Abstract
The vaccinia virus G3L/WR079 gene encodes a conserved protein with a predicted transmembrane domain. Our proteomic analyses of vaccinia virus revealed that G3L protein is incorporated into intracellular mature virus; however, the function of G3L protein in the vaccinia virus life cycle has not been investigated. In this study, a recombinant vaccinia virus, viG3L, expressing G3L protein under IPTG (isopropyl-β-d-thiogalactopyranoside) regulation was constructed. Under permissive conditions when G3L protein was expressed, the vaccinia virus life cycle proceeded normally, resulting in plaque formation in BSC40 cells. In contrast, under nonpermissive conditions when G3L protein expression was repressed, no plaques were formed, showing that G3L protein is essential for vaccinia virus growth in cell cultures. In infected cells when G3L protein was not expressed, the formation of intracellular mature virus (IMV) and cell-associated enveloped virus occurred normally, showing that G3L protein is not required for virion morphogenesis. IMV particles containing (G3L+) or lacking (G3L−) G3L protein were purified and were found to be indistinguishable on microscopic examination. Both G3L+ and G3L− IMV bound to HeLa cells; however, G3L− IMV failed to enter the cells, showing that G3L protein is required for IMV penetration into cells. Finally, G3L protein was required for fusion of the infected cells under low-pH treatment. Thus, our results provide direct evidence that G3L is an essential component of the vaccinia virus fusion complex, in addition to the previously reported A28, H2, L5, A21, and A16 proteins.
Vaccinia virus, the prototype of the orthopoxvirus genus of the family Poxviridae, infects many cell lines and animals (9) and produces several forms of infectious particles, namely, intracellular mature virus (IMV), intracellular enveloped virus (IEV), cell-associated enveloped virus (CEV), and extracellular enveloped virus (EEV) (25). The membrane structure of IMV is different from that of IEV, which, again, is different from that of CEV and EEV (40). Although IMV and EEV were originally shown to enter cells through different mechanisms (22, 35, 39), recent studies have revealed that a conserved viral fusion complex is required for the penetration of both viruses (26, 27, 31, 32, 37, 38). Although no cellular coreceptor has been identified for vaccinia virus, some data suggest that cell-bound IMV initiates plasma membrane fusion to deliver viral cores into cells (1, 2, 8, 22, 39). In addition, IMV entry has been shown to trigger cell signaling pathways that involve Rac, MEK, extracellular signal-regulated kinase, protein kinase A, and protein kinase C activation, but the molecular mechanism is unknown (7, 20, 22).
IMV represents the majority of the infectious progeny produced in cells, and our recent proteomic study revealed that it contains 75 viral proteins (4). Of these, nine have been shown to play a role in vaccinia virus entry (see below). The viral envelope proteins, H3L and A27L, bind to heparan sulfates, whereas D8L binds to chondroitin sulfates (12, 13, 21). L1R is also implicated in virus penetration, since a monoclonal antibody (MAb) recognizing L1R blocks IMV entry at the penetration step (15, 43). Furthermore, five viral envelope proteins—A16L, A21L, A28L, H2R, and L5R—have been shown to be essential for vaccinia virus penetration into mammalian cells (27, 31, 33, 37, 38). All five proteins are conserved in poxviruses, have common features including a transmembrane domain, are rich in cysteine, and are required for cell-cell fusion. The functions of six novel virion proteins G3L, C6L, E6R, A6L, A15L, and A31R, which were identified in the vaccinia IMV proteome, remain unknown (4). Recently, two viral proteins, J5L and G3L, were shown to associate with these viral penetration proteins, but their role in virus entry has not been investigated (32). This study examined the function of G3L in the vaccinia virus life cycle and its role in IMV entry into cells.
MATERIALS AND METHODS
Reagents, cells, and viruses.
Mycophenolic acid, hypoxanthine, and xanthine (Sigma, Inc.) were dissolved at a concentration of 10 mg/ml in 0.1 N NaOH and stored at −20°C. Cytosine β-d-arabinofuranoside (araC) was purchased from Sigma, Inc. BSC40 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum (CS). Wild-type WR strain vaccinia virus was supplied by S. Pennathur. VT7LacOI was obtained from B. Moss (42). IMV virions were purified on a 36% sucrose cushion, followed by 25 to 40% sucrose gradient centrifugation as described previously (17) and stored at −70°C. Rabbit antibodies recognizing the vaccinia virus protein H3L were described previously (13, 22). Rabbit antibodies to viral cores were provided by J. Krijnse Locker (16, 28, 30). The anti-B5R MAb 206C5 is a mouse MAb from a hybridoma generated by immunization of mice with a recombinant B5 protein expressed as a glutathione S-transferase fusion protein in Escherichia coli. Rabbit anti-G3L antibodies were generated by using a synthetic peptide, NGKKHTFNLYDDNDIRTL, derived from the G3L sequence.
Generation of viG3L virus. (i) Plasmid construction.
To construct pMITEO-G3L containing an inducible copy of the G3L gene, the full-length G3L open reading frame (ORF) was generated by PCR using the primers 5′-AAACCATGGCATCTTTATTATATCTT-3′ and 5′-CCCGGATCCTCATTTACTAAGGAGTAAAAT-3′ (the NcoI and BamHI restriction sites are underlined) and the genomic DNA of vT7LacOI as the template (14). The PCR product was digested with NcoI and BamHI and cloned into pMITEOlac.20/3 to produce pMITEO-iG3L.
