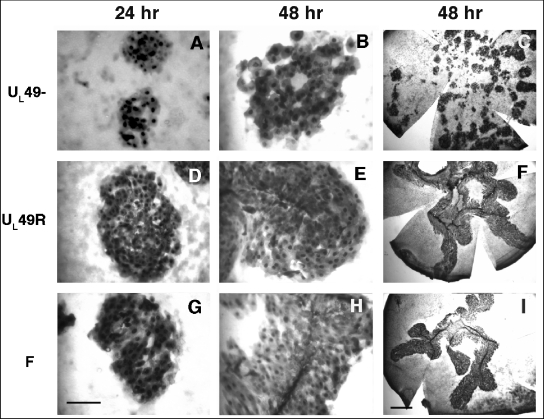
Many HSV-1 proteins exhibit functional redundancy and are therefore deemed nonessential when studied in cultured cells, but they exhibit more dramatic phenotypes in live animals. To determine whether VP22 is essential for viral growth in vivo, we analyzed viral spread in the mouse cornea. Corneas in live mice were scarified and infected with ∼100 PFU of WT, UL49−, or UL49R virus. At 24 or 48 hpi, the mice were sacrificed, the corneas were dissected, and the presence of infectious virus was revealed by immunohistochemistry with an HSV-specific antibody. At both time points the UL49− virus spread only ∼60% of the distance spread by the WT and UL49R viruses (Fig. 9 and Table 1). The far-right panels of Fig. 9 show whole flat-mounted corneas photographed at lower magnification and illustrate the decreased size of corneal lesions produced upon infection with the UL49-null virus (Fig. 9C) compared to the lesions produced by the WT and UL49R viruses (Fig. 9I and F, respectively). The UL49− virus formed isolated rosettes of infected cells, whereas the WT and UL49R viruses formed extensive borders of infected cells along the length of the scarification. These data identify a role for VP22 in HSV-1 viral spread in vivo as well as in cultured cells.
FIG. 9.
Analysis of UL49−, UL49R, and WT HSV-1 viral spread in the mouse cornea. The corneas of live mice were scarified and infected with ∼1,000 PFU of either UL49− (A to C), UL49R (D to F), or WT HSV-1 (G to I) virus. At 24 or 48 hpi, the mice were sacrificed, and the corneas were dissected and prepared for immunohistochemistry using an antibody directed against HSV-1 proteins. Photographs of corneas flat mounted on glass slides are shown with the left and middle panels at the same magnification (bar = 100 μm) and at lower magnification in the right panels (bar = 1 mm).
TABLE 1.
Spread of the WT, UL49-null, and UL49R viruses in corneal epithelium
| Time (hpi) | Mean distance spread ± SD (μm)a
|
||
|---|---|---|---|
| WT (HSV-1F) | UL49− | UL49R | |
| 24 | 179 ± 81.7 (81) | 105.9 ± 75.6 (78) | 171.7 ± 112.2 (70) |
| 48 | 193.8 ± 79.3 (64) | 128.4 ± 81.9 (77) | 206.4 ± 78.0 (64) |
The number of measurements is given in parentheses.
DISCUSSION
Although many studies have focused on identifying the biological properties of HSV-1 VP22 and its homologs in other herpesviruses, the role this protein plays in HSV-1 infection is unclear. To address this problem, we generated two recombinant viruses, one lacking the entire UL49 gene and one containing a restored UL49 gene, and characterized their growth. In this report we show that VP22 is required for efficient viral spread during infection of both cultured cells and the mouse cornea.
Role of VP22 in viral spread in cultured cells.
While single-step intracellular and extracellular growth analyses did not indicate a defect in virus assembly for the UL49-null virus, plaques produced by this virus in cultured cells were reduced in size by an average of 95% compared to the WT and UL49-repaired viruses. A similar, though less dramatic, phenotype was previously observed with an HSV-1 UL49 truncation mutant that expresses low levels of the amino-terminal 212 residues of VP22 (37). This virus, HSV-1 RF177, produced plaques in cultured cells that were reduced in size by nearly 40% compared to those produced by the parental WT virus. It is currently not known whether the reduced plaque size observed with this virus is due to the lack of the carboxy-terminal 89 residues of VP22 or to the low expression level of the truncated protein, and, therefore, we cannot attribute the function of VP22 in viral spread to a particular domain of the protein. In any case, the two studies are in agreement in defining a role for VP22 in viral spread of HSV-1.
