Abstract
Human immunodeficiency virus type 1 (HIV-1) clade C causes >50% of all HIV infections worldwide, and an estimated 90% of all transmissions occur mucosally with R5 strains. A pathogenic R5 simian-human immunodeficiency virus (SHIV) encoding HIV clade C env is highly desirable to evaluate candidate AIDS vaccines in nonhuman primates. To this end, we generated SHIV-1157i, a molecular clone from a Zambian infant isolate that carries HIV clade C env. SHIV-1157i was adapted by serial passage in five monkeys, three of which developed peripheral CD4+ T-cell depletion. After the first inoculated monkey developed AIDS at week 137 postinoculation, transfer of its infected blood to a naïve animal induced memory T-cell depletion and thrombocytopenia within 3 months in the recipient. In parallel, genomic DNA from the blood donor was amplified to generate the late proviral clone SHIV-1157ipd3. To increase the replicative capacity of SHIV-1157ipd3, an extra NF-κB binding site was engineered into its 3′ long terminal repeat, giving rise to SHIV-1157ipd3N4. This virus was exclusively R5 tropic and replicated more potently in rhesus peripheral blood mononuclear cells than SHIV-1157ipd3 in the presence of tumor necrosis factor alpha. Rhesus macaques of Indian and Chinese origin were next inoculated intrarectally with SHIV-1157ipd3N4; this virus replicated vigorously in both sets of monkeys. We conclude that SHIV-1157ipd3N4 is a highly replication-competent, mucosally transmissible R5 SHIV that represents a valuable tool to test candidate AIDS vaccines targeting HIV-1 clade C Env.
Simian-human immunodeficiency virus (SHIV) strains are chimeric viruses constructed from the pathogenic simian immunodeficiency virus (SIV) clone SIVmac239, in which the env, tat, and rev genes had been replaced by the corresponding regions of human immunodeficiency virus type 1 (HIV-1); most SHIVs also contain vpu (7, 21, 22, 25, 27, 31, 35, 47). SHIV infection of rhesus macaques has been an invaluable in vivo model to study the role of HIV-1 envelopes in transmission and pathogenesis as well as to evaluate the efficacy of candidate vaccines based on HIV-1 envelope glycoproteins, which specify cell tropism and coreceptor usage and are also primary targets of the immune response. However, the majority of current SHIV strains utilize envelope genes derived from HIV-1 clade B strains, which represent less than 10% of all global infections. Therefore, the available SHIV chimeras do not reflect the genetic diversity of the HIV-1 epidemic, which is dominated by non-B clades, especially HIV-1 clade C.
HIV-1 clade C, the dominant subtype in the world, is estimated to comprise more than 50% of all infections in the pandemic and is the most prevalent clade in sub-Saharan Africa and parts of Asia, where the AIDS epidemic is growing fastest (http://www.unaids.org). The rapid spread of this particular subtype in these heavily populated regions has resulted in over 5 million infections in Asia alone.
Over 90% of all HIV transmission events worldwide involve mucosal transmission, including most sexual and mother-to-child transmissions (45). HIV-1 strains isolated from individuals soon after infection (14, 34, 59) preferentially use CCR5 as the coreceptor for cell entry (2, 12, 15, 18). Such viruses are referred to as R5 HIV-1 isolates. Therefore, a highly replication-competent SHIV that is mucosally transmissible in rhesus monkeys and that encodes a non-clade B HIV-1 env gene would be an important tool in HIV/AIDS research.
Although several non-clade B HIV-1 envelope-based SHIV chimeric constructs have been described so far (8, 10, 30, 41, 58), none of them has been reported to be mucosally transmissible or to induce signs of disease in rhesus macaques, the most commonly used nonhuman primate in AIDS research. Here we report the construction of SHIV-1157ipd3N4. This virus was isolated from a rhesus monkey, RPn-8, which had been inoculated as an infant with a parental SHIV construct that expresses the envelope glycoprotein of a relatively recently transmitted R5 HIV-1 clade C isolate from a 6-month-old Zambian infant. SHIV-1157ipd3N4 was derived from this same animal, RPn-8, after it developed AIDS approximately 2.7 years postinoculation. SHIV-1157ipd3N4 exclusively uses CCR5 as a coreceptor and could be intrarectally transmitted to rhesus monkeys of both Indian and Chinese origin.
MATERIALS AND METHODS
Original virus isolates and nomenclature.
HIV1157i is a biological isolate obtained from a Zambian infant at 6 months of age. At birth, this infant was PCR positive for HIV-1. The designation “i” indicates a virus strain (or env gene) isolated from an infant. SHIV-1157i is the original infectious molecular clone, not yet adapted to rhesus monkeys. SHIV-1157ip is an early biological isolate obtained after passage through five rhesus monkeys; “p” designates a passaged (or monkey-adapted) virus. SHIV-1157ipd is a late biological isolate; “d” indicates that the virus was reisolated from an infected animal with disease (AIDS as defined by persistent depletion of CD4+ T cells to <200 cells/μl). SHIV-1157ipd3 designates the late-stage infectious molecular clone #3; the 3′ half of this provirus was derived from the biological isolate SHIV-1157ipd. SHIV-1157ipd3N4 is identical to SHIV-1157ipd3 except that the 3′ long terminal repeat (LTR) was engineered to contain two rather than the usual one NF-κB site. This NF-κB site duplication is copied into the 5′ LTR during the reverse transcription steps occurring in the course of the subsequent retroviral propagation (13).
Cell lines and antibodies.
CEMx174-GFP cells, provided by B. Felber (National Cancer Institute, Frederick, MD), contain the green fluorescent protein gene under HIV-1 LTR regulation and express CXCR4 but not CCR5. U87 or GHOST cell lines, which express CD4 only or CD4 with different chemokine receptors, were provided by the AIDS Research and Reference Reagents Program (ARRRP; Germantown, MD). TZM-bl cells (also called JC53-bl [clone 13] cells; ARRRP) (16) are derived from a HeLa cell line (JC.53) that stably expresses CD4 and CCR5. TZM-bl cells also express luciferase and β-galactosidase under control of the HIV-1 LTR. The neutralizing monoclonal antibody (NMAb) 2G12 (57) was a gift of Hermann Katinger (Polymune Scientific, Vienna, Austria).
Animals and animal care.
Rhesus monkeys (Macaca mulatta) of Indian and Chinese origin were used in this study. The animals were kept according to National Institutes of Health guidelines on the care and use of laboratory animals at the Yerkes National Primate Research Center (Emory University, Atlanta, GA) and the Centers for Disease Control and Prevention (CDC; Atlanta, GA). These facilities are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Animal experiments were approved by the Animal Care and Use Committees of the Yerkes National Primate Research Center, the Centers for Disease Control and Protection, and the Dana-Farber Cancer Institute.
Construction of SHIV-1157ipd molecular clones and sequence analysis.