Three DNA fragments were used to replace the endogenous G3L gene with a gpt expression cassette. The 531-bp 5′ flanking fragment containing the G2R promoter and coding sequences was generated by PCR with the primers 5′-GCGGCCGCGGATAATATGTAAAATAA-3′ (the NotI site is underlined) and 5′-TAAATGTAACTTGAGAAA-3′ and vT7lacOI genomic DNA as the template. The 558-bp 3′ flanking fragment containing the G1L promoter and the coding sequences was generated by PCR using the primers 5′-GCGGCCGCGTAAAATTATAATGTCAC-3′ (the NotI site is underlined) and 5′-CTCGAGTATTAAGATTATCTATCA-3′ (XhoI site underlined) and vT7lacOI genomic DNA as the template. The 5′ and 3′ flanking DNA fragments were cloned into the pCRII-Topo vector (Invitrogen) to create pCRII-Topo-G2/G1. The 2.2-kb G3L expression cassette was purified from NotI-digested pMITEO-iG3L and cloned into pCRII-Topo-G2/G1 to obtain pCRII-G2/iG3L/G1. The sequences of the PCR fragments were confirmed by DNA sequencing.
(ii) Construction of the recombinant viG3L virus.
The recombinant viG3L virus was constructed based on previously described established protocols (24, 42). In brief, 3 × 105 BSC40 cells were seeded in a 60-mm dish, incubated for 1 day, and infected for 1 h at 37°C with vT7LacOI at a multiplicity of infection (MOI) of 1 PFU per cell. The cells were then washed three times with DMEM and transfected with 4 μg of pCRII-G2/iG3L/G1 in 40 μl of Arrest-In transfection reagent (Open Biosystems, Inc.). After 5 h, the transfection mixture was removed and replaced with DMEM containing 10% CS and 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside). Lysates were prepared at 2 days postinfection (p.i.) and used to infect BSC40 monolayer cells in the presence of 25 μg of mycophenolic acid/ml, 250 μg of xanthine/ml, 15 μg of hypoxanthine/ml, and 50 μM IPTG to select for plaques formed by viG3L, which expresses xanthine-guanine phosphoribosyltransferase (Gpt) and G3L. Pure recombinant viG3L viruses were obtained after three rounds of plaque purification. The insertion of Gpt and the inducible G3L gene into the endogenous G3L locus was confirmed by PCR.
One-step virus growth curve analysis.
BSC40 monolayer cells were infected for 1 h at 37°C with viG3L or vT7LacOI at an MOI of 5 PFU per cell. The cells were washed, incubated in complete DMEM containing 10% CS with or without 50 μM IPTG, harvested at various times after infection (0, 8, 12, 16, 24, and 48 h), subjected to three freeze-thaw cycles, and sonicated, and the virus titers were determined by plaque assay on BSC40 cells in the presence of 50 μM IPTG. The experiments were repeated three times.
Immunoblot analysis.
Viral proteins from purified IMV virions or extracts from virus-infected cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes, which were then blocked by incubation in I-block (2 μg/ml; Tropix) in phosphate-buffered saline (PBS) containing 0.5% Tween 20, and then incubated with primary antibodies to viral proteins, followed by alkaline phosphatase-conjugated secondary antibody. Bound antibody was then detected by using a chemiluminescence method according to the manufacturer's protocol (Tropix).
Membrane protein extraction from IMV.
Vaccinia IMVs were extracted with detergent and separated into membrane and core fractions essentially as described previously (3). Proteins in the pellet and supernatant were analyzed by SDS-PAGE on 12.5 or 15% polyacrylamide gels and transferred to nitrocellulose for immunoblot analyses with various antibodies as described above.
Electron microscopy of virion morphogenesis and purified IMV particles.
The experiments to monitor virion morphogenesis in cells were done as previously described (3). In brief, BSC40 cells were infected at an MOI of 5 PFU per cell, cultured in medium with or without IPTG, fixed at 12 or 24 h p.i., treated with 1% OsO4, dehydrated, and embedded as described previously (36). After embedding, the cells were stained with uranyl acetate and lead citrate and analyzed under a Zeiss 902 transmission electron microscope (29). For negative staining of IMV, 1-μl portions of serial dilutions of purified vaccinia IMV virions were spotted onto 300 mesh Parlodion-coated grids and stained with 2% phosphotungstic acid for 15 s, and then the virion particles were photographed with a Zeiss 902 transmission electron microscope as described previously (21).
Confocal immunofluorescence microscopy.
For virion entry assays, the amounts of cell surface-attached virions and uncoated cores within the cells were measured by confocal microscopy as described previously (41). In brief, HeLa cells (105 per well) were seeded on coverslips in 12-well plates. The cells were then infected for 1 h at 4°C with G3L+ virus at an MOI of 40 PFU per cell. Alternatively, the cells were infected with equal amounts of G3L− virus, as measured by determining the optical density at 260 nm. These cells were subsequently washed three times with PBS and either fixed immediately or first incubated for 2 h at 37°C in the presence of cycloheximide (30 μg/ml) and then fixed. Cells were fixed by incubation with 4% paraformaldehyde for 5 min at 4°C and then for 15 min at room temperature. The cells were then permeabilized in PBS-0.2% saponin and stained with rabbit anti-A4L antibody or mouse anti-L1R MAb, followed by fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG) or Cy5-conjugated goat anti-mouse IgG antibody, respectively. DNA was visualized by staining with 0.5 μg of DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride; Molecular Probes)/ml in mounting solution. Cell images were collected with a LSM510 META confocal laser scanning microscope (Carl Zeiss, Germany) using a ×63 objective lens and confocal microscopy software (release 2.8; Carl Zeiss). The number of fluorescent-staining particles was counted from multiple photos, and the averaged numbers of surface-bound virions and uncoated cores per cell were determined.
