Abstract
The mechanisms by which reoviruses induce apoptosis have not been fully elucidated. Earlier studies identified the mammalian reovirus S1 and M2 genes as determinants of apoptosis induction. However, no published results have demonstrated the capacities of the proteins encoded by these genes to induce apoptosis, either independently or in combination, in the absence of reovirus infection. Here we report that the mammalian reovirus μ1 protein, encoded by the M2 gene, was sufficient to induce apoptosis in transfected cells. We also found that μ1 localized to lipid droplets, endoplasmic reticulum, and mitochondria in both transfected cells and infected cells. Two small regions encompassing amphipathic α-helices within a carboxyl-terminal portion of μ1 were necessary for efficient induction of apoptosis and association with lipid droplets, endoplasmic reticulum, and mitochondria in transfected cells. Induction of apoptosis by μ1 and its association with lipid droplets and intracellular membranes in transfected cells were abrogated when μ1 was coexpressed with σ3, with which it is known to coassemble. We propose that μ1 plays a direct role in the induction of apoptosis in infected cells and that this property may relate to the capacity of μ1 to associate with intracellular membranes. Moreover, during reovirus infection, association with σ3 may regulate apoptosis induction by μ1.
The mammalian orthoreoviruses (reoviruses) are nonenveloped viruses that carry 10 genomic segments of double-stranded RNA within two concentric capsid layers. Reovirus infection has become a well-studied viral model of apoptosis in a variety of cultured cells as well as in animals, where virus-induced apoptosis is a primary pathogenetic mechanism of tissue damage (reviewed in reference 17). Differences among reovirus strains in their capacities to induce apoptosis have allowed genetic determinants to be mapped to two viral genes: S1, encoding both outer capsid protein σ1 and nonstructural protein σ1s, and M2, encoding outer capsid protein μ1 (51, 60, 61).
Much work has focused on the role of the S1-encoded σ1 protein in reovirus-induced apoptosis. σ1 is a homotrimeric attachment protein (15, 56) that binds to junctional adhesion molecule A (JAM-A) and to α-linked sialic acid residues on the surface of susceptible cells (3, 4, 14, 46). Both of these binding events are required for apoptosis induction by type 3 (T3) reovirus strains (4, 19), and these findings have led to the proposal that binding of T3 reovirus σ1 to cell surface JAM-A and α-sialic acid transduces signals that are required for apoptosis induction in cultured cells (4). The role of sialic acid binding in animals may be complex, however, in that it plays a role in apoptosis induction in the central nervous system but not in myocardial tissues of infected mice (32). Recently, the hypothesis that σ1 binding of JAM-A and sialic acid initiates signals required for apoptosis induction has been seemingly disproven by findings that these binding activities of σ1 are dispensable for reovirus-induced apoptosis; specifically, virions with monoclonal antibodies (MAbs) bound to σ1 can infect and induce apoptosis in CHO cells that express Fc receptor but not JAM-A or sialic acid; moreover, reassortant mapping shows that the M2-encoded μ1 protein is the sole proapoptotic virus determinant in these cells (23).
The S1 gene is bicistronic and encodes nonstructural protein σ1s in an overlapping reading frame relative to σ1. σ1s is responsible for reovirus-induced cell cycle arrest at G2/M (47) but is not required for reovirus-induced apoptosis in cultured cells (52). Again, however, the situation in animals appears to be more complex, as σ1s-null viruses are less virulent and induce lower levels of apoptosis in the heart and central nervous system of infected mice (32).
At high multiplicity of infection (MOI) in cultured cells, reoviruses can induce apoptosis without the need for viral transcription or replication (61). Apoptosis induced in this way appears to require both binding to JAM-A and α-sialic acid followed by conversion of virions to intermediate subviral particles (ISVPs) by endo/lysosomal proteolysis (20). Little is known about the roles played by μ1 in reovirus-induced apoptosis. μ1 is a major outer capsid protein (600 copies per virion) that undergoes proteolytic processing within endo/lysosomes following viral uptake from cell surfaces, and multiple lines of evidence suggest that one or more of the μ1 cleavage products interact with cellular membranes to effect penetration of a partially disassembled, subviral particle into the cytoplasm (7, 9, 10, 42, 44). Thus, a cleavage product of μ1 might provide the postbinding signal required for induction of apoptosis during cell entry. Indeed, we have shown that a large fragment of virion-derived μ1 enters the cytosol and nucleus of infected cells early in infection (11).
Although high-multiplicity infection by reovirus in cultured cells can induce apoptosis without the need for viral transcription or replication, the latter processes appear to be required for efficient apoptosis induction in cells infected at lower multiplicity (61). In addition, as reovirus-induced apoptosis occurs relatively late in the infectious cycle (17), it seems likely that replicative events are needed to amplify the proapoptotic signals that accompany cell entry by reovirus. Thus, de novo expression of the S1-encoded σ1, S1-encoded σ1s, and/or M2-encoded μ1 proteins might play a role in apoptosis induction during reovirus infection.
In this study, we tested the hypothesis that μ1 can induce apoptosis independently of σ1 and σ1s when expressed in cultured cells. We show that μ1 induced apoptosis and localized to lipid droplets, endoplasmic reticulum (ER), and mitochondria in transfected cells and had similar distributions in infected cells. We further show that regions encompassing two amphipathic α-helices within a carboxyl (C)-terminal portion of μ1 were key determinants of apoptosis induction and localization to lipid droplets, ER, and mitochondria. Apoptosis induction and localization to lipid droplets and intracellular membranes by μ1 in transfected cells were inhibited by coexpression of its assembly partner σ3, suggesting that in infected cells the levels of free μ1 may regulate apoptosis. Based on these findings, we conclude that the μ1 protein and, in fact, specific small regions of this protein play a major role in apoptosis induction during reovirus infection.
MATERIALS AND METHODS
Cells and viruses.
CHO and CV-1 cells were grown in Ham's F-12 medium (CellGro) supplemented with 10% fetal bovine serum (HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and nonessential amino acids (CellGro). Reoviruses T1L and T3D were laboratory stocks of the isolates previously identified as T1/human/Ohio/Lang/1953 and T3/human/Ohio/Dearing/1955, respectively, (27). The superscript N in T3DN differentiates a laboratory stock obtained from the Nibert laboratory from a T3D clone obtained from L. W. Cashdollar (Medical College of Wisconsin), denoted T3DC. The T3DC clone differs from the T3DN clone in viral factory morphology and in the nucleotide sequence of its M1 genome segment (45). Viruses were plaque isolated and amplified in murine L929 cells in Joklik's modified minimal essential medium (Gibco) supplemented with 2% fetal bovine serum, 2% bovine calf serum (HyClone), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Antibodies and reagents.