Using a DNAzol genomic DNA isolation kit (Molecular Research Center Inc., Cincinnati, OH), chromosomal DNA was extracted from 106 peripheral blood mononuclear cells (PBMC) from animal RPn-8, the monkey first exposed to SHIV-1157i, after its absolute CD4+ T-cell counts fell below 200 cells/μl. A pair of specific primers was designed to amplify the entire 3′ half of SHIV-1157ipd. The primers were designed to incorporate SphI or NotI restriction enzyme sites with the following sequences: 1157ipd-SphI, 5′-CCGCCCTCTAGAAGCATGCTGTAG-3′; and 1157ipd-NotI, 5′-AAAGTTGAATGCGGCCGCTACTTCTAAAATGGCAGCTTTATTGAAGAGG-3′. The 5′ half of SHIV-vpu+ (31) was digested with the restriction enzymes EcoRI and SphI and cloned into vector pSP73-N, which was modified with the introduction of an extra NotI site at the multiple cloning sites of pSP73. The PCR products were digested with SphI and NotI and cloned into the pSP73-N vector that has the 5′ half of SHIV-vpu+ to form the full-length proviral DNA SHIV-1157ipd. The env gene of SHIV-1157ipd was also amplified using primers 1157ipd-forward (5′-TACAAAGAGGAAATGGATAAA-3′) and 1157ipd-reverse (5′-ATCCATGTGTGTACTATTGTC-3′) and cloned into TOPO sequencing vector (Invitrogen, Carlsbad, CA). Five clones were randomly picked for plasmid preparation and DNA sequencing. An additional NF-κB element was added to the 3′ LTR of SHIV-1157ipd3 using the Quikchange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) and a pair of primers (N2-1, 5′-ACTCGCTGAAACAGCAGGGACTTTCCACAAAGGGACTTTCCACAAGGGGATGTTACGGGGAGG-3′; and N2-2, 5′-CCTCCCCGTAACATCCCCTTGTGGAAAGTCCCTTTGTGGAAAGTCCCTGCTGCTGTTTCAGCGAGT-3′).
Construction of SIV LTR pLuc mutants.
The pLuc reporter construct (a gift of J. Clements, Johns Hopkins University, Baltimore, MD) (50) contains a truncated portion of the SIVmac239 LTR (−225→+149), upstream of the firefly luciferase reporter gene. HIV-1 or SIV Tat is required for the activation of pLuc in order to drive expression of the luciferase gene by the LTR. The Quikchange Site-Directed Mutagenesis kit (Stratagene) was used to introduce mutations into the SIVmac239 LTR in the pLuc construct.
Coreceptor usage of SHIV constructs.
The U87 or GHOST cell lines expressing CD4 and/or HIV-1 or SIV coreceptors were used to study virus tropism. U87CD4, U87CCR1, U87CCR2, U87CCR3, U87CXCR4, U87CCR5, GHOST.BOB, and GHOST.BONZO were infected with 0.5 ml of virus stock. SHIVSF162P3 was obtained from ARRRP. Cells were washed and resuspended in 1 ml of fresh medium. On days 1, 2, 3, 4, and 5, supernatants were collected for p27 measurement.
Measurement of plasma viral RNA levels.
Plasma viral RNA was isolated by use of the QiaAmp Viral Mini-kit (QIAGEN), and viral RNA levels were measured by quantitative reverse transcriptase PCR (RT-PCR) for SIV gag sequences (24) at weeks 0, 1, 2, 4, and 8 and monthly thereafter. The assay sensitivities were 50 viral RNA copies/ml of plasma.
Generation of large-scale SHIV-1157ipd3N4 stock.
A large-scale stock of the infectious molecular clone was prepared by infecting concanavalin A (ConA)-stimulated naïve rhesus monkey PBMC in the presence of human interleukin-2 (IL-2; 20 U/ml) and tumor necrosis factor alpha (TNF-α; 10 ng/ml) with virus harvested from transiently transfected 293T cells. This rhesus PBMC-grown stock has a p27 concentration of 227 ng/ml and 4 × 106 50% tissue culture infectious doses (TCID50) per ml as titrated in TZM-bl cells.
TCID50 determination of virus stock in TZM-bl cells.
Viruses were added to TZM-bl cells (16) at serial 1:4 dilutions in the presence of 40 μg/ml of DEAE-dextran hydrochloride (Sigma, St. Louis, MO). Virus infectivity was determined 48 h postinoculation by measuring the level of luciferase activity expressed in infected cells. Each experiment was performed in triplicate. The TCID50 was calculated as the dilution point at which 50% of the cultures were infected.
Intrarectal inoculation of SHIV-1157ipd3N4.
Chinese-origin rhesus monkeys received 1 ml of the original large-scale virus stock intrarectally at various dilutions: undiluted stock (one monkey), 1:5.5 (one monkey), 1:10 (one monkey), and 1:50 (three monkeys; only two became systemically infected). Higher dilutions of the stock did not result in systemic infection. Eight Indian-origin rhesus macaques received 0.5 ml of the same large-scale virus stock intrarectally. These monkeys were part of a DNA prime/protein boost vaccine study and served as vector-only controls (R. A. Rasmussen, H. Ong, R. Song, E. Shai-Kolber, A.-L. Chenine, S.-L. Hu, P. Policano, J. McKenna, J. Moon, B. Travis, H. M. McClure, E. Strobert, F. J. Novembre, J. G. Else, and R. M. Ruprecht, unpublished data); they had received 200 μg of the empty DNA vector pJW4303 twice intradermally at a 6-week interval as well as 0.1 ml of incomplete Freund's adjuvant (IFA) twice intramuscularly at a 6-week interval 1 year later. Two weeks after the second inoculation of IFA, the animals underwent the intrarectal virus challenge. All animals were monitored prospectively for viral loads and T-cell subsets at different time points postinoculation.
Neutralization assay.
The neutralization assay was performed in triplicate in human PBMC, as described elsewhere (29). NMAb 2G12 was not washed away but rather was diluted 1:1 with fresh medium daily, starting on day 3 of the experiment. Due to this assay condition, which takes into account the long half-lives of antibodies, neutralization titers may differ slightly from titers measured by other methods (6, 36). Antibody-mediated neutralization is expressed as a percentage of neutralization of virus infectivity (29); antibody concentrations during the preincubation of 2G12 with virus ranged from 0.32 μg/ml to 40 μg/ml.
Lymphocyte immunophenotyping.
PBMC isolated from Indian-origin rhesus monkeys were stained for flow cytometric analysis using combinations of the following fluorochrome-conjugated MAbs: anti-CD3-Alexa Fluor 700 (SP34-2; BD Biosciences, San Jose, CA), anti-CD4-PerCP-Cy5.5 (L200; BD Biosciences), anti-CD28-PE (28.2; BD Biosciences), and anti-CD95-PE (DX2; BD Biosciences). The samples were analyzed by four-color flow cytometry (FACSCalibur; BD Biosciences Immunocytometry Systems). Data analysis was performed by using FLOWJO (TreeStar, San Carlos, CA). CD3+CD4+ T cells were gated based on CD28 and CD95 expression to define memory CD4+ T-cell subpopulations: naive (CD28+CD95−), central memory (CD28+CD95+), and effector memory (CD28−CD95+).
Nucleotide sequence accession number.
The nucleotide sequence determined in the course of this work was deposited in GenBank under accession number DQ779174.
RESULTS
SHIV-1157i infection and disease progression in rhesus monkey RPn-8.