To visualize actin tails on CEV, HeLa cells (7 × 104) were seeded on coverslips in 12-well plates and infected with viG3L at an MOI of 5 PFU per cell. The infected cells were cultured in medium with or without 50 μM IPTG for 17 h p.i., fixed for 20 min at room temperature with 4% paraformaldehyde in PBS-0.5 μM Taxol (Molecular Probes), washed three times, permeabilized with 0.2% saponin-PBS, and incubated for 1 h at room temperature with anti-B5R MAb (1:2,500) and Alexa Fluor 647-phalloidin (Molecular Probes) and then for 30 min with Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (1:1,000; Molecular Probes). After three washes with PBS, the cells were stained for 5 min with 0.5 μg of DAPI (Molecular Probes)/ml and washed, and images were collected on a LSM510 META confocal laser scanning microscope as described above.
Cell fusion assay (fusion-from-within) induced by low-pH treatment.
Freshly confluent BSC-1 cells were infected with viG3L virus at an MOI of 5 PFU per cell and incubated at 37°C for 21 h in medium with or without IPTG. Cells were washed three times with PBS (pH 7.2), treated with PBS (pH 7.2) or PBS (pH 4.7) for 3 min at room temperature, washed again, and replaced with normal medium. These cells were incubated for another 3 h and photographed with a Nikon inverted microscope.
RESULTS
The conserved vaccinia G3L gene encodes a late viral envelope protein associated with IMV particles.
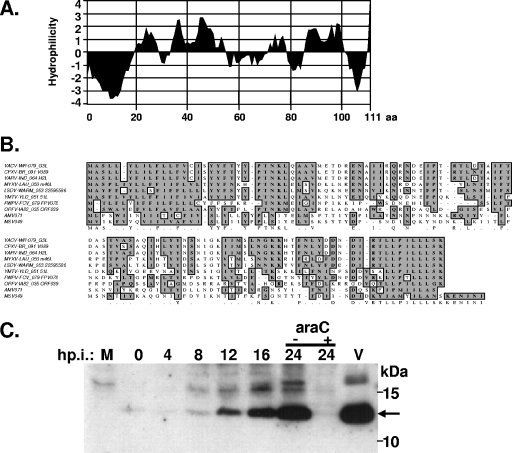
The vaccinia G3L gene encodes a conserved polypeptide of 111 amino acids with a predicted molecular mass of 12.8 kDa (10). The G3L transcript was previously detected at the late phase of virus infection, indicating that G3L belongs to the viral late gene family (23). Hydropathy analysis (18) predicted that G3L has two hydrophobic domains at the N and C termini (Fig. 1A). Alignment of the amino acid sequences of vaccinia G3L and its orthologues in the poxvirus family (Fig. 1B) revealed a high level of homology between these proteins (>46% conserved residues), suggesting that G3L might play an important function in the poxvirus life cycle. The internal region of G3L was less conserved than the N- and C-terminal regions.
FIG. 1.
(A). Hydropathy plot of G3L protein. The numbers at the bottom of the figure are the amino acid residues. (B) Alignment of the deduced amino acid sequences for vaccinia G3L and G3L orthologues in other poxviruses. VACV, vaccinia virus (WR strain); CPXV, cowpox virus (Brighton Red strain); VARV, variola virus (INDIA-1967/isolate IND3); MYXV, myxoma virus; LSDV, lumpy skin disease virus; YMTV, Yaba-like monkey tumor virus; FWPV, fowlpox virus; AMV, Amsacta moorei entomopoxvirus; MSV, Melanoplus sanguinipes entomopoxvirus. The boxed sequences in gray are conserved amino acid sequences. (C) Expression of G3L protein in infected cells and purified IMV virions. BSC40 cells were infected with vT7LacOI at an MOI of 5 PFU per cell and harvested at the indicated times postinfection; the lysates were then separated by SDS-12.5% PAGE and transferred to nitrocellulose for immunoblotting with the anti-G3L antiserum. M, mock-infected cells; V, purified IMV particles. When araC treatment was required, the drug (40 μg/ml) was added to the cells immediately after infection. The arrow shows the position of G3L protein.
Anti-G3L antibody was used to study the expression of G3L during vaccinia virus infection. Rabbits were immunized with a synthetic peptide derived from the G3L amino acid sequence (see Materials and Methods), and the antiserum produced was tested on immunoblots of lysates prepared from virus-infected cells. The antiserum did not recognize any protein in mock-infected cells but recognized a 12.8-kDa protein in virus-infected cells that was detected at 8 h p.i. and increased in abundance until 24 h p.i. (Fig. 1C). The 12.8-kDa protein was expressed at late phase in virus-infected cells, since araC, which inhibits viral DNA replication, blocked G3L expression. The antiserum also recognized a 12.8-kDa protein in purified IMV, demonstrating that G3L protein is present in IMV particles, a finding consistent with our proteomic analysis of vaccinia IMV (4).