Mouse MAbs to μ1 (4A3) and σ3 (5C3) and rabbit polyclonal antiserum to μNS and reovirus cores have been described previously (8, 59, 62). MitoTracker Red CMXros (Molecular Probes), MAbs to human golgin-97 (Molecular Probes), adipose differentiation-related protein (ADRP) (Progen), CD63 (LAMP1) (BD Pharmingen), calnexin (Affinity BioReagents), and protein disulfide isomerase (PDI) (Molecular Probes) were used as organelle markers for mitochondria, Golgi, lipid droplets, lysosomes, and ER, respectively. Rabbit polyclonal antiserum to activated caspase-3 was obtained from Cell Signaling Technologies. The FLAG epitope tag was detected with anti-FLAG M2 MAb (Stratagene or Sigma). Secondary antibodies for immunofluorescence (IF) microscopy were goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG conjugated to Alexa 488 or Alexa 594 (Molecular Probes). All antibodies were titrated to optimize signal-to-noise ratios.
Plasmid construction.
The reovirus M2 and S4 genes from T1L were subcloned from plasmids pBKS-M2L and pcDNAI-S4L (12) into the mammalian expression vector pCI-neo (Promega) to produce pCI-M2(T1L) and pCI-S4(T1L), respectively.
To generate in-frame truncations of μ1, as well as in-frame fusions with enhanced green fluorescent protein (EGFP), fragments of the M2 gene were PCR amplified using pCI-M2(T1L) as the template. The primers used for amplification are shown in Tables 1 and 2. PCR products were purified and cut with the appropriate restriction enzymes (Tables 1 and 2) and then ligated into the indicated vectors cut with the same enzymes. The pEGFP-N1 and pEGFP-C1 vectors were purchased from Clontech.
TABLE 1.
Primers used to prepare plasmids expressing M2 truncations
| Constructa | Primer sequence (5′ to 3′)b
|
|
|---|---|---|
| Forward | Reverse | |
| pCI-M2(1-675) | TATGCTAGCATGGGGAACGCTTCCTCTATC | ATCTCGAGTCAAGGCGCTGGAAAGTGCGTCGA |
| pCI-M2(1-582) | TATGCTAGCATGGGGAACGCTTCCTCTATC | AAACTCGAGTCAACCATACCCAGTCTCAAG |
| pCI-M2(43-582) | TTTGCTAGCATGCCTGGAGGAGTACCATG | AAACTCGAGTCAACCATACCCAGTCTCAAG |
| pCI-M2(43-708) | TTTGCTAGCATGCCTGGAGGAGTACCATG | AAACTCGAGTCAGCGTGTATACCCACGCTTAACCAC |
Each construct was designed to contain the portion of the M2 gene encoding the indicated amino acid residues of μ1. The pCI-neo vector and all PCR products were digested with NheI and XhoI for cloning.
Forward primers are in the same orientation as the coding strand; reverse primers are in the reverse orientation to the coding strand. Restriction enzyme sites are in italics, and the inserted start and stop codons are underlined.
TABLE 2.
Primers used to prepare plasmids expressing EGFP-M2 truncations
| Constructa | Primer sequence (5′ to 3′)b
|
|
|---|---|---|
| Forward | Reverse | |
| pEGFP-N-M2(1-41) | CGATTCCTCGAGGCAAAGATGGGGAACGC | CGATCGAAGCTTATTTAACATTCCAGGTG |
| pEGFP-C-M2(582-708) | ATACTCGAGGAATGGGTGTACGGATATTCAATC | AAGCTTTCAGCGTGTATACCCACGCTTAACC |
| pEGFP-C-M2(585-708) | ATCTCGAGGTATATTCAATCCCAAAGGAATTCTC | ATAAGCTTTCAGCGTGTATACCCACGCTTAACCAC |
| pEGFP-C-M2(610-675) | ATCTCGAGCAATTACGCAGGCAGCACCGGT | TATAAGCTTTCAAGGCGCTGGAAAGTGCGTCGA |
| pEGFP-C-M2(582-611) | ATACTCGAGGAATGGGTGTACGGATATTCAATC | TAAAGCTTTCAAATAATTGTACTTGGATCACC |
| pEGFP-N-M2(582-611) | ATACTCGAGGAATGGGTGTACGGATATTCAATC | TAAAGCTTAATAATTGTACTTGGATCACC |
| pEGFP-C-M2(582-643) | ATACTCGAGGAATGGGTGTACGGATATTCAATC | ATAAGCTTCACCCCGCCGATAAACTTTTAGTACG |
| pEGFP-C-M2(582-675) | ATACTCGAGGAATGGGTGTACGGATATTCAATC | TATAAGCTTTCAAGGCGCTGGAAAGTGCGTCGA |
| pEGFP-C-M2(582-675ΔH2)c | (i) ACCCCAACGAGAAGCGCGATCAC | (ii) ACCCCCGCCGATAAACTTTTAGTAATAATTGTACTTGGATCACC |
| (iii) TGATCCAAGTACAATTATTACTAAAAGTTTATCGGCGGGGGTG | (iv) TGGACAAACCACAACTAGAATGCAGTG | |
Each construct was designed to express an EGFP fusion including the indicated amino acid residues of μ1. The pEGFP-C1 and -N1 vectors and all PCR products were digested with XhoI and HindIII for cloning.
Forward primers are in the same orientation as the coding strand; reverse primers are in the reverse orientation to the coding strand. Restriction enzyme sites are in italics, and the inserted start and stop codons are underlined.
Primer pairs i and ii amplified a region in pEGFP-C-M2(582-675) from within EGFP to M2 residue 611. Additional sequence in reverse primer ii (bold type) was complementary to the 5′ sequence of M2(637-675). Primer pairs iii and iv amplified a region in pEGFP-C-M2(582-675) from M2 residue 637 to a region in the vector downstream of the M2 gene. Additional sequence in forward primer iii (bold type) was complementary to sequence at the 3′ end of M2(582-611).
To generate the FLAG-M2(582-708) plasmid, the pEGFP-C-M2(582-708) plasmid was digested with XhoI and ApaI and the cut fragment was ligated into the pCMV-3Tag-1B (Stratagene) vector cut with the same enzymes.