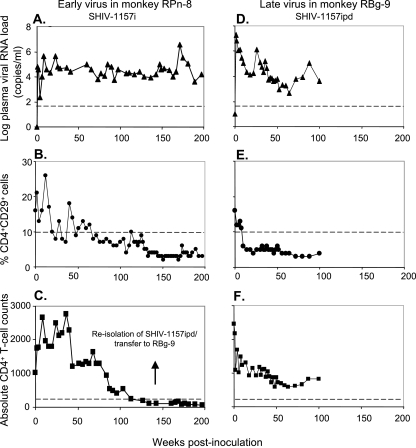
We modified the SHIV-vpu+ backbone to express env of a relatively recently transmitted pediatric HIV isolate (HIV1157i) from a 6-month-old Zambian infant born to an HIV-positive mother. SHIV-1157i contains most of gp120 as well as the entire extracellular domain and transmembrane region of gp41 of HIV1157i (Fig. 1A). An infant macaque, RPn-8, was inoculated intravenously with 6 ml of SHIV-1157i stock. Persistent viremia (Fig. 2A) and signs of pathogenicity were observed in this monkey. The memory CD4+CD29+ T-cell subset (Fig. 2B) (4, 51) as well as absolute CD4+ T cells (Fig. 2C) in this monkey became depleted, with absolute CD4+ T-cell counts below 200 cells/μl for more than 1 year. By the above criterion, this monkey developed AIDS at week 137 postinoculation. Monkey RPn-8 also developed thrombocytopenia at week 46 postinoculation.
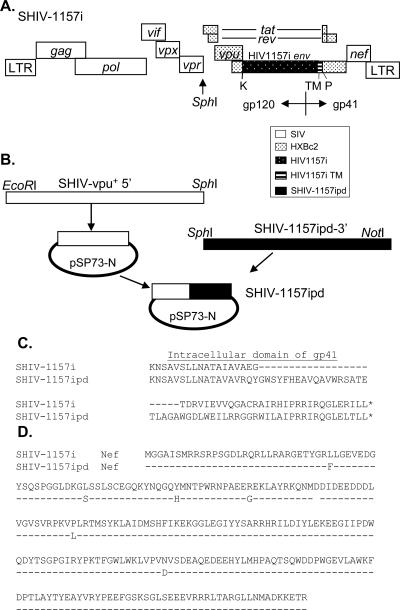
FIG. 1.
Schematic representation of SHIV-1157ipd3 construction. (A) Structure of SHIV-1157i. SHIV-1157i contains env of a relatively recently transmitted pediatric HIV isolate from a 6-month-old Zambian infant. A unique restriction site, PuvI (P), was introduced into the 3′ half of SHIV-vpu+ proviral DNA. The 2.0-kb KpnI (K)-PvuI fragment of HIV1157i (spanning most of gp120 as well as the entire gp41 extracellular domain and the transmembrane region [TM]) was amplified to replace the corresponding region of SHIV-vpu+ env. The modified 3′ half was ligated with the 5′ half of SHIV-vpu+ proviral DNA to form full-length SHIV-1157i. (B) Construction of SHIV-1157ipd3. The entire 3′ half of SHIV-1157ipd3 (SphI-NotI fragment) was amplified from the genomic DNA of PBMC isolated 4 weeks after animal RPn-8 developed AIDS, as defined by persistent CD4+ T-cell counts of <200 cells/μl. This fragment was ligated with the 5′ half of SHIV-vpu+, which was previously cloned into a modified pSP-73 vector, to form proviral DNA SHIV-1157ipd3. (C) Predicted amino acid sequences of the C-terminal domains of gp41 in SHIV-1157i and SHIV-1157ipd. (D) Predicted amino acid sequences of Nef in SHIV-1157i and SHIV-1157ipd.
FIG. 2.
Plasma viremia and CD4+ T-cell loss due to SHIV-1157i (monkey RPn-8) and SHIV-1157ipd (monkey RBg-9). Infant macaque RPn-8 was inoculated intravenously with 6 ml of early-stage SHIV-1157i stock (left panels). Rhesus monkey RBg-9 received 10 ml of blood intravenously, transferred from monkey RPn-8 4 weeks after the latter had CD4+ T-cell counts of <200 cells/μl; the transferred blood contained the late-stage virus, SHIV-1157ipd (right panels). (A and D) Plasma viral RNA loads. The dashed lines indicates the lower level of sensitivity of the RT-PCR assay (50 copies/ml) (24). (B and E) CD4+ memory T-cell counts (CD4+ CD29+). The dashed lines denote the lowest normal value (10%) for the percent CD4+ CD29+ cells. (C and F) Absolute CD4+ T-cell counts. The dashed lines denote 200 CD4+ T cells. By definition, a persistent peripheral CD4+ T-cell count of <200 cells/μl is indicative of AIDS. The arrow denotes the time of blood transfer of SHIV-1157ipd to animal RBg-9 and also the time of reisolation of the SHIV-1157ipd biological isolate.
Passage of uncloned, late-stage virus.
To test whether more aggressive progeny viruses had emerged during the protracted chronic infection and gradual progression to AIDS in monkey RPn-8, we transferred 10 ml of blood containing the late-stage virus (SHIV-1157ipd) from RPn-8 intravenously to another naïve rhesus monkey, RBg-9. Compared to the early-stage virus SHIV-1157i, SHIV-1157ipd induced peak viremia (week 1 postinoculation) that was >2 logs higher (Fig. 2D). RBg-9 developed persistent thrombocytopenia (data not shown) and became relatively depleted of its CD4+CD29+ memory T-cell subset (Fig. 2E) 12 weeks after transfer of late-stage virus.
Isolation and characterization of late-stage virus.
Our results in recipient animal RBg-9 are in agreement with previous observations that SIV variants emerging during late-stage disease have a higher replicative capacity and increased pathogenicity (28). Therefore, in an effort to obtain a more virulent SHIV clade C strain, uncloned virus (SHIV-1157ipd) was reisolated from animal RPn-8 4 weeks after the onset of AIDS. Genetic analysis of SHIV-1157ipd env sequences exhibited a number of changes resulting in single-amino-acid substitutions in gp120 (especially in V1, V2, and V4) and gp41. In addition, a 118-bp deletion in the 3′ end of gp41 was seen in all five clones that were sequenced. This deletion led to a loss of the last 35 amino acids in the original HIV-1 gp41 and a gain of the last 57 amino acids of the SIVmac239 gp41 through frameshift (Fig. 1C) relative to the parental SHIV-1157i provirus. These changes are similar to, yet distinct from, the env changes that occurred in SHIV89.6P during its serial passage (47). These changes in gp41 represent a striking example of convergent molecular evolution of different SHIV constructs in independently inoculated rhesus monkeys. Compared with the parental SHIV-1157i, nef sequences in SHIV-1157ipd exhibited six changes resulting in single-amino-acid substitutions (Fig. 1D).
We next determined coreceptor usage of late-stage virus. SHIV-1157ipd replicated neither in CEMx174-GFP cells nor in U87.CD4, U87.CD4.CCR1, U87.CD4.CCR2, U87.CD4.CCR3, U87.CD4.CXCR4, GHOST-BOB, and GHOST-BONZO cells. Productive infection occurred only in U87.CD4.CCR5 cells (data not shown), indicating that SHIV-1157ipd exclusively uses CCR5 as its coreceptor.