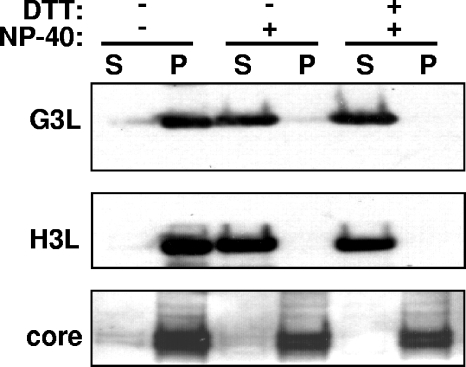
To determine whether G3L is present in the membrane fraction of IMV, purified IMVs were extracted with 1% NP-40 with or without 50 mM dithiothreitol (DTT), and the virion membrane proteins (supernatant) were separated from the insoluble core components (pellet) by centrifugation. As shown in Fig. 2, G3L was extracted from purified vaccinia IMV by buffer containing 1% NP-40; inclusion of 50 mM DTT during extraction did not result in greater release of G3L into the supernatant. These results showed that G3L is associated with membranes. Another IMV membrane protein, H3L, served as a control and was similarly extracted into the supernatant fraction (Fig. 2) (3). In contrast, the viral core proteins 4a/4b were resistant to detergent extraction. Our results showed that G3L is a late protein that is present in the membrane of vaccinia IMV.
FIG. 2.
Membrane and core proteins after NP-40-DTT extraction of vaccinia IMV. Sucrose-purified vaccinia IMVs were incubated with buffer containing 1% NP-40 with or without 50 mM DTT. After centrifugation, the supernatant (S) and pellet (P) were analyzed on immunoblots with antibodies to G3L, H3L, or core proteins as indicated in Materials and Methods.
Construction of a recombinant vaccinia virus expressing the inducible G3L gene under the control of the Lac operator.
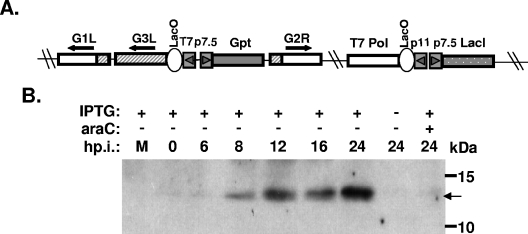
The role of G3L during the vaccinia virus life cycle in cell culture was explored using a recombinant vaccinia virus, viG3L, that expressed G3L conditionally regulated by IPTG (Fig. 3). As described in Materials and Methods, viG3L was generated from the vT7LacOI parental virus (42) with an inducible G3L cassette containing the E. coli gpt marker gene inserted into its endogenous locus (Fig. 3A). Recombinant virus, viG3L, was isolated in the presence of mycophenolic acid and purified after three rounds of plaque purification.
FIG. 3.
(A) Schematic diagram of viG3L virus. The G3L and J2R (tk) loci in the viG3L recombinant virus are indicated. The J2R locus contains T7 RNA polymerase and the lacI repressor gene as described previously (42). The inducible G3L (shaded box)/Gpt selectable marker (black box) gene cassette is also shown. The arrows indicate the transcription direction. Abbreviations: T7 Pol, bacteriophage T7 RNA polymerase; LacO, E. coli lac operator; LacI, E. coli lac repressor gene; Gpt, E. coli xanthine guanine phosphoribosyltransferase gene; p7.5 and p11, viral promoters; T7, promoter for T7 RNA polymerase. (B) Expression of G3L in viG3L-infected cells. BSC40 cells were infected with viG3L at an MOI of 5 PFU per cell, incubated in culture medium with or without 50 μM IPTG, then harvested at the indicated times, and subjected to SDS-PAGE and immunoblot analyses with the anti-G3L antiserum.
BSC40 cells were infected with viG3L and cultured in medium with or without IPTG for 24 h and harvested for immunoblot analysis. Abundant G3L was only detected in the infected cells from 8 to 24 h p.i. in the presence of IPTG, and its production was blocked by araC (Fig. 3B), demonstrating that expression of the G3L gene was tightly regulated at the late phase by IPTG.
G3L is required for plaque formation and IMV production in cell culture.
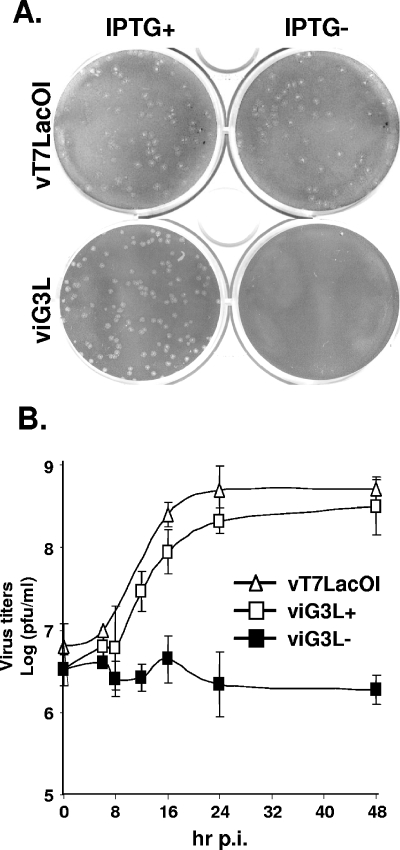
The role of G3L during vaccinia viral infection was examined in BSC40 cells infected with viG3L in the presence or absence of IPTG. As shown in Fig. 4A, at 3 days p.i. the control parental virus, vT7LacOI, formed similar plaques on BSC40 cells in the presence or absence of IPTG, whereas viG3L only formed plaques in the presence of IPTG, thus showing that G3L expression is required for plaque formation.
FIG. 4.
(A) ViG3L virus does not form plaques in BSC40 cells when the expression of G3L protein is repressed. BSC40 cells were infected with viG3L and incubated in medium with or without IPTG for 3 days, fixed, stained with crystal violet, and photographed. (B) One-step growth curve analysis of viG3L. BSC40 cells were infected with vT7LacOI or viG3L at an MOI of 5 PFU per cell and then incubated in the presence (viG3L+) or absence (viG3L− and vT7LacOI) of 50 μM IPTG for 0, 8, 12, 16, 24, or 48 h p.i. Virus titers in the lysates were determined by plaque formation on BSC40 cells. These experiments were performed three times.