To generate pEGFP-C-M2(582-675ΔH2), we used overlap extension PCR to splice M2 fragments 582 to 611 and 637 to 675 (31). We PCR amplified the M2 fragments (582 to 611) and (637 to 675) from the pEGFP-C-M2(582-675) template using primer pair i and ii and primer pair iii and iv, respectively (Table 2). The two PCR products were then mixed in equimolar amounts and further PCR amplified with primers i and iv (Table 2). The resulting PCR product was digested with XhoI and HindIII and ligated into pEGFP-C1 cut with the same enzymes. All recombinant plasmids were recovered from transformed Escherichia coli, and the inserts were sequenced for correctness (Cornell University BioResource Center).
Infections and transfections.
Cells were seeded the day before transfection or infection at a density of 1 × 104 to 2 × 104 per cm2 in six-well plates containing 18-mm-diameter round coverslips. Infections were begun by adsorbing virus stocks to cells at an MOI of 5 to 100 PFU/cell, as indicated, for 1 h at room temperature in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1 mM KH2PO4 [pH 7.5]) containing 2 mM MgCl2. Cells were then overlaid with growth medium and incubated at 37°C for 6 to 48 h, as indicated. All cells were transfected using FuGENE 6 transfection reagent (Roche) according to the manufacturer's instructions. In brief, the following ratios of FuGENE reagent to DNA (μl of FuGENE/μg of DNA) were used: CV-1, 3:2; CHO, 6:1; HeLa and L929, 3:1. The complexes were incubated for 30 min then added to cells and incubated at 37°C for 18 to 48 h, as indicated.
IF microscopy.
Cells on coverslips were fixed for 10 min at room temperature in 2% paraformaldehyde in PBS, washed three times in PBS, and then permeabilized for 5 min in PBS containing 1% bovine serum albumin (BSA) and 0.1% Triton X-100 (PBSA-T) or 0.5% saponin. All antibody incubations were carried out for 25 to 40 min at room temperature in PBSA-T or PBS containing 1% BSA and 0.05% saponin. Coverslips were washed three times in PBS between primary and secondary antibody incubations. Cell nuclei were labeled by incubation of coverslips with 300 nM DAPI (4′,6′-diamidino-2-phenylindole) (Molecular Probes). Coverslips were mounted on glass slides with Prolong reagent (Molecular Probes). Fluorescence and phase images were obtained with a Nikon TE2000 inverted microscope equipped with fluorescence and phase optics through a 60× 1.4 NA oil objective with 1.5× optical zoom. Images were collected digitally with a Coolsnap HQ charge-coupled-device camera (Roper) and Openlab software (Improvision) and then were prepared for publication with Photoshop and Illustrator software (Adobe Systems).
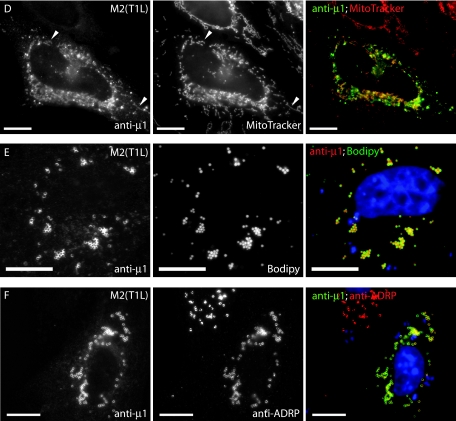
Mitochondria or the ER was labeled with MitoTracker Red CMXros (Molecular Probes) or an anti-protein disulfide isomerase (PDI) MAb (SelectFX Alexa Fluor 488 ER labeling kit; Molecular Probes), respectively, according to the manufacturer's instructions.
Lipid droplets were detected by incubating coverslips with Bodipy 493/503 (20 μg/ml) for 20 min prior to mounting or with Oil Red O as described previously (55).
SDS-PAGE and immunoblot analysis of steady-state levels of μ1 and φ truncation constructs.
CHO cells in 60-mm dishes were transfected in duplicate with each of the constructs shown in Fig. 6 by using FuGene6 (Roche) according to the manufacturer's instructions. Immediately after transfection, 50 μM z-VAD-fmk (Biomol) or an equivalent volume of dimethyl sulfoxide (DMSO) carrier was added to the transfected cells. At 24 h posttransfection (p.t.), cells were scraped into ice-cold buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.2]) and pelleted at 500 × g for 5 min at 4°C. The cell pellets were then lysed on ice in the preceding buffer that also included 1% Nonidet P-40, 1% desoxycholate, 0.1% sodium dodecyl sulfate (SDS), protease inhibitor cocktail (Roche), and 1 mM phenylmethylsulfonyl fluoride (Sigma). DNA was sheared by passing the cell lysate through a 27-gauge needle five times. Protein concentrations were determined by the DC Protein Assay (Bio-Rad Laboratories), and 50 μg protein from each lysate was suspended in 1× sample buffer, boiled for 10 min, and subjected to 10 or 15% SDS-polyacrylamide gel electrophoresis (PAGE), as indicated. Proteins were transferred from gels to nitrocellulose membranes, and the expressed proteins were detected with polyclonal rabbit anti-T1L virion serum (to detect μ1) or MAb anti-GFP (Clontech) in 0.1 M Tris-buffered saline (pH 7.4) containing 0.05% Tween 20, 1% BSA, and 2% powdered milk. Binding of antibodies was detected with goat anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase (HRP) and developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce) followed by fluorography. As a loading control, the blots were also probed with MAb anti-β-actin (Sigma) either concurrently with the anti-T1L serum or after the immunoblot had been stripped of previous antibodies with Restore Western Blot Stripping Buffer (Pierce) for 15 min at room temperature.
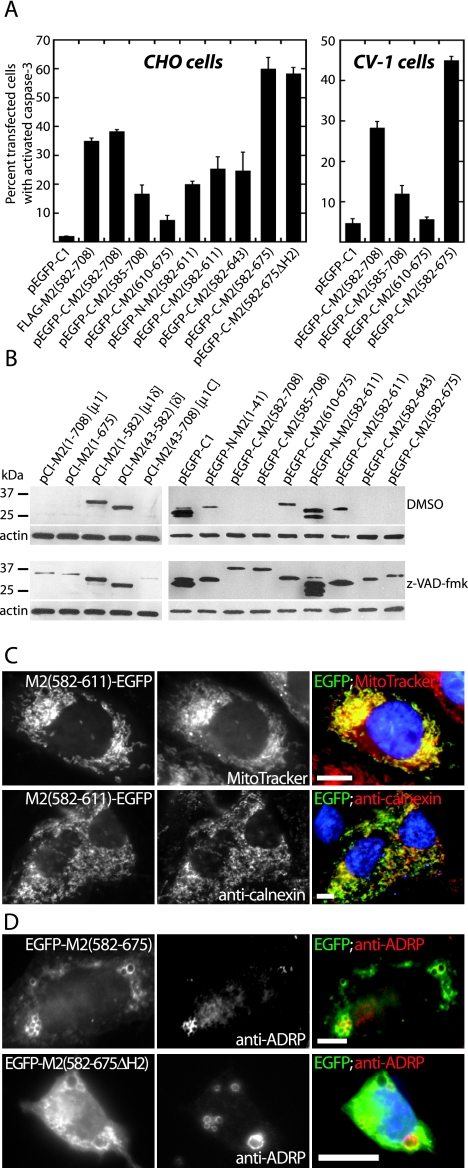
FIG. 6.