Construction and characterization of SHIV-1157ipd3.
A molecular clone of SHIV-1157ipd, termed SHIV-1157ipd3, was constructed using SHIV-vpu+ as the backbone and containing the entire 3′ half, including env and nef, of SHIV-1157ipd. Genomic DNA from RPn-8 PBMC collected at week 141 after virus exposure (4 weeks after AIDS was diagnosed) was used to amplify the SHIV-1157ipd 3′ half, which was ligated with the 5′ half of SHIV-vpu+ (31) to form the full-length proviral DNA of SHIV-1157ipd clones (Fig. 1B). A total of 23 full-length clones were produced and screened for their infectivity on TZM-bl using the supernatant of transfected 293T cells. SHIV-1157ipd3 was picked due to its highest infectivity among these clones. SHIV-1157ipd3 replicated well in the PBMC of all six randomly selected naïve macaques (data not shown).
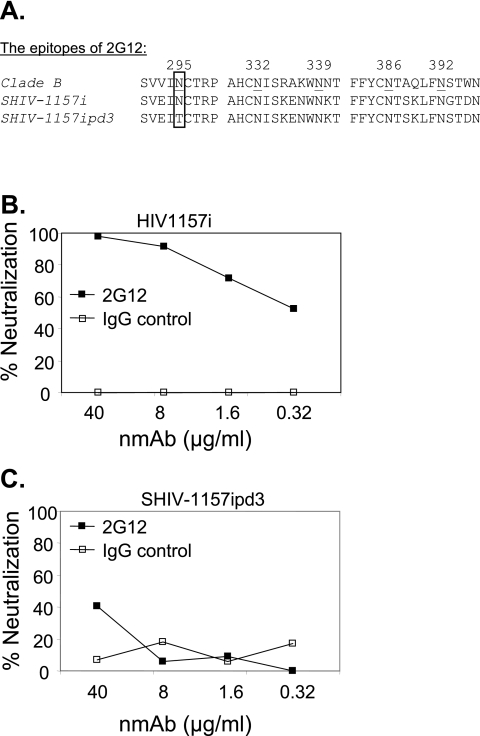
SHIV-1157ipd3 lacks the 2G12 epitope contained in HIV1157i.
The carbohydrate-dependent epitope of the human NMAb 2G12 includes N-linked mannan moieties associated with the five residues N295, N332, N339, N386, and N392. The core epitope consists of glycans attached to N295, N332, and N392 (52, 53). HIV1157i and SHIV-1157i have all five of these N-linked glycosylation sites, but after almost 3 years of replication in monkey RPn-8, N295 was mutated to T295 in SHIV-1157ipd3 (Fig. 3A). The other four asparagines associated with the 2G12 epitope were retained in SHIV-1157ipd3. When we evaluated the susceptibility of HIV1157i and SHIV-1157ipd3 to neutralization by human NMAb 2G12 in vitro, infection of human PBMC by HIV1157i was inhibited by 2G12 in a dose-dependent manner (Fig. 3B). However, SHIV-1157ipd3 was not sensitive to 2G12 (Fig. 3C).
FIG. 3.
Neutralization of HIV1157i and SHIV-1157ipd3 with neutralizing monoclonal antibody 2G12. (A) Amino acid sequence alignment of the 2G12 epitope. The boxed amino acid residues show the critical mutation at position 295. (B) Neutralization of HIV1157i by NMAb 2G12. (C) Neutralization of SHIV-1157ipd3 by 2G12. Neutralization assays were performed in phytohemagglutinin (PHA)-activated human PBMC in triplicate. Virus was incubated with the NMAb 2G12 or an isotype control NMAb at the indicated concentrations for 1 h at 37°C and then added to PBMC with IL-2; NMAbs were not washed out. Virus production was assessed by measuring p24 or p27 levels in the culture supernatants. IgG, immunoglobulin G.
Increase in the virulence of SHIV-1157ipd3 by introduction of an additional NF-κB binding site.
One to three NF-κB binding sites may be present in the enhancer regions of SIV or different HIV-1 subtypes (9, 38, 46). We hypothesized that the number of such sites in the viral LTR may affect viral replication and virulence. For instance, SIVmac239, the backbone normally used for the construction of SHIV chimeras, has one NF-κB binding element. However, duplication of NF-κB binding sites in monkey-passaged SIVs can result in acutely lethal variants such as SIVpbj14 (17) or pathogenic progeny that had evolved from live attenuated SIVmac239Δ3 (1). In the latter case, progeny virus containing two NF-κB sites had replaced the original virus, implying better in vivo replicative capacity of virus that had emerged late in the course of the infection (1). In addition, other investigators have described a direct correlation between the number of NF-κB sites and LTR-driven gene expression (26).
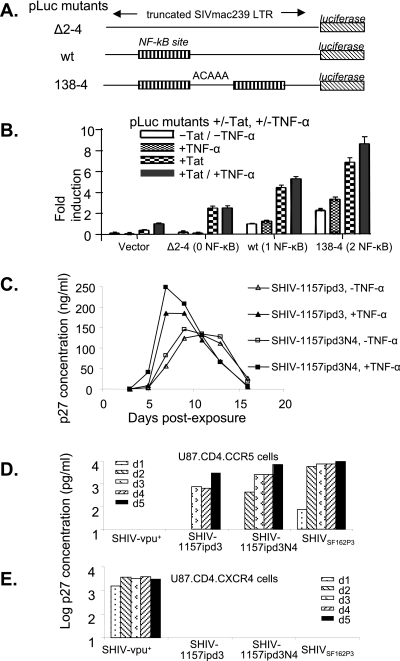
To directly test our hypothesis, we first evaluated a set of solo-LTR constructs. The pLuc reporter plasmid with a truncated portion of the SIVmac239 LTR (−225→+149; one NF-κB binding site) upstream of the firefly luciferase reporter gene was engineered by site-directed mutagenesis to produce a total of three constructs containing different numbers of NF-κB binding sites (Fig. 4A). Mutants Δ2-4 and 138-4 contain zero and two NF-κB binding sites, respectively, in their LTR regions. Luciferase activity was measured in 293T cells transiently transfected with one of the three mutant constructs with or without cotransfecting a plasmid containing SHIV89.6 tat. The transfected cells were also cultured in the presence or absence of TNF-α, a known inducer of NF-κB activation, to determine the level of transcription driven by each of the mutant LTRs (Fig. 4B). The level of luciferase activity produced in 293T cells transfected with each of the solo-LTR constructs was associated with the number of NF-κB binding sites in the LTR enhancer region. The differences were most pronounced in the presence of Tat and/or TNF-α. The highest luciferase activity was observed in 293T cells transfected with the mutant that has two NF-κB binding sites, 138-4, in the presence of Tat and TNF-α. These results suggest that a viral LTR with two NF-κB binding sites may be more responsive to Tat and TNF-α than LTRs with zero or only one NF-κB binding site.
FIG. 4.