The viG3L titer was measured in cell culture by using one-step growth analysis. BSC40 cells were infected with viG3L at an MOI of 5 PFU per cell and cultured in the presence or absence of IPTG, and then cell lysates were collected and the virus titers were determined. As shown in Fig. 4B, viG3L showed a 2-log increase in titer at 24 and 48 h p.i., similar to the vT7lacOI parental virus, whereas viG3L grew poorly in the absence of IPTG, with no increase in titer at 24 or 48 h p.i., showing that G3L is required for vaccinia virus growth in cell culture.
G3L is not required for virion morphogenesis of IMV and CEV.
Experiments were carried out to determine more precisely when and where G3L is required during the vaccinia virus life cycle. When viral protein synthesis was monitored by pulse-labeling, the results showed that G3L was not required for viral gene expression (data not shown). Moreover, pulse-chase experiments, performed to monitor the proteolytic conversion of the viral core p4a/p4b to mature 4a/4b proteins, demonstrated that processing of 4a/4b occurred normally during virion morphogenesis in the absence of G3L, showing that G3L is not required for the conversion of IV to IMV (data not shown).
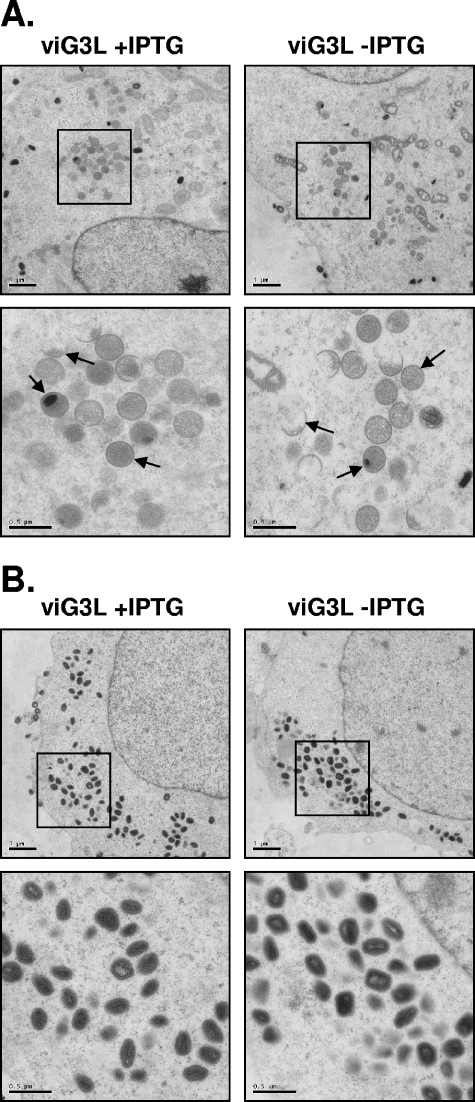
To demonstrate that mature virions were produced in cells, BSC40 cells infected with viG3L in the presence or absence of IPTG were analyzed at 12 and 24 h p.i. by electron microscopy. At 12 h p.i., viral crescents, immature virions, and other intermediate membrane structures were detected in cells infected with viG3L in the presence or absence of IPTG (Fig. 5A). At 24 h p.i., a large number of dense mature IMV particles were detected in the cytoplasm of cells infected with viG3L in the presence or absence of IPTG (Fig. 5B). These IMV particles in both cells appeared to be indistinguishable from each other. We therefore concluded that G3L is not required for the formation of IMV.
FIG. 5.
Electron micrographs of vaccinia virion morphogenesis in cells infected by viG3L. BSC40 cells were infected with viG3L at an MOI of 5 PFU per cell in the presence or absence of IPTG and then fixed at 12 h (A) or 24 h (B) p.i. for electron microscopy. Photographs were taken at magnifications of ×3,000 (top rows in panels A and B) and ×9,300 (lower rows in panels A and B). The arrows in panel A represent typical viral intermediate structures such as crescents and IV.
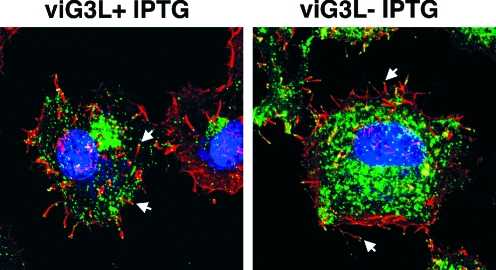
A fraction of IMV in cells is transported to the Golgi, where they are wrapped in additional membranes to form IEV, which, upon fusion with the plasma membrane, form CEV, with localized actin polymerization that can be readily detected as an “actin tail” by confocal microscopy (6, 34). HeLa cells were infected with viG3L in the presence or absence of IPTG, fixed at 17 p.i., and stained with phalloidin to examine actin tail formation. As shown in Fig. 6, formation of actin tails with CEV (labeled with anti-B5R MAb) at the tips was seen in the infected cells in the presence or absence of IPTG. The kinetics and the extent of actin tail formation appeared comparable in these cells, showing that G3L is not required for CEV formation.
FIG. 6.
Actin tail formation in association with CEV in infected cells. HeLa cells were infected with viG3L at an MOI of 5 PFU per cell, cultured in medium with (+IPTG) or without (−IPTG) 50 μM IPTG for 17 h p.i., fixed with 4% paraformaldehyde, permeabilized, and stained for 1 h at room temperature with anti-B5R MAb (1:2,500) (green) and Alexa Fluor 647-phalloidin (Molecular Probes) (red) and then for 30 min with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:1,000) (Molecular Probes). DNA was visualized by staining with DAPI (Molecular Probes) (blue); the cells were then washed, and images were collected on an LSM510 META confocal laser scanning microscope (Carl Zeiss, Germany) using a ×63 objective lens. The white arrows indicate representative CEV with actin tails on their tips.