Recently, an HSV-1 recombinant virus in which the UL49 gene was replaced with the gene encoding green fluorescent protein was reported (13). Elliott et al. constructed their UL49-null virus on a VP22-complementing cell line because the recombinant virus could not be generated on noncomplementing cells, and it was therefore suggested that the WT virus maintained a growth advantage over the UL49-null virus. However, once the UL49-null virus was generated, the authors observed that VP22 was not necessary for virus growth on noncomplementing Vero cells and, thus, performed all further experiments with virus stocks propagated on noncomplementing cells. A decrease in plaque size was not reported for this virus.
We have also observed that VP22 is dispensable for growth on Vero cells. However, we have noted with interest that while UL49− stocks propagated on VP22-complementing cells consistently produced plaques ∼5% the size of those produced by the WT and UL49R viruses, some plaques produced by the UL49-null virus were comparable in size to those produced by the wild-type virus (http://www.vet.cornell.edu/labs/baines/) after as few as two to three passages on noncomplementing cells. The reproducibility of these observations suggests that the UL49-null virus somehow compensates for the lack of VP22 when propagated on noncomplementing cells. The mechanism of this phenotypic change is unknown but may reflect the acquisition of secondary mutations that compensate for the lack of VP22. Because phenotypic change may further define the function of VP22 during virus infection, as well as identify the basic mechanisms of HSV-1 spread, we are intrigued by this phenotypic morphogenesis, and studies are under way to identify its cause.
In the present work, studies were undertaken to determine the mechanism by which VP22 promotes viral spread. During multistep growth, both intracellular and extracellular UL49− PFU were reduced 11-fold compared to the WT and UL49R viruses. Multistep growth experiments performed in the presence and absence of neutralizing antibody together with VP5 immunoblot-PFU and particle-PFU analyses of UL49− extracellular particle infectivity showed that this reduction was largely attributable to decreased levels of extracellular UL49− virus rather than to a defect in either UL49− virus assembly, cell-cell spread, or particle infectivity. At this time it is unclear whether the decreased extracellular virus observed during UL49− infection is due to a defect in viral exit from infected cells or decreased release of extracellular UL49− virions from intercellular spaces. The answer to this question and the role of VP22 in the mechanism(s) of viral exit and/or intercellular release will be the topics of future studies. Our present work shows that at least in some culture systems and tissue types, extracellular virus contributes greatly to viral spread, and VP22 plays a role in the accumulation of extracellular virus.
VP22 may promote extracellular virus accumulation and spread through a direct or indirect mode of action, and several possibilities exist for either scenario. For example, VP22 may affect the cytoskeleton of infected cells and promote efficient transport of virion-containing vesicles to the cell surface through its ability to stabilize microtubules (15). Alternatively, VP22 may function indirectly through its interactions with and/or optimal incorporation of other viral proteins such as ICP0, gE, or gD into virions (7, 13, 20). Elliott et al. have reported that in the absence of VP22 there is both a delay in the synthesis of the transactivator ICP0 and a complete loss of ICP0 virion incorporation (13). If the decreased viral spread observed upon UL49− infection was due solely to these effects on ICP0, one would expect to observe decreased production of intracellular virions, especially over several rounds of infection as measured by multistep growth, and/or decreased infectivity of UL49− virions. However, we did not observe either of these phenomena (Fig. 6, lower panel). Another possibility is that VP22 mediates viral spread through incorporation of gD into virions. However, because HSV-1 gD is required for virus entry and our particle-PFU analyses did not show a measurable decrease in UL49− virion infectivity, it is unlikely that the decreased incorporation of gD into UL49− virions can fully explain the UL49-null phenotype. Likewise, we observed decreased incorporation of gE into UL49− virions. However, gE is required for normal cell-cell spread (12) which was not decreased upon infection with the UL49− virus. Thus, while both gE and VP22 contribute to HSV-1 viral spread, VP22 appears to contribute through extracellular virus, whereas gE facilitates cell-cell spread.
Contribution of VP22 to herpetic disease.
The spread of HSV between corneal epithelial cells is critical for two phases of herpetic eye disease. In the initial, primary phase, aerosolized HSV that has been released from one infected individual may enter a break in the corneal epithelium of a second individual. The virus replicates and spreads between epithelial cells of the second host, and it ultimately reaches the basal cell layer, where it can invade the fine axon endings of trigeminal ganglion cells. Within these axons HSV is transported retrograde to the ganglion cell bodies, where it can establish a latent infection. In the second phase following reactivation, new virus is transported anterograde within the nerve endings to the corneal epithelium (33). The corneal epithelial cells again become infected, and the resulting humoral and cellular immunological responses may result in scarring and recurrent inflammation in the epithelium and deeper tissues (for review, see reference 41).