Number of NF-κB sites and LTR-mediated gene expression, replication kinetics of SHIV-1157ipd3 and SHIV-1157ipd3N4, and coreceptor usage of the late viruses SHIV-1157ipd3 and SHIV-1157ipd3N4. (A) Schematic representation of pLuc mutant constructs containing various numbers of NF-κB sites. The pLuc reporter plasmid with a truncated portion of the SIVmac239 LTR upstream of the firefly luciferase reporter gene was engineered to yield three constructs with different numbers of NF-κB binding sites. Isolates Δ2-4, wild-type (wt), and 138-4 contain zero, one, and two NF-κB binding sites, respectively, in their LTR enhancer regions. (B) Luciferase expression of each of the constructs in transfected 293T cells with or without TNF-α or a plasmid containing SHIV89.6 tat (pTat). Luciferase activity was measured to determine the level of gene expression driven by each LTR. The amount of luciferase activity produced in 293T cells transfected with the indicated plasmid was divided by that of 293T cells transfected with wt plasmid only (without TNF-α or pTat) to calculate fold induction. Error bars represent standard deviations of the experiments. (C) PBMC (2 × 106) from a randomly selected rhesus monkey (CF-37) were activated with ConA and exposed to either SHIV-1157ipd3 (one NF-κB site per LTR) or SHIV-1157ipd3N4 (two NF-κB sites per LTR) produced from transiently transfected 293T cells, each at 30 ng of p27 Gag protein in the presence (closed symbols) or absence (open symbols) of human TNF-α (10 ng/ml). Levels of p27 were measured in the PBMC supernatants on the indicated days following virus exposure. U87.CD4.CCR5 cells (D) and U87.CD4.CXCR4 cells (E) were exposed to SHIV-vpu+, SHIV-1157ipd3, SHIV-1157ipd3N4, or SHIVSF162P3. Supernatants were harvested and replaced daily. Levels of p27 Gag were measured in the supernatants on the indicated days following virus exposure.
We next sought to test this concept in the context of replication-competent, isogenic viruses and to evaluate whether this strategy could be used to enhance the replicative capacity of SHIV-1157ipd3. To this end, a modified molecular clone, SHIV-1157ipd3N4, was constructed by introducing an additional NF-κB binding site into the 3′ LTR of SHIV-1157ipd3 by site-directed mutagenesis. We evaluated the growth of SHIV-1157ipd3 and SHIV-1157ipd3N4 in PBMC from randomly selected rhesus monkey donors in the presence or absence of TNF-α. Although both viruses replicated well in PBMC from a naive donor (Fig. 4C), the highest peak p27 concentration was observed with SHIV-1157ipd3N4 in the presence of TNF-α, suggesting that the latter virus may have a replicative advantage over SHIV-1157ipd3 due to its extra NF-κB binding site. The same pattern was observed in PBMC from all three macaque donors tested (data not shown).
Coreceptor usage of late-stage SHIV strains.
We next assessed coreceptor usage of SHIV-1157ipd3 and SHIV-1157ipd3N4. Neither virus replicated in any cell line lacking CCR5, including CEMx174-GFP, U87.CD4, U87.CD4.CCR1, U87.CD4.CCR2, U87.CD4.CCR3, U87.CD4.CXCR4, GHOST-BOB, and GHOST-BONZO cells (Fig. 4E and data not shown). The observation that productive infection occurred only in U87.CD4.CCR5 cells (Fig. 4D) suggests that late-stage SHIV-1157ipd3 and SHIV-1157ipd3N4, like the uncloned virus SHIV-1157ipd, exclusively use CCR5 as their coreceptor.
Comparison of the infectivity of SHIV-1157ipd3N4 in rhesus monkeys of Chinese or Indian origin.
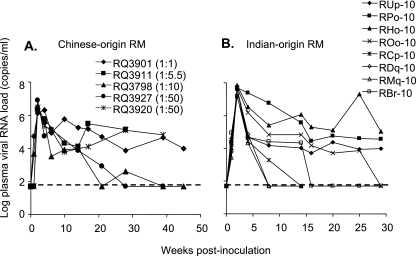
A large stock of SHIV-1157ipd3N4 was generated in Indian-origin rhesus monkey PBMC. Chinese rhesus monkeys were inoculated intrarectally with various dilutions of the viral stock to determine the in vivo infectivity. Undiluted stock and dilutions of 1:5.5 and 1:10 resulted in systemic infection of each of the exposed animals, whereas a dilution of 1:50 led to infection in two out of three animals. Dilutions of 1:60 or higher did not yield systemic infection. Among the five monkeys that became systemically infected, no correlation between the viral inocula and peak viral RNA loads was seen, consistent with earlier observations (11). The monkey with the highest peak viral RNA load of 8.2 × 106 copies/ml had received 1 ml of a 1:50 dilution of the original stock (Fig. 5A). The statistical method of Spouge (55) was used to determine the 50% animal infectious dose of the SHIV-1157ipd3N4 stock for the intrarectal route in Chinese-origin rhesus monkeys; the value was 2.46 × 10−2 ml. During the 40-week follow-up period, three out of the five Chinese rhesus monkeys have maintained high viral loads (Fig. 5A).
FIG. 5.
Intrarectal inoculation of SHIV-1157ipd3N4. Five Chinese-origin rhesus monkeys (A) and eight Indian-origin rhesus monkeys (B) were inoculated intrarectally with SHIV-1157ipd3N4 stock with concentrations noted in the text. Viral loads were measured at indicated time points postinoculation. The dashed lines indicate the lower level of sensitivity of the RT-PCR assay (50 copies/ml).
We next assessed intrarectal transmission in eight Indian rhesus monkeys (Fig. 5B). These animals were inoculated intrarectally with 0.5 ml of the same large-scale SHIV-1157ipd3N4 stock and monitored for infection. All eight macaques showed robust viral replication during the first 2 weeks postinoculation, with peak viral RNA loads ranging from 1.4 × 107 to 7.1 × 107 copies/ml of plasma. The average peak viral RNA load of SHIV-1157ipd3N4 in the eight Indian-origin rhesus monkeys was 11 times higher than that in the five Chinese-origin rhesus monkeys (two-sided P = 0.002; Wilcoxon rank-sum test). During the 29-week follow-up period, three out of the eight Indian rhesus monkeys have maintained high viral loads. Five more Indian rhesus monkeys were also inoculated intrarectally with various dilutions of the SHIV-1157ipd3N4 stock. Dilutions of 1:10, 1:50, and 1:60 resulted in systemic infection of the exposed animals, whereas a dilution of 1:100 led to infection in one out of two animals (unpublished data). These data indicate that intrarectal transmission of our SHIV-1157ipd3N4 stock is reproducible.
We then estimated the replicative capacity of SHIV-1157ipd3N4, the late virus, compared to that of SHIV-1157ip, the early virus, after mucosal inoculation of Indian-origin rhesus macaques. Mean peak viral RNA load in SHIV-1157ipd3N4-infected animals (n = 8) was 4.05 × 107 copies/ml, compared to 9.90 × 106 viral RNA copies/ml in SHIV-1157ip-infected animals (n = 19; P = 0.000284; Student's t test). These results indicate that SHIV-1157ipd3N4 has a significantly higher replicative capacity than SHIV-1157ip.
Early signs of pathogenicity of SHIV-1157ipd or SHIV-1157ipd3N4.