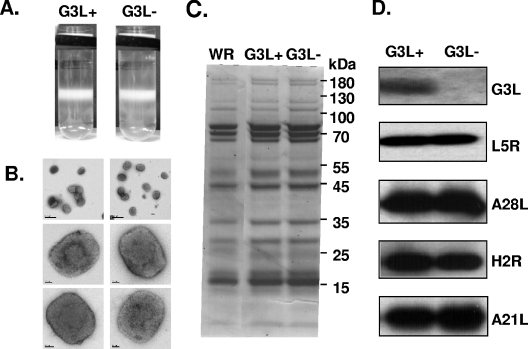
IMV particles devoid of G3L have a normal morphology and major protein content.
IMV particles containing G3L (G3L+) or lacking G3L (G3L−) were purified, respectively, from cells infected with viG3L cultured in the presence or absence of IPTG. The two types of IMV particles (G3L+ and G3L−) showed the same sedimentation profile on sucrose gradient centrifugation (Fig. 7A). When both types of IMV virions were analyzed by electron microscopy, no detectable differences in morphology or size were detected (Fig. 7B). SDS-PAGE analysis of these IMV showed an identical major protein pattern (Fig. 7C). Although G3L was not incorporated into purified IMV, the abundance of the envelope proteins, L5R, A28L, H2R, and A21L, all of which interact with G3L, was not affected (Fig. 7D).
FIG. 7.
Structural analyses of G3L+ and G3L− IMV by electron microscopy and SDS-PAGE. (A) Purified G3L+ and G3L− IMV on 25 to 40% sucrose gradients. (B) Electron micrographs of negatively stained G3L+ and G3L− IMV. Virions were deposited on grids, washed with water, and stained with 7% uranyl acetate in 50% ethanol for 30 s. (C) SDS-PAGE of sucrose gradient-purified wild-type (WR) vaccinia, G3L+, or G3L− IMV. The numbers of particles were determined from the optical density at 260 nm, and equal amounts of the three types of virions were analyzed by SDS-PAGE with Coomassie blue staining. (D) Immunoblot of G3L+ and G3L− IMV. Equivalent amounts of G3L+ and G3L− IMV were separated on SDS-PAGE and analyzed by immunoblots with antibodies recognizing the indicated envelope proteins.
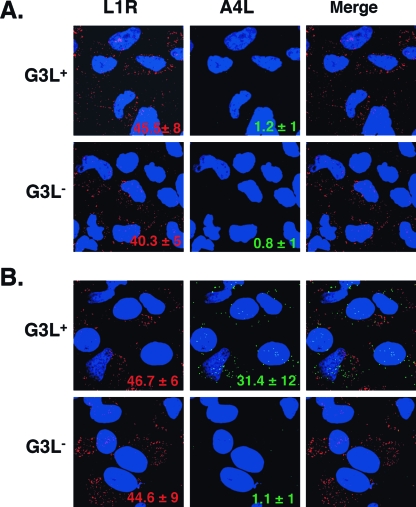
G3L is essential for IMV penetration into cells.
To investigate the role of G3L in the IMV entry process, we used a previously described virus entry assay (41). HeLa cells were infected for 1 h at 4°C with purified G3L+ and G3L− IMVs and then examined by confocal microscopy to determine the number of IMV particles bound to cells (Fig. 8A). Alternatively, HeLa cells were infected as described above and then, after being washed, were cultured at 37°C for 2 h to allow IMV penetration into cells before harvesting (Fig. 8B). The infected cells were stained with anti-L1R MAb to detect bound virions on the cell surface or with anti-A4L antiserum to detect uncoated cores of penetrated virus in the cytoplasm. As shown in Fig. 8A, similar numbers of G3L+ ( 45.5 ± 8) and G3L− (40.3 ± 5) IMV particles bound to cells, indicating that G3L is not required for IMV binding to cells. In contrast, as shown in Fig. 8B, significant A4L staining was only seen in cells infected with G3L+ virus (31.4 ± 12) and not with G3L− virus (1.1 ± 1), indicating that G3L+ viruses penetrated into the cells and successfully uncoated inside the cells. These results showed that G3L plays an essential role in IMV penetration into cells.
FIG. 8.
G3L is required for virion penetration to release cores into the cytosol. HeLa cells were infected for 1 h at 4°C with equivalent amounts of G3L+ and G3L− virions (determined from the optical density at 260 nm) and then the cells were washed extensively and either immediately fixed (A) or incubated for a further 2 h at 37°C in the presence of cycloheximide before fixation (B). Cells were stained with anti-L1R MAb, followed by Cy5-conjugated goat anti-mouse IgG antibody (red) or with rabbit anti-A4L antiserum, followed by FITC-conjugated goat anti-rabbit IgG antibody (green). DNA was visualized by staining with DAPI. Optical sections were obtained by confocal microscopy and are displayed as maximum-intensity projections as described previously (5). The numbers of fluorescent-staining particles were counted from multiple photos and averaged.
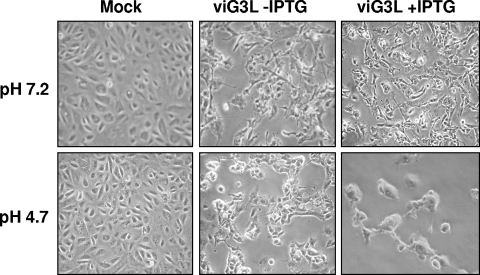
G3L is essential for cell-cell fusion induced by low-pH treatment.