Apoptosis induction and subcellular localization by φ truncation mutants in transfected cells examined by fluorescence microscopy. (A) CHO cells were transfected with the indicated constructs, fixed at 48 h p.t., and then immunostained with rabbit anti-activated caspase-3 polyclonal antibody followed by goat anti-rabbit IgG conjugated to Alexa 594. The FLAG-tagged construct was detected by immunostaining with an anti-FLAG MAb followed by goat anti-mouse IgG conjugated to Alexa 488. Cells were scored for caspase-3 activation as in Fig. 1. The means and standard deviations of three determinations are shown; Kruskal-Wallis (P) values of the group for CHO and CV-1 cells are 0.0014 and 0.01, respectively. (B) Immunoblots showing expression of μ1 and φ truncation constructs in CHO cells at 24 h p.t. in the absence or presence of the broad-spectrum caspase inhibitor z-VAD-fmk. Cells were transfected with 1 μg of each construct. Immediately after transfection, cells were treated with DMSO or 50 μM z-VAD-fmk. At 24 h p.t., cell lysates were collected, and samples were subjected to 10% or 15% SDS-PAGE, followed by protein transfer to nitrocellulose. Expression of μ1 constructs was detected with polyclonal rabbit anti-virion serum followed by goat anti-mouse IgG conjugated to HRP. Expression of EGFP-fused constructs was detected with MAb anti-GFP (Clontech) followed by goat anti-mouse IgG conjugated to HRP. In all cases, β-actin was used as a loading control and detected (after the blot was stripped and reprobed) with MAb anti-β-actin followed by HRP-conjugated goat anti-mouse IgG. Positions of molecular weight markers are indicated. (C) CHO cells were transfected with pEGFP-N-M2(582-611) and then at 48 h p.t. either were stained with MitoTracker CMXros to detect mitochondria and then fixed (top row) or were fixed and then immunostained with anticalnexin followed by goat anti-mouse IgG conjugated to Alexa 594 to detect ER (bottom row). Nuclei were stained with DAPI. Right panels show colored merges of the different staining patterns, with labels in matching colors. Scale bars, 5 μm. (D) CHO cells were transfected with pEGFP-C-M2(582-675) or CV-1 cells were transfected with pEGFP-C-M2(582-675ΔH2), fixed at 48 h p.t., and then immunostained with anti-ADRP followed by goat anti-mouse IgG conjugated to Alexa 594. Right panels show colored merges of the different staining patterns, with labels in matching colors. Scale bars, 5 μm.
Statistical analyses.
Comparisons of apoptosis levels induced by groups of three or more different constructs or conditions were analyzed by the Kruskal-Wallis rank sum test. Graphs were prepared with Kaleidagraph (Synergy Software).
RESULTS
Induction of apoptosis in transfected cells expressing reovirus μ1 protein.
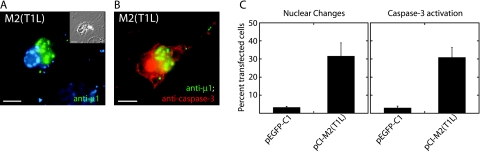
To test the hypothesis that μ1 can induce apoptosis in cultured cells independently of σ1 and σ1s, we examined transfected cells that expressed this protein intracellularly. The μ1-encoding M2 gene derived from T1L reovirus was cloned into the expression vector pCI-neo, under the control of a cytomegalovirus immediate-early promoter. Upon examining the M2-transfected CHO cells by phase-contrast and fluorescence microscopy, we saw that many of the cells expressing μ1 had morphological changes characteristic of apoptosis: the cells were rounded up, partially detached from the coverslip, and/or spiculated, and DAPI staining revealed that many nuclei were small and had condensed, marginated chromatin (Fig. 1A). In addition, many of the cells expressing μ1 stained positive for activated caspase-3 (Fig. 1B), an effector caspase whose activation denotes commitment to apoptosis (48). At 48 h p.t., ∼30% and ∼20% of M2-transfected CHO and CV-1 cells, respectively, had nuclear changes and/or activated caspase-3 (Fig. 1C; also see Fig. 5C). From these data, we conclude that expression of μ1 induces apoptosis in a subset of transfected cells.
FIG. 1.
Apoptosis induction by μ1 in transfected cells examined by fluorescence microscopy. CHO cells were transfected with pEGFP-C1 to express EGFP, as a control, or pCI-M2(T1L) to express μ1. Cells were fixed at 48 h p.t. and then immunostained with anti-μ1 (MAb 4A3) and rabbit anti-activated caspase-3 serum followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. Nuclei were stained with DAPI. (A and B) Marginated chromatin of an apoptotic nucleus (A) and activated caspase-3 (B) are present in a representative μ1-expressing cell. The inset in panel A shows a phase-contrast image. Scale bars, 5 μm. (C) Percentage of transfected cells showing apoptotic nuclear changes or activated caspase-3. At least 100 cells expressing either μ1 or EGFP were scored for the presence of apoptosis indicators. Means (± standard deviations) of three replicates are shown.
FIG. 5.