The uncloned late virus SHIV-1157ipd, which was transferred to juvenile rhesus macaque RBg-9 by intravenous blood transfer from RPn-8, induced memory CD4+ CD29+ T-cell depletion (Fig. 2E) and thrombocytopenia (data not shown) in the new host within 3 months of inoculation. Among Indian-origin rhesus macaques inoculated with SHIV-1157ipd3N4, a sharp reduction in CD4+ central memory T cells (CD28+CD95+) was observed in two animals (monkeys RPo-10 and RCp-10) out of the five animals tested (Table 1). The central memory T-cell population in animal RCp-10 dropped from 17% at week 0 to 5.8% at week 8. However, in all eight Indian-origin and five Chinese-origin animals, absolute peripheral CD4+ T-cell counts remained within the normal range during the early period of observation (data not shown). Longer term pathogenicity studies are needed to assess the rate of progression to immunodeficiency for these late-stage virus strains.
TABLE 1.
T-cell populations of Indian-origin rhesus monkeys inoculated with SHIV-1157ipd3N4
| Macaque | T-cell population at week:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0
|
8
|
|||||||
| CD4+ naïve (%) | CD4+ EMa (%) | CD4+ CMb (%) | CD4/CD8 ratio | CD4+ naïve (%) | CD4+ EMa (%) | CD4+ CMb (%) | CD4/CD8 ratio | |
| RCp-10 | 51 | 1.13 | 46 | 1.95 | 72 | 0.23 | 26c | 1.46 |
| RPo-10 | 81 | 0.42 | 17 | 1.63 | 94 | 0.26 | 5.8c | 0.88 |
| RUp-10 | 79 | 0.09 | 18 | 1.95 | 83 | 0.09 | 15 | 1.68 |
| RHo-10 | 78 | 0.74 | 18 | 0.83 | 66 | 1.13 | 30 | 0.81 |
| ROo-10 | 65 | 0.32 | 34 | 1.71 | 69 | 0.41 | 30 | 1.11 |
EM, effector memory T cells.
CM, central memory T cells.
Values in boldface are abnormal.
DISCUSSION
The overall goal of this study was to generate a highly relevant R5 SHIV that encodes HIV-1 clade C env. To this end, we have generated a series of related SHIVs that carry env of a pediatric HIV-1 clade C strain. The early SHIV forms, SHIV-1157i and SHIV-1157ip, are exclusively R5 tropic and induced progressive disease after 2.6 to almost 5 years in three out of the five rhesus monkeys used in the initial virus adaptation. While this rate of disease progression is consistent with lentiviral biology, faster disease progression rates would render the SHIV clade C/rhesus monkey model more practical. To generate more virulent versions of the virus, we employed a dual approach: first, we reisolated late-stage virus from the first monkey that had progressed to AIDS, and second, we engineered an extra NF-κB site into the LTR of the molecularly cloned late-stage virus to boost its replicative capacity and responsiveness to TNF-α. The resulting infectious molecular clone, SHIV-1157ipd3N4, has a number of biologically relevant characteristics: (i) it is exclusively R5 tropic; (ii) it is mucosally transmissible in rhesus macaques; and (iii) infection with SHIV-1157ipd3N4 induces high peak viral RNA loads in all animals tested and high viral set points in 10 out of 13 infected animals. Although SHIV-1157ipd3N4 lacks the acute pathogenicity of typical X4 or X4R5 SHIVs, it induced abnormalities in immune parameters, such as central memory CD4+ T-cell depletion and decreased CD4/CD8 ratios, during a relatively short period of postexposure followup.
SHIV-1157ipd3N4 carries the envelope gene of the HIV clade C, the most prevalent substrain worldwide. Although four SHIV strains (SHIVCHN19, SHIVMJ4, SHIV-MCGP1.3, and SHIV-XJ02170) encoding clade C envelopes have been created thus far, they either are dual tropic (SHIV-MCGP1.3) (8), are unable to replicate in rhesus macaque PBMC (SHIVCHN19) (10), or show no evidence thus far of mucosal transmissibility or pathogenicity (SHIVMJ4 and SHIV-XJ02170) (41, 58). In contrast, SHIV-1157ipd3N4 was mucosally transmissible and exhibited uniform robust viral replication kinetics during acute viremia; in fact, a high-titer stock has been titrated by the intrarectal route. Thus, this new R5 clade C SHIV can be used to assess vaccine efficacy, with prevention of infection, lowering of peak viral loads, and lowering of postacute viremia levels as read-out parameters. Longer term follow-up studies of SHIV-1157ipd3N4-infected monkeys will determine whether protection from disease progression will be an additional criterion.
Genetic analysis of our data showed a major deletion in the cytoplasmic tail of the SHIV-1157ipd env gene that ended immediately proximal to the start codon of nef compared with the parental SHIV-1157i. This deletion led to the removal of the original stop codon of HIV-1 env and the overlapping of env and nef in the resultant SHIV-1157ipd. Therefore, the new cytoplasmic tail of gp41 contains both HIV-1 and SIVmac239 sequences, similar to the pattern of the cytoplasmic tail of SHIV89.6P env (47). This suggests that the carboxy-terminal cytoplasmic tail of SIVmac239 gp41 plays a very important role in the pathogenesis of SHIVs in rhesus monkeys.
The exclusive R5 tropism of SHIV-1157ipd3N4 represents a more biologically relevant coreceptor usage for mucosally transmitted viruses than the dual tropism of SHIV89.6P (47, 49), which has been widely used as the challenge virus in macaque vaccine trials (3, 5, 54). It is noteworthy that SHIV-1157ipd3N4 infection does not induce the acute, severe pathogenicity typically seen within 2 weeks of SHIV89.6P infection, which does not reflect the biology of acute HIV-1 infection in humans. Of note, SHIV89.6P infects and destroys naïve CD4+ T cells, whereas typical R5 viruses, such as SIVmac239 and SIVsmE543, affect predominantly the central memory T-cell subset (43). Indeed, the biological relevance of the SHIV89.6P challenge model has been called into question for these reasons (20, 42, 43). In contrast to SHIV89.6P, the gradual pathogenicity of SHIV-1157i and SHIV-1157ip (early form) more closely reflects the pattern of HIV-1 disease progression in humans, which is characterized by years of clinically stable, chronic infection before immune exhaustion sets in. While we have ruled out acute, severe pathogenicity of the late forms SHIV-1157ipd as well as SHIV-1157ipd3N4, the currently ongoing prospective studies will reveal the rates of disease progression in rhesus monkeys.
SHIV-1157ipd3N4 is the first highly replication-competent, mucosally transmissible R5 clade C SHIV. The fact that it induces reproducibly high peak viral RNA loads with high viral set points will allow viremia to be used as a readout in vaccine challenge studies (Rasmussen et al., unpublished).
Our late-stage virus, SHIV-1157ipd3N4, models another aspect of most HIV-1 clade C strains: it has an extra NF-κB site in the LTR compared to the standard version of the viral LTR (as in SIVmac239). Most HIV-1 clade B isolates are characterized by two NF-κB binding sites, whereas the majority of HIV-1 clade C isolates have an extra NF-κB binding site (23, 39). The number of NF-κB binding elements is associated with the efficacy of transcriptional initiation from the proviral genome, and TNF-α leads to the up-regulation of NF-κB (19, 44). In addition, a correlation between HIV-1 LTR subtype configuration of the NF-κB promoter region and responsiveness to TNF-α was reported earlier (37). The data derived from the in vitro pLuc expression controlled by a truncated LTR with zero to two NF-κB binding elements support the previously held notion (26, 37, 40) that the number of NF-κB binding sites in the LTR directly correlates with the level of gene expression driven by the LTRs. As expected, the additional NF-κB binding element in the SHIV-1157ipd3N4 LTRs rendered our virus more responsive to the effects of TNF-α than SHIV-1157ipd3.