Cells infected by vaccinia virus undergo cell fusion when they are briefly incubated in acidic buffer with a pH of <6 (8, 11). Although the parameters mediating virus and cell fusion (fusion from without) are not necessarily identical to those for the fusion of infected cells (fusion from within), both cell fusion assays have been widely used to investigate virus-mediated cell fusion. Moreover, several components of viral entry-fusion proteins—A28L, H2R, L5R, and A21L—were shown to be required for low pH-induced cell fusion from within or fusion from without (31, 33, 37, 38). Since G3L protein is essential for IMV penetration, its role in cell fusion from within was examined (Fig. 9). BSC-1 cells infected with viG3L virus and maintained in medium containing IPTG did not develop cell fusion at neutral pH at 24 h p.i. However, when these cells were briefly treated with acidic buffer they developed into gigantic fused cells. In the absence of IPTG when G3L protein is not expressed no cell fusion was observed at either neutral or acidic buffer. We thus conclude that G3L protein is required for low-pH-triggered cell fusion.
FIG. 9.
G3L is required for cell fusion. Freshly confluent BSC-1 cells were infected with viG3L virus at an MOI of 5 PFU per cell and incubated at 37°C for 21 h in medium with or without IPTG. Cells were washed three times with PBS (pH 7.2) and treated with PBS (pH 7.2) or PBS (pH 4.7) for 3 min at room temperature, and washed again, and the PBS was then replaced with normal medium. These cells were incubated for another 3 h and photographed with a Nikon inverted microscope.
DISCUSSION
The vaccinia G3L ORF encodes a conserved protein of 111 amino acids in size and shares significant homology with its orthologues in the Poxviridae (10). The G3L sequence is not homologous to that of any known cellular protein in the current database. A previous attempt to identify gene defects in a collection of vaccinia virus temperature-sensitive mutants showed that the temperature-sensitive mutant Cts60 was rescued by the G3L ORF, suggesting that G3L is important for virus growth in cell culture (19). In the present study, we demonstrated that G3L plays an essential role in the IMV penetration step. The phenotype of the G3L− virus was identical to that previously reported for mutant viruses defective in A28L, H2R, L5R, A21L, or A16L (27, 31, 33, 37, 38). Thus, vaccinia virus contains at least six proteins that are essential for IMV penetration into cells.
During construction of the viG3L virus, the inducible G3L gene cassette was inserted into its endogenous locus between G1 and G2 ORFs in the virus genome. To be sure that insertion of the inducible G3L gene cassette did not interrupt expression of the neighboring G2R ORF, we included 50-nucleotide upstream sequences that contain putative early promoters for G2R ORF in the flanking region. That viG3L virus was successfully isolated and did not exhibit the G2R mutant phenotype (23) suggested that G2R expression is intact in viG3L virus and that early promoter for G2R resides within the upstream 50 nucleotides as expected. On the other hand, Meis and Condit determined the G2 early transcript (23) and concluded that the G2R promoter is 600 nucleotides upstream of the G2R ORF, which means that G2R transcription is initiated in G1L and transcribed through the entire G3L ORF and into G2R. However, this raised an interesting puzzle regarding how G2R is transcribed in viG3L. Based on our rationale, we think that the most likely possibility is that G2R transcript is initiated from viral early promoter resides within the 50 nucleotides upstream of the G2R ORF. Alternatively, although less likely, G2R transcript could be generated from the p7.5 promoter that is situated even farther upstream of the Gpt ORF. We cannot exclude the possibility that an active G2R promoter exists 600 nucleotides upstream, as described by Meis and Condit, although it is difficult to comprehend how that transcription is achieved in viG3L. At this point we have not investigated this issue yet, and this will require more experiments in the future.
G3L was detected in association with the virion membrane, possibly through the N-terminal transmembrane region. In contrast to several envelope proteins that are rich in cysteine (31, 33, 37, 38), G3L contains only one cysteine and does not appear to form intra- or intermolecular disulfide bonds, a finding consistent with our finding that G3L was efficiently extracted from purified IMV by detergent even in the absence of DTT. How G3L interacts with other viral fusion proteins during the complex formation will be investigated in the future.
Assembly and disassembly of viral fusion complex needs to be tightly regulated during the virus life cycle. The fusion activity of these viral fusion proteins must be suppressed during virus assembly in cells. The mature virion progeny also need to be protected from aberrant fusion activation. In contrast, during virus entry into cells, activation of the multimeric fusion proteins on virus particles is often achieved through receptor interaction and conformational alteration, resulting in fusion of the virus envelope with cell membrane. The requirement of six or more envelope proteins for vaccinia virus penetration and fusion is unprecedented compared to other viruses. Whether poxvirus evolves a novel fusion mechanism that is distinct from other viral fusion proteins remains to be determined.
Acknowledgments
We thank Sue-Ping Lee for excellent technical support for the electron microscopy. We also thank J. Krijnse Locker for providing the anti-core antibodies.
This study was supported by grants from the Academia Sinica and the National Science Council (NSC96-2627-M-001-004) of the Republic of China.