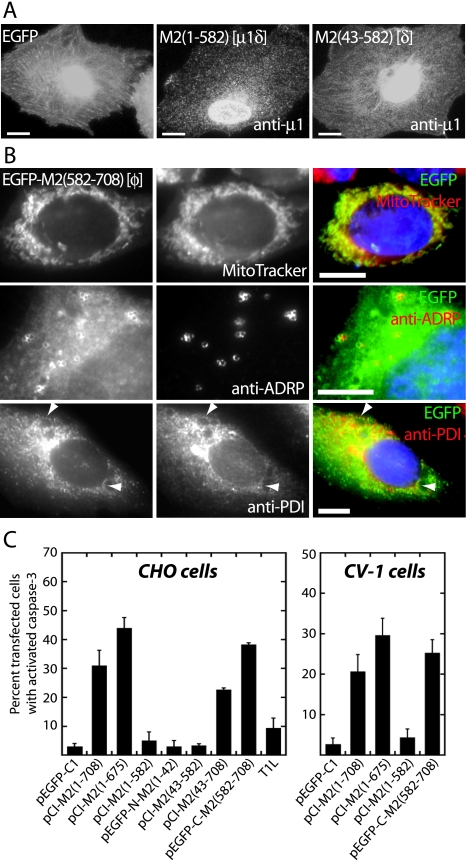
Subcellular localization and apoptosis induction by μ1 truncation mutants in transfected cells examined by fluorescence microscopy. (A) CHO cells were transfected with pEGFP-C1 to express EGFP as control, pCI-M2(1-582) to express the equivalent of μ1δ, or pCI-M2(43-582) to express the equivalent of δ. The cells were fixed at 48 h p.t. and then immunostained with anti-μ1 (MAb 4A3) followed—except for pEGFP-C1—by goat anti-mouse IgG conjugated to Alexa 488. The apparent concentration of μ1δ and δ in the nuclear region is likely because the images are not confocal, and therefore the nuclear region is the thickest part of the cell that is imaged. Scale bars, 5 μm. (B) CHO cells (top rows) and CV-1 cells (bottom row) were transfected with pEGFP-C-M2(582-708) to express the equivalent of φ tagged with EGFP. At 48 h p.t., the cells either were stained with MitoTracker CMXros to detect mitochondria (top row, middle panel) and then fixed or were fixed and then immunostained with either anti-ADRP to detect lipid droplets (middle row, middle panel) or anti-PDI to detect ER (bottom row, middle panel), followed in each of the last two cases by goat anti-mouse IgG conjugated to Alexa 594. Nuclei were stained with DAPI in each case. Left panels show EGFP-based fluorescence. Right panels show colored merges of the different staining patterns, with labels in matching colors. Arrowheads in the bottom panels indicate areas of colocalization between EGFP-M2(582-708) and ER. Scale bars, 5 μm. (C) CHO and CV-1 cells were transfected with the indicated constructs [including pCI-M2(1-708) to express full-length μ1] or infected with T1L (MOI = 100). The cells were fixed at 48 h p.t. or p.i. and then immunostained with anti-μ1 (MAb 4A3) and/or a rabbit anti-activated caspase-3 polyclonal antibody followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. Cells were scored for caspase-3 activation as in Fig. 1. The means and standard deviations of three determinations are shown; Kruskal-Wallis (P) values of the group for CHO and CV-1 cells are 0.0025 and 0.015, respectively.
Association of μ1 with annular rings and tubulovesicular structures in transfected cells.
We next examined the distribution of μ1 in transfected cells by immunostaining with μ1-specific MAbs. Despite the effects in the nucleus described above, the distribution of μ1 in M2-transfected CHO cells (Fig. 1A and B) and CV-1 cells (Fig. 2A and C) was largely cytoplasmic. Moreover, although much of μ1 appeared diffuse in the cytoplasm, two distinctive patterns suggested that it might also be associated with cytoplasmic organelles. First, in many M2-transfected CV-1 cells, μ1 localized to annular ring-like structures (Fig. 2A) which surrounded phase-dense globules (Fig. 2B). These structures may correspond to the “discrete particles” seen at lower magnification by Yue and Shatkin (65) following expression of μ1 in transfected HeLa cells. Second, though less frequently, μ1 distributed in a tubulovesicular pattern in transfected CV-1 cells (Fig. 2C). Similar patterns of μ1 distribution were observed in M2-transfected CHO, HeLa, and L929 cells (data not shown).
FIG. 2.
Distributions of μ1 in transfected cells examined by fluorescence microscopy. CV-1 cells were transfected with pCI-M2(T1L) to express μ1, fixed at 24 h p.t., and then immunostained with anti-μ1 (MAb 4A3) followed by goat anti-mouse IgG conjugated to Alexa 488. Scale bars, 10 μm. (A) Annular ring distribution of μ1 (arrowheads; enlarged in inset). (B) Phase-dense globules in a phase-contrast image of the cells shown in panel A (arrowheads; enlarged in inset). (C) Tubulovesicular distribution of μ1 (arrowheads; enlarged in inset). This cell also contains μ1-staining annular rings, which are indistinct and brighter as a result of the longer exposure needed to image the tubulovesicular structures.
Association of μ1 with lipid droplets, ER, and mitochondria in transfected cells.
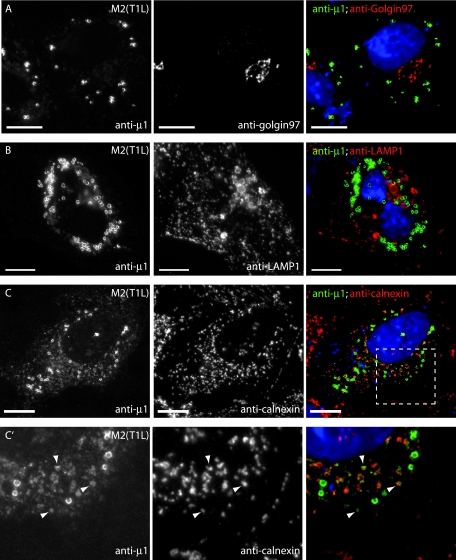
To identify the cytoplasmic organelles with which μ1 may associate in transfected cells, we tested for μ1 colocalization with various cellular markers in M2-transfected CV-1 cells (Fig. 3) and HeLa cells (data not shown). We found no colocalization between μ1 and antibody markers for the Golgi body or lysosomes (Fig. 3A and B). However, μ1 staining of annular ring structures was often closely juxtaposed to ER, as shown by immunostaining for the integral ER membrane protein calnexin (Fig. 3C), and some μ1 staining appeared to colocalize directly with calnexin (Fig. 3C′). The tubulovesicular distribution of μ1 was faint in both CV-1 and HeLa cells but was often more prominent in the HeLa cells. Although much of this tubulovesicular μ1 colocalized with the ER marker calnexin (Fig. 3C), some was also present on tubulovesicular structures that did not appear to be ER and instead colocalized with MitoTracker, which stains mitochondria (Fig. 3D).
FIG.3.