In our intrarectal transmission studies of SHIV-1157ipd3N4, significantly higher peak viral RNA loads (week 2) were observed in Indian-origin rhesus macaques than in Chinese-origin rhesus macaques (Fig. 5A and B). Since we reisolated our late-stage virus after it replicated for 141 weeks in the Indian-origin monkey RPn-8, which had been given the original construct, we postulate that the late-stage virus was better adapted and more replication competent in Indian-origin monkeys than in Chinese-origin monkeys. Although some Chinese-origin rhesus monkeys received lower viral inocula, we and others have demonstrated a lack of any correlation between inoculum size, peak viremia levels (11), viral set points, and disease progression (7). This observation was made not only with intrarectal inoculation but also with oral administration (11). This lack of a correlation is not surprising for outbred populations of primates and is consistent with the idea that systemic infection occurs once the mucosal virus inoculum exceeds a minimal dose (11). Although previous studies have shown that the set points of SIV or SHIV in Indian-origin rhesus monkeys are generally higher than in Chinese-origin rhesus monkeys, conflicting results on the peak viremia have been observed (32, 33, 48, 56). Our data showed that the peak viremia of SHIV-1157ipd3N4 was significantly higher in Indian-origin rhesus monkeys than in Chinese-origin rhesus monkeys. Despite the differences in viral parameters postinoculation, our data show that SHIV-1157ipd3N4 can be used for intrarectal challenges in rhesus macaques regardless of their origin.
Our in vivo data indicate that SHIV-1157ipd3N4 is a highly replication-competent R5 clade C SHIV that can reproducibly infect rhesus macaques mucosally. Because its coreceptor usage reflects the tropism of the mucosally transmitted forms of HIV-1, SHIV-1157ipd3N4 may represent a practical tool to test the efficacy of vaccine candidates targeting Env of the world's most prevalent clade, HIV clade C, in a relevant primate model.
Acknowledgments
We thank Joseph Sodroski (Dana-Farber Cancer Institute, Boston, MA) for the gift of the SHIV-vpu+ proviral clones, Hermann Katinger (Polymune Scientific, Vienna, Austria) for NMAb 2G12, K. A. Buckley for technical support, Susan Sharp for assistance in the preparation of the manuscript, Daniel Anderson for pathology support, Stephanie Ehnert for coordinating sample collections, and Patrick Autissier for technical support in fluorescence-activated cell sorter analysis.
This work was supported in part by NIH grants R01 DE12937, R01 DE0160354, and R37 AI34266 to R.M.R., HD39620 and RR15635 to C.W., P01 AI48240 to R.M.R., C.W., and H.M.M., and RR00165, which provided base grant support to the Yerkes National Primate Research Center. J.B.W. was supported by a postdoctoral fellowship from the Canadian Institutes for Health Research.
REFERENCES
- 1.Alexander, L., P. O. Illyinskii, S. M. Lang, R. E. Means, J. Lifson, K. Mansfield, and R. C. Desrosiers. 2003. Determinants of increased replicative capacity of serially passaged simian immunodeficiency virus with nef deleted in rhesus monkeys. J. Virol. 77:6823-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T. W., Y. S. Jeong, D. Pennick, R. Bronson, M. F. Greene, and R. M. Ruprecht. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820-1825. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 6.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogers, W. M., R. Dubbes, P. ten Haaft, H. Niphuis, C. Cheng-Mayer, C. Stahl-Hennig, G. Hunsmann, T. Kuwata, M. Hayami, S. Jones, S. Ranjbar, N. Almond, J. Stott, B. Rosenwirth, and J. L. Heeney. 1997. Comparison of in vitro and in vivo infectivity of different clade B HIV-1 envelope chimeric simian/human immunodeficiency viruses in Macaca mulatta. Virology 236:110-117. [DOI] [PubMed] [Google Scholar]
- 8.Cayabyab, M., D. Rohne, G. Pollakis, C. Mische, T. Messele, A. Abebe, B. Etemad-Moghadam, P. Yang, S. Henson, M. Axthelm, J. Goudsmit, N. L. Letvin, and J. Sodroski. 2004. Rapid CD4+ T-lymphocyte depletion in rhesus monkeys infected with a simian-human immunodeficiency virus expressing the envelope glycoproteins of a primary dual-tropic Ethiopian clade C HIV type 1 isolate. AIDS Res. Hum. Retrovir. 20:27-40. [DOI] [PubMed] [Google Scholar]
- 9.Centlivre, M., P. Sommer, M. Michel, R. Ho Tsong Fang, S. Gofflo, J. Valladeau, N. Schmitt, F. Thierry, B. Hurtrel, S. Wain-Hobson, and M. Sala. 2005. HIV-1 clade promoters strongly influence spatial and temporal dynamics of viral replication in vivo. J. Clin. Investig. 115:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Z., Y. Huang, X. Zhao, E. Skulsky, D. Lin, J. Ip, A. Gettie, and D. D. Ho. 2000. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina). J. Virol. 74:6501-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chenine, A. L., F. Ferrantelli, R. Hofmann-Lehmann, M. G. Vangel, H. M. McClure, and R. M. Ruprecht. 2005. Older rhesus macaque infants are more susceptible to oral infection with simian-human immunodeficiency virus 89.6P than neonates. J. Virol. 79:1333-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 13.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 14.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 16.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewhurst, S., J. E. Embretson, D. C. Anderson, J. I. Mullins, and P. N. Fultz. 1990. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature 345:636-640. [DOI] [PubMed] [Google Scholar]
- 18.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 19.Duh, E. J., W. J. Maury, T. M. Folks, A. S. Fauci, and A. B. Rabson. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-κB sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA 86:5974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinberg, M. B., and J. P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207-210. [DOI] [PubMed] [Google Scholar]
- 21.Harouse, J. M., A. Gettie, T. Eshetu, R. C. Tan, R. Bohm, J. Blanchard, G. Baskin, and C. Cheng-Mayer. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIVSF162P3. J. Virol. 75:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harouse, J. M., A. Gettie, R. C. Tan, T. Eshetu, M. Ratterree, J. Blanchard, and C. Cheng-Mayer. 2001. Pathogenic determinants of the mucosally transmissible CXCR4-specific SHIV(SF33A2) map to env region. J. Acquir. Immun. Defic. Syndr. 27:222-228. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, A. J., X. Zou, and K. L. Calame. 1995. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J. Virol. 69:5337-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann-Lehmann, R., R. K. Swenerton, V. Liska, C. M. Leutenegger, H. Lutz, H. M. McClure, and R. M. Ruprecht. 2000. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res. Hum. Retrovir. 16:1247-1257. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, M., S. H. Ho, P. Balfe, A. Gettie, J. Harouse, J. Blanchard, and C. Cheng-Mayer. 2005. A CCR5-tropic simian-HIV molecular clone capable of inducing AIDS in rhesus macaques. J. Acquir. Immun. Defic. Syndr. 40:383-387. [DOI] [PubMed] [Google Scholar]