Understanding the pathogenesis of both phases of herpetic epithelial disease requires knowledge of the role(s) that viral genes play in viral spread. Because productive infection in animals may require specific viral proteins that are dispensable for replication in cultured cells, we considered it important to study the contribution of VP22 to HSV-1 infection in the context of the whole-animal model. Corneal lesions produced by the UL49-null virus were dramatically reduced in size compared to those produced by the WT and UL49R viruses. Thus, we have identified a role for VP22 in HSV-1 corneal viral spread in vivo.
Studies on the contribution of VP22 to infection by other herpesviruses have shown variable importance for VP22 in viral growth and virulence. While a bovine herpesvirus UL49 deletion virus exhibited a significant delay in single-step growth analyses and reduced virulence in cattle (26, 27), a PRV UL49 deletion virus showed single-step growth profiles identical to WT PRV and did not promote virulence or neuroinvasiveness in the rat eye infection model (11). Together, these data indicate that the presence of VP22 is of variable importance to different herpesviruses and experimental systems.
In summary, we have characterized the growth of an HSV-1 UL49-null virus and have identified a role for VP22 in viral spread during HSV-1 infection both in vitro and in vivo. Future studies will identify how this protein functions to promote viral spread and determine whether VP22 plays a role in virion exit and/or intercellular release. In addition, the use of a UL49-null virus with its genetically repaired partner should greatly facilitate investigations of the involvement of VP22 in HSV-1 tegument formation, pathogenesis, and virion egress.
Acknowledgments
These studies were supported by NIH R01 grants GM 50740 and AI 52341 to J.D.B. and AI 38873 to J.A.B., NIH NRSA F32 grant GM067519 to C.D., PHS grant EY 13867, and funds from That Man May See, Inc., to J.H.L.
We thank Yasuchi Kawaguchi for the gift of the HSV-1F BAC. We thank Klaus Osterrieder, B. Karsten Tischer, Jens von Einem, Daniel Schumacher, Laura Goodman, and Cristina Rosas for their generous sharing of reagents, techniques, and helpful advice. We thank David Johnson for the gift of the anti-gE antibody and Elisabeth Schlegel (Mount Sinai School of Medicine) for assistance with the V49 cells.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. The DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerli. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Eschericia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, M. E. M., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 7.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen.Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Rio, T., C. J. DeCoste, and L. W. Enquist. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, O. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, G., D. O'Reilly, and P. O'Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140-145. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, G. D., and D. M. Meredith. 1992. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J. Gen. Virol. 73:723-726. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss, B. J., J. E. Tavis, L. M. Metzger, D. A. Leib, and L. A. Morrison. 2001. Temporal regulation of herpes simplex virus type 2 VP22 expression and phosphorylation. J. Virol. 75:10721-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, D. C., M. C. Frame, M. W. Ligas, A. m. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knopf, K. W., and H. C. Kaerner. 1980. Virus-specific basic phosphoproteins associated with herpes simplex virus type a (HSV-1) particles and the chromatin of HSV-1-infected cells. J. Gen. Virol. 46:405-414. [DOI] [PubMed] [Google Scholar]
- 25.Lee, E. C., D. Yu, J. Martinez De Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 26.Liang, X., B. Chow, and L. A. Babiuk. 1997. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine 15:1057-1064. [DOI] [PubMed] [Google Scholar]
- 27.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lumb, W. V. 1963. Small animal anesthesia. Lea and Febiger, Philadelphia, Pa.
- 29.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 30.Meredith, D. M., J. A. Lindsay, I. W. Halliburton, and G. R. Whittaker. 1991. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J. Gen. Virol. 72:2771-2775. [DOI] [PubMed] [Google Scholar]
- 31.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohara, P. T., M. S. Chin, and J. H. LaVail. 2000. The spread of herpes simplex virus type 1 from trigeminal neurons to the murine cornea: an immunoelectron microscopy study. J. Virol. 74:4776-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 82:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinard, M. F., R. Simard, and V. Bibor-Hardy. 1987. DNA-binding proteins of herpes simplex virus type 1-infected BHK cell nuclear matrices. J. Gen. Virol. 68:727-735. [DOI] [PubMed] [Google Scholar]
- 36.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious Herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potel, C., and G. Elliott. 2005. Phosphorylation of the herpes simplex virus tegument protein VP22 has no effect on incorporation of VP22 into the virus but is involved in optimal expression and virion packaging of ICP0. J. Virol. 79:14057-14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Bovine herpesvirus 1 tegument protein VP22 interacts with histones, and the carboxyl terminus of VP22 is required for nuclear localization. J. Virol. 75:8251-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritterband, D. C., and D. N. Friedberg. 1998. Virus infections of the eye. Rev. Med. Virol. 8:187-201. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40:191-197. [DOI] [PubMed] [Google Scholar]