Subcellular localizations of μ1 in transfected cells examined by fluorescence microscopy. CV-1 cells transfected with pCI-M2(T1L) were fixed at 18 h p.t. and stained as described below for each row of panels. Right panels show colored merges of the different staining patterns, with labels in matching colors. Nuclei were stained with DAPI in each case. Scale bars, 10 μm. (A to C, C′, and F) After fixation, cells were immunostained with the following MAbs as markers for organelles—anti-golgin-97 for Golgi complex (A), anti-LAMP1 for lysosomes (B), anticalnexin for ER (C and C′), and anti-ADRP for lipid droplets (F)—followed by goat anti-mouse IgG conjugated to Alexa 594. Cells were then fixed again and immunostained with anti-μ1 (MAb 4A3) conjugated to Cy2. The area outlined in panel C is enlarged in panel C′ and shows colocalization between calnexin and μ1 (arrowheads). (D) Mitochondria were labeled with MitoTracker CMXros prior to fixation. After fixation, cells were immunostained with anti-μ1 (MAb 4A3) conjugated to Cy2. Arrowheads indicate areas of colocalization between μ1 and mitochondria. (E) After fixation, cells were immunostained with anti-μ1 (MAb 4A3) followed by goat anti-mouse IgG conjugated to Alexa 594. Neutral lipids were then stained with Bodipy 493/503.
FIG. 3—
Continued.
The annular ring pattern was the most common and prominent distribution of μ1 observed in M2-transfected cells (present in ∼82% of transfected CV-1 cells). These rings surrounded lipid droplets that were labeled with Bodipy 493/503 (Fig. 3E) or Oil Red O (data not shown) dye, both of which stain for neutral fatty acids. Lipid droplets are storage organelles for cholesterol esters and triglycerides and are believed to be metabolic organelles involved in the synthesis and trafficking of cellular lipids (reviewed in reference 39). They are surrounded by a protein-encrusted monolayer of phospholipid (57), and in most cells the major protein associated with this monolayer is ADRP (38). Strong colocalization between μ1 and ADRP confirmed that μ1 was localizing to the periphery of lipid droplets (Fig. 3F).
The C-terminal, φ region of μ1 determines both targeting to intracellular membranes and induction of apoptosis in transfected cells.
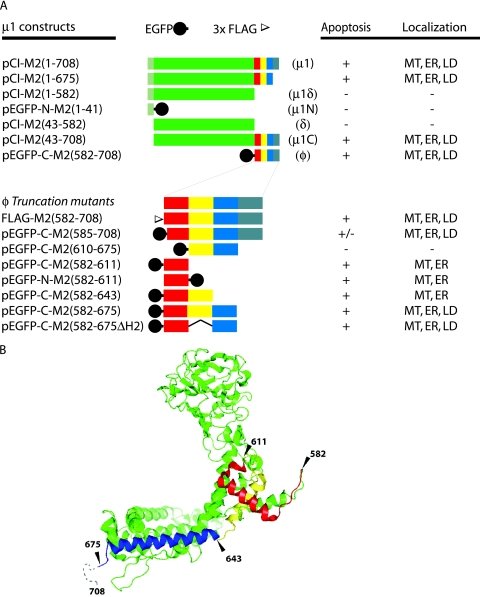
Three major regions of the μ1 protein, divided by proteolytic cleavage sites, are commonly identified: the amino (N)-terminal, myristoylated fragment μ1N (residues 2 to 41); the central fragment δ (residues 42 to 582); and the C-terminal fragment φ (residues 582 to 708) (40). In addition, pairwise combinations of these fragments can yield two other species: μ1δ (residues 2 to 582 plus the N-terminal, N-myristoyl group) and μ1C (residues 42 to 708) (40). To identify the region(s) of μ1 responsible for its activities in the preceding Results sections, we constructed a series of M2 gene truncations that encode the near-equivalents of each of these fragments (as some constructs have the addition of an initiating methionine residue at the N terminus). In addition, we prepared a construct to encode full-length μ1 lacking only residues 676 to 708 [μ1(1-675)], which are disordered in the μ1:σ3 crystal structure and genetically absent from the μ1 homologs of avian reoviruses and aquareoviruses (1, 37, 43, 68). Because none of the available anti-μ1 MAbs (62) recognize the μ1N or φ fragment, we prepared constructs to express EGFP-tagged versions of those regions. The constructs and results are summarized in Fig. 4A.
FIG. 4.
Summary of μ1 constructs. (A) Amino acid residues of μ1 represented in each construct are indicated in the name. Full-length μ1 is indicated by a bar spanning residues 1 to 708, with colors corresponding to regions denoted in panel B, the structure of a μ1 monomer. Each truncation mutant is represented by a bar spanning the approximate portion of μ1. A black circle indicates EGFP fused to the N or C terminus of μ1 or φ. An open triangle indicates three repeats of the FLAG epitope (3× FLAG) fused to the N terminus of φ(582-708). The name of the proteolytic fragments represented by some of the constructs is shown in parentheses following the bar. Fine mapping of the φ fragment of μ1 is represented similarly. A deletion mutant lacking the second (yellow) amphipathic α-helix of φ is denoted by ΔH2. The capacity of each construct to induce apoptosis in CHO cells is indicated by +, −, or +/−. A summary of each mutant's subcellular localization is also indicated: diffuse (—), to mitochondria (MT), to lipid droplets (LD), and/or to ER. (B) Ribbon diagram of the μ1 monomer X-ray crystal structure. Green represents the μ1N (lighter) and δ (darker) regions of μ1. The φ region is colored red, yellow, and blue to represent subregions encompassing three consecutive amphipathic α-helices and gray to represent the hydrophilic C-terminal tail. Amino acid residues at the boundaries of these subregions are numbered. The ribbon diagram was prepared with Pymol (Delano Software) from Protein Data Bank coordinates for the μ1:σ3 heterohexamer, 1JMU (37).
When expressed in transfected CHO cells, the three truncations lacking the φ region (μ1δ, δ, and μ1N-EGFP), as well as EGFP alone, were distributed diffusely through the cytosol and nucleus (Fig. 5A and data not shown). In contrast, the three truncations containing most or all of the φ region [μ1(1-675), μ1C, and EGFP/φ] were targeted to mitochondria, lipid droplets, and ER (Fig. 5B and data not shown). The association of EGFP/φ with mitochondria was more prominent, and its association with lipid droplets less so (Fig. 5B), than seen with full-length μ1 (Fig. 3), μ1(1-675), or μ1C. EGFP/φ was associated with lipid droplets in ∼25% of transfected CHO cells compared to ∼87% of CHO cells expressing full-length μ1. The reason(s) for these differences is not yet known. We also tested truncated versions of μ1 for the capacity to induce apoptosis. At 48 h p.t. in CHO and CV-1 cells, only the truncations containing most or all of the φ region [μ1(1-675), μ1C, and EGFP/φ], and not EGFP alone, induced apoptosis in a substantial percentage of cells, similarly to full-length μ1 (Fig. 5C).