- 26.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joag, S. V., Z. Li, L. Foresman, E. B. Stephens, L. J. Zhao, I. Adany, D. M. Pinson, H. M. McClure, and O. Narayan. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70:3189-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 29.Kitabwalla, M., F. Ferrantelli, T. Wang, A. Chalmers, H. Katinger, G. Stiegler, L. A. Cavacini, T. C. Chou, and R. M. Ruprecht. 2003. Primary African HIV clade A and D isolates: effective cross-clade neutralization with a quadruple combination of human monoclonal antibodies raised against clade B. AIDS Res. Hum Retrovir. 19:125-131. [DOI] [PubMed] [Google Scholar]
- 30.Klinger, J. M., S. Himathongkham, H. Legg, P. A. Luciw, and S. W. Barnett. 1998. Infection of baboons with a simian immunodeficiency virus/HIV-1 chimeric virus constructed with an HIV-1 Thai subtype E envelope. AIDS 12:849-857. [DOI] [PubMed] [Google Scholar]
- 31.Li, J. T., M. Halloran, C. I. Lord, A. Watson, J. Ranchalis, M. Fung, N. L. Letvin, and J. G. Sodroski. 1995. Persistent infection of macaques with simian-human immunodeficiency viruses. J. Virol. 69:7061-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling, B., R. S. Veazey, A. Luckay, C. Penedo, K. Xu, J. D. Lifson, and P. A. Marx. 2002. SIV− pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS 16:1489-1496. [DOI] [PubMed] [Google Scholar]
- 33.Ling, B., R. S. Veazey, C. Penedo, K. Xu, J. D. Lifson, and P. A. Marx. 2002. Longitudinal follow up of SIVmac pathogenesis in rhesus macaques of Chinese origin: emergence of B cell lymphoma. J. Med. Primatol. 31:154-163. [DOI] [PubMed] [Google Scholar]
- 34.Long, E. M., S. M. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrovir. 18:567-576. [DOI] [PubMed] [Google Scholar]
- 35.Luciw, P. A., E. Pratt-Lowe, K. E. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. USA 92:7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCann, C. M., R. J. Song, and R. M. Ruprecht. 2005. Antibodies: can they protect against HIV infection? Curr. Drug Targets Infect. Disord. 5:95-111. [DOI] [PubMed] [Google Scholar]
- 37.Montano, M. A., C. P. Nixon, T. Ndung'u, H. Bussmann, V. A. Novitsky, D. Dickman, and M. Essex. 2000. Elevated tumor necrosis factor-alpha activation of human immunodeficiency virus type 1 subtype C in southern Africa is associated with an NF-κB enhancer gain-of-function. J. Infect. Dis. 181:76-81. [DOI] [PubMed] [Google Scholar]
- 38.Montano, M. A., V. A. Novitsky, J. T. Blackard, N. L. Cho, D. A. Katzenstein, and M. Essex. 1997. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J. Virol. 71:8657-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munkanta, M., R. Handema, H. Kasai, C. Gondwe, X. Deng, A. Yamashita, T. Asagi, N. Yamamoto, M. Ito, F. Kasolo, and H. Terunuma. 2005. Predominance of three NF-κB binding sites in the long terminal repeat region of HIV type 1 subtype C isolates from Zambia. AIDS Res. Hum. Retrovir. 21:901-906. [DOI] [PubMed] [Google Scholar]
- 40.Naghavi, M. H., S. Schwartz, A. Sonnerborg, and A. Vahlne. 1999. Long terminal repeat promoter/enhancer activity of different subtypes of HIV type 1. AIDS Res. Hum. Retrovir. 15:1293-1303. [DOI] [PubMed] [Google Scholar]
- 41.Ndung'u, T., Y. Lu, B. Renjifo, N. Touzjian, N. Kushner, V. Pena-Cruz, V. A. Novitsky, T. H. Lee, and M. Essex. 2001. Infectious simian/human immunodeficiency virus with human immunodeficiency virus type 1 subtype C from an African isolate: rhesus macaque model. J. Virol. 75:11417-11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimura, Y., C. R. Brown, J. J. Mattapallil, T. Igarashi, A. Buckler-White, B. A. Lafont, V. M. Hirsch, M. Roederer, and M. A. Martin. 2005. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc. Natl. Acad. Sci. USA 102:8000-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura, Y., T. Igarashi, O. K. Donau, A. Buckler-White, C. Buckler, B. A. Lafont, R. M. Goeken, S. Goldstein, V. M. Hirsch, and M. A. Martin. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. USA 101:12324-12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osborn, L., S. Kunkel, and G. J. Nabel. 1989. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. USA 86:2336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pope, M., and A. T. Haase. 2003. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat. Med. 9:847-852. [DOI] [PubMed] [Google Scholar]
- 46.Rabson, A. B., and H. C. Lin. 2000. NF-kappa B and HIV: linking viral and immune activation. Adv. Pharmacol. 48:161-207. [DOI] [PubMed] [Google Scholar]
- 47.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I. W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimann, K. A., R. A. Parker, M. S. Seaman, K. Beaudry, M. Beddall, L. Peterson, K. C. Williams, R. S. Veazey, D. C. Montefiori, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J. Virol. 79:8878-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimann, K. A., A. Watson, P. J. Dailey, W. Lin, C. I. Lord, T. D. Steenbeke, R. A. Parker, M. K. Axthelm, and G. B. Karlsson. 1999. Viral burden and disease progression in rhesus monkeys infected with chimeric simian-human immunodeficiency viruses. Virology 256:15-21. [DOI] [PubMed] [Google Scholar]
- 50.Roos, J. W., M. F. Maughan, Z. Liao, J. E. Hildreth, and J. E. Clements. 2000. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology 273:307-315. [DOI] [PubMed] [Google Scholar]
- 51.Sanders, M. E., M. W. Makgoba, S. O. Sharrow, D. Stephany, T. A. Springer, H. A. Young, and S. Shaw. 1988. Human memory T lymphocytes express increased levels of three cell adhesion molecules (LFA-3, CD2, and LFA-1) and three other molecules (UCHL1, CDw29, and Pgp-1) and have enhanced IFN-gamma production. J. Immunol. 140:1401-1407. [PubMed] [Google Scholar]
- 52.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 55.Spouge, J. L. 1992. Statistical analysis of sparse infection data and its implications for retroviral treatment trials in primates. Proc. Natl. Acad. Sci. USA 89:7581-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trichel, A. M., P. A. Rajakumar, and M. Murphey-Corb. 2002. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J. Med. Primatol. 31:171-178. [DOI] [PubMed] [Google Scholar]
- 57.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, Y., K. Hong, A. L. Chenine, J. B. Whitney, W. Xu, Q. Chen, Y. Geng, R. M. Ruprecht, and Y. Shao. 2005. Molecular cloning and in vitro evaluation of an infectious simian-human immunodeficiency virus containing env of a primary Chinese HIV-1 subtype C isolate. J. Med. Primatol. 34:101-107. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]