REFERENCES
- 1.Armstrong, J. A., D. H. Metz, and M. R. Young. 1973. The mode of entry of vaccinia virus into L cells. J. Gen. Virol. 21:533-537. [DOI] [PubMed] [Google Scholar]
- 2.Chang, A., and D. H. Metz. 1976. Further investigations on the mode of entry of vaccinia virus into cells. J. Gen. Virol. 32:275-282. [DOI] [PubMed] [Google Scholar]
- 3.Chiu, W. L., and W. Chang. 2002. Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis. J. Virol. 76:9575-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, C. S., C. Y. Huang, and W. Chang. 2005. Vaccinia virus penetration requires cholesterol and results in specific viral envelope proteins associated with lipid rafts. J. Virol. 79:1623-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 7.de Magalhaes, J. C., A. A. Andrade, P. N. Silva, L. P. Sousa, C. Ropert, P. C. Ferreira, E. G. Kroon, R. T. Gazzinelli, and C. A. Bonjardim. 2001. A mitogenic signal triggered at an early stage of vaccinia virus infection: implication of MEK/ERK and protein kinase A in virus multiplication. J. Biol. Chem. 276:38353-38360. [DOI] [PubMed] [Google Scholar]
- 8.Doms, R. W., R. Blumenthal, and B. Moss. 1990. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 64:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenner, F. 1990. Poxviruses, p. 2113-2133. In B. Fields and D. M. Knipe (ed.), Virology. Raven Press, New York, N.Y.
- 10.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virol. 179:247-266. [DOI] [PubMed] [Google Scholar]
- 11.Gong, S. C., C. F. Lai, and M. Esteban. 1990. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N terminus of the 14-kDa virus envelope protein. Virology 178:81-91. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao, J.-C., C.-S. Chung, and W. Chang. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycans (GAGs)-binding domain. J. Virol. 72:8374-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiao, J.-C., C.-S. Chung, and W. Chang. 1999. Vaccinia envelope D8L protein binds to cell surface chondroitin sulfate and mediates intracellular mature virions adsorption to cells. J. Virol. 73:8750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, X., L. J. Carroll, E. J. Wolffe, and B. Moss. 1996. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J. Virol. 70:7669-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichihashi, Y., and M. Oie. 1996. Neutralizing epitope on penetration protein of vaccinia virus. Virology 220:491-494. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 18:9-18. [DOI] [PubMed] [Google Scholar]
- 18.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 19.Lackner, C. A., S. M. D'Costa, C. Buck, and R. C. Condit. 2003. Complementation analysis of the dales collection of vaccinia virus temperature-sensitive mutants. Virology 305:240-259. [DOI] [PubMed] [Google Scholar]
- 20.Leao-Ferreira, L. R., R. Paes-De-Carvalho, F. G. De Mello, and N. Moussatche. 2002. Inhibition of vaccinia virus replication by adenosine in BSC-40 cells: involvement of A(2) receptor-mediated PKA activation. Arch. Virol. 147:1407-1423. [DOI] [PubMed] [Google Scholar]
- 21.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locker, J. K., A. Kuehn, S. Schleich, G. Rutter, H. Hohenberg, R. Wepf, and G. Griffiths. 2000. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 11:2497-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meis, R. J., and R. C. Condit. 1991. Genetic and molecular biological characterization of a vaccinia virus gene which renders the virus dependent on isatin-beta-thiosemicarbazone (IBT). Virology 182:442-454. [DOI] [PubMed] [Google Scholar]
- 24.Mohamed, M. R., and E. G. Niles. 2004. Transient and inducible expression of vaccinia/T7 recombinant viruses. Methods Mol. Biol. 269:41-50. [DOI] [PubMed] [Google Scholar]
- 25.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 26.Moss, B. 2006. Poxvirus entry and membrane fusion. Virology 344:48-54. [DOI] [PubMed] [Google Scholar]
- 27.Ojeda, S., T. G. Senkevich, and B. Moss. 2006. Entry of vaccinia virus and cell-cell fusion require a highly conserved cysteine-rich membrane protein encoded by the A16L gene. J. Virol. 80:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. K. Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, E. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 55:541-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. K. Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senkevich, T. G., and B. Moss. 2005. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 79:4744-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senkevich, T. G., S. Ojeda, A. Townsley, G. E. Nelson, and B. Moss. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. USA 102:18572-18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senkevich, T. G., B. M. Ward, and B. Moss. 2004. Vaccinia virus A28L gene encodes an essential protein component of the virion membrane with intramolecular disulfide bonds formed by the viral cytoplasmic redox pathway. J. Virol. 78:2348-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, G. L., B. J. Murphy, and M. Law. 2003. Vaccinia virus motility. Annu. Rev. Microbiol. 57:323-342. [DOI] [PubMed] [Google Scholar]
- 35.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus: entry, egress, and evasion. Adv. Exp. Med. Biol. 440:395-414. [PubMed] [Google Scholar]
- 36.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 37.Townsley, A. C., T. G. Senkevich, and B. Moss. 2005. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J. Virol. 79:10988-10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Townsley, A. C., T. G. Senkevich, and B. Moss. 2005. Vaccinia virus A21 virion membrane protein is required for cell entry and fusion. J. Virol. 79:9458-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanderplasschen, A., M. Hollinshead, and G. L. Smith. 1998. Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J. Gen. Virol. 79(Pt. 4):877-887. [DOI] [PubMed] [Google Scholar]
- 40.Vanderplasschen, A., E. Mathew, M. Hollinshead, R. B. Sim, and G. L. Smith. 1998. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 95:7544-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanderplasschen, A., and G. L. Smith. 1997. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 71:4032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, G. A., C. K. Stover, B. Moss, and T. R. Fuerst. 1995. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc. Natl. Acad. Sci. USA 92:6773-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolffe, E. J., S. Vijaya, and B. Moss. 1995. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 211:53-63. [DOI] [PubMed] [Google Scholar]