Although the M2 gene we used for creating the μ1-expressing constructs was derived from the T1L reovirus, T1L infection of CHO cells at an MOI of 100 induced a low level of apoptosis (∼10% of infected cells) (Fig. 5C). This finding is in agreement with those of others showing that T1L is a poor inducer of apoptosis (51, 60). To address the possibility that virus strain differences in the μ1 protein were responsible for different levels of apoptosis induction, we examined the capacities of μ1 derived from the T3DN and T3DC reoviruses (44) to induce apoptosis in transfected CHO cells. We found no differences in the capacities of μ1 derived from the T1L, T3DN, or T3DC strain to induce apoptosis; they induced apoptosis in ∼27%, ∼30%, and ∼29% of transfected CHO cells, respectively, as assessed by nuclear changes. These findings were somewhat surprising, as previous genetic studies have shown that strain differences in the capacity to induce apoptosis are determined at least in part by the μ1-encoding M2 gene (23, 50, 59). One likely explanation is that in the context of viral infection, μ1-induced apoptosis is modulated indirectly by how, or the extent to which, it interacts with other viral factors in a strain-dependent manner (see Discussion).
In summary, we conclude that residues 582 to 675 in the φ region of μ1 contain determinants for both inducing apoptosis and targeting to lipid droplets, ER, and mitochondria in transfected cells. Moreover, the determinants in the φ region appear to be necessary and sufficient for the same activities exhibited by full-length μ1. Possible differences in the levels of apoptosis induced by the different φ-containing proteins are addressed in the Discussion.
Two regions encompassing amphipathic α-helices in the φ region of μ1 are major determinants for inducing apoptosis in transfected cells.
To identify specific determinants within the φ region for inducing apoptosis in transfected CHO cells, we constructed a further series of truncation or deletion mutants for expressing this region fused to the N or C terminus of EGFP. The constructs and results are summarized in Fig. 4A. Considering that the weak capacity of EGFP to dimerize (67) might influence the proapoptotic activity of φ, we also prepared a FLAG epitope-tagged version of the φ region (Fig. 4A). FLAG/φ behaved like EGFP/φ with regard to both induction of apoptosis and localization to intracellular membranes (Fig.and data not shown), suggesting that EGFP was not involved in these activities.
As for further truncations in the φ region, we first created a construct to express a version of φ lacking μ1 residues 676 to 708 [construct pEGFP-C-M2(582-675)], i.e., lacking the C-terminal region that is disordered in the μ1:σ3 crystal structure and genetically absent from the μ1 homologs of related viruses, as indicated above. Expression of this protein induced higher levels of apoptosis than those of full-length φ [construct pEGFP-C-M2(582-708)] (Fig. 6A). Thus, this C-terminal region of φ is dispensable for apoptosis induction and may even serve to downregulate it (see Discussion).
The truncation junctions of additional φ mutants were designed to fall between three amphipathic α-helices observed in the μ1:σ3 crystal structure (37) (Fig. 4B). Extended truncations from the C terminus were designed to remove either a region encompassing helix 3 [construct pEGFP-C-M2(582-643)] or a region encompassing both helix 3 and helix 2 [construct pEGFP-C-M2(582-611)]. Expression of each of these mutants led to moderate levels of apoptosis, albeit somewhat lower than those of full-length φ (Fig. 6A). Thus, the 582-to-611 region encompassing helix 1 alone retained most of the activity at inducing apoptosis, and this activity was similar when the 582-to-611 region was fused to either the N or C terminus of EGFP (Fig. 6A). The reduction in levels of apoptosis induced by the 582-to-643 region relative to those by the 582-to-675 region and full-length φ (Fig. 6A) suggests that helix 3 may represent another determinant of apoptosis induction. However, a truncation designed to express a protein containing only helix 2 and helix 3 [construct pEGFP-C-M2(610-675)] showed limited activity at inducing apoptosis (Fig. 6A), suggesting that helix 3 may exhibit its proapoptotic activity predominantly in concert with helix 1. We obtained further evidence that the region encompassing helix 3 contributes to proapoptotic activity, whereas the region encompassing helix 2 does not, by replacing the helix 2 region with a short linker in a protein otherwise containing the regions encompassing helix 1 and helix 3 [construct pEGFP-C-M2(582-675ΔH2)]. Expression of this mutant induced high levels of apoptosis, similar to those induced by the 582-to-675 region containing all three helices (Fig. 6A). These results appear to identify the regions encompassing helix 1 and helix 3 as the minimal determinants for inducing maximal levels of apoptosis. The importance of the region encompassing helix 1 was further suggested by reduced levels of apoptosis induced by a mutant in which only the N-terminal three residues of this region were missing from full-length φ [construct pEGFP-C-M2(585-675)] (Fig. 6A) (see Discussion).
In summary, we conclude that regions encompassing the first and third amphipathic α-helices of φ mediate its full proapoptotic activity. Moreover, including evidence from the previous section, these two helical regions of φ are probably responsible for the full proapoptotic activity of full-length μ1. We also note further evidence that residues 675 to 708 at the C terminus of μ1 and φ may serve to downregulate this activity.
Steady-state levels of μ1 constructs and φ truncation mutants in transfected CHO cells differ in the presence or absence of the broad-spectrum caspase inhibitor z-VAD-fmk.
Although we were able to detect expression of all μ1 constructs and φ truncation mutants in transfected cells by fluorescence microscopy, we had limited success at detecting those constructs that induced apoptosis by immunoblotting. We also observed that constructs that induced apoptosis often appeared to have lower levels of relative fluorescence compared to constructs that did not induce apoptosis. As general translation is inhibited in cells undergoing apoptosis (28), we hypothesized that the proapoptotic constructs downregulated their own translation. To test this hypothesis, we compared expression of the different constructs in transfected CHO cells incubated with either the broad-spectrum caspase inhibitor z-VAD-fmk (50 μM) or DMSO control (Fig. 6B). We found that in the presence of z-VAD-fmk, those constructs that induced apoptosis had notably increased expression levels compared to untreated controls (e.g., compare expression of μ1, μ1C, and EGFP/φ), whereas there was little change in the relative expression levels of those constructs that did not induce apoptosis. We conclude that in the presence of the broad-spectrum caspase inhibitor, most of the constructs had similar steady-state levels of expression. In addition, we were able to confirm that each construct was of the appropriate size.
The regions of φ that induce apoptosis also determine targeting to intracellular membranes.
As described above, the full-length φ region fused to either EGFP or FLAG localized to lipid droplets, ER, and mitochondria (Fig. 5B and data not shown). The localizations of the various truncation and deletion mutants in the φ region are summarized in Fig. 4A. All mutants containing residues 582 to 611 localized to ER and mitochondria (Fig. 6C and data not shown), identifying this region encompassing helix 1 as a minimal determinant of these activities. Only mutants containing both the helix 1 and helix 3 regions, however, localized to lipid droplets (Fig. 6D). The truncation containing residues 610 to 675 did not associate with intracellular membranes, suggesting that the helix 3 region is not sufficient for membrane targeting and identifying the helix 1 and helix 3 regions together as minimal determinants for targeting to lipid droplets. In general, all mutants that targeted to ER and mitochondria also induced apoptosis. This correlation appeared weakest in the case of the 585-to-708 protein, which showed clear membrane targeting (data not shown) but induced lower levels of apoptosis. Targeting to lipid droplets, in contrast, did not correlate strongly with apoptosis induction. In summary, we conclude that similar regions of φ determine both membrane targeting and apoptosis induction and that localization to ER and/or mitochondria may be part of the mechanism by which these determinants induce apoptosis.
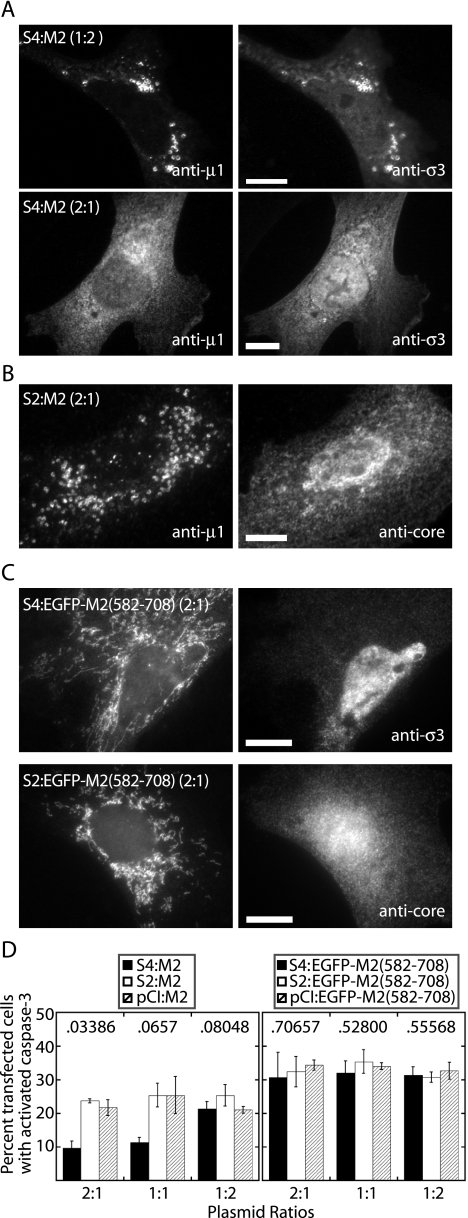
Membrane association and apoptosis induction by μ1, but not φ, are abrogated by coexpression of σ3.
Previous reports have noted that coexpression of μ1 leads to a redistribution of σ3 in cells (58, 64). We therefore hypothesized that coexpression of σ3 would reciprocally lead to a redistribution of μ1. To test this hypothesis, we cotransfected μ1- and σ3-expressing plasmids into CV-1 or CHO cells at different relative molar ratios. As a control, we cotransfected the μ1-expressing plasmid with one encoding the reovirus σ2 protein. As the ratio of σ3-to-μ1 plasmid increased, the distribution of μ1 became more diffuse throughout the cytosol and less associated with lipid droplets, ER, or mitochondria (Fig. 7A). At a molar ratio of 14:1 (S4:M2), essentially all of μ1 was diffuse in the cytosol and the nucleus in the vast majority of transfected cells (data not shown). In contrast, coexpression of σ2 with μ1 had no substantive effect on the subcellular distribution of μ1, i.e., μ1 remained strongly associated with intracellular membranes (Fig. 7B). We also found that as the σ3-to-μ1 ratio increased, apoptosis levels in CHO cells decreased, whereas σ2 coexpression with μ1 had little or no effect on apoptosis levels (Fig. 7D, left panel). In other words, increasing σ3, but not σ2, had an increasingly antiapoptotic effect. The σ3 and μ1 proteins are known to coassemble into soluble heterohexameric oligomers when coexpressed (37); thus, one possible explanation for these results is that progressive sequestration of μ1 into μ1:σ3 heterohexamers decreases the amount of free μ1 able to associate with intracellular membranes and to induce apoptosis.
FIG. 7.
Effect of coexpressing σ3 on the subcellular localization and ability to induce apoptosis of μ1 or EGFP/φ in transfected cells examined by fluorescence microscopy. (A) CV-1 cells were transfected with pCI-S4(T1L) to express σ3 plus pCI-M2(1-708) to express μ1 at plasmid DNA ratios of 1:2 and 2:1 (S4:M2). Cells were fixed at 48 h p.t. and then immunostained with Cy2-conjugated anti-μ1 (4A3) and Alexa 594-conjugated anti-σ3 (MAb 5C3). Representative examples of the predominant distribution patterns are shown. Scale bars, 5 μm. (B) CV-1 cells were transfected with pCI-S2(T1L) to express σ2 plus pCI-M2(1-708) to express μ1 at a plasmid DNA ratio of 2:1 (S2:M2). Cells were fixed at 48 h p.t. and then immunostained with Cy2-conjugated anti-μ1 (4A3) and rabbit anti-core serum (to detect σ2) followed by goat anti-rabbit IgG conjugated to Alexa 594. Scale bar, 5 μm. (C) CV-1 cells were transfected with pEGFP-C-M2(582-708) to express the equivalent of φ tagged with EGFP plus either pCI-S4(T1L) or pCI-S2(T1L). Again in this experiment, each plasmid pair was transfected at a ratio of 1:2, respectively. Cells were fixed at 48 h p.t. and then immunostained for σ3 and σ2 as in panel A. Representative examples of the predominant distribution patterns are shown. Scale bars, 5 μm. (D) CHO cells were transfected with the indicated plasmid pairs at ratios of 2:1, 1:1, or 1:2 and then immunostained with anti-activated caspase-3 followed by goat anti-rabbit IgG conjugated to Alexa 594. For samples expressing full-length μ1, cells were also immunostained with anti-μ1 (MAb 4A3) followed by goat anti-mouse IgG conjugated to Alexa 488. Cells were scored for caspase-3 activation as in Fig. 1. The means and standard deviations of three determinations are shown. Also shown are the Kruskal-Wallis (P) values for the differences within each group.
Another possible explanation for the preceding results for μ1-σ3 coexpression is that the antiapoptotic effect of σ3 is independent of its interaction with μ1. σ3 is known to interact with double-stranded RNA and to prevent activation of protein kinase R, an effect that could be antiapoptotic (24, 64). To address the possibility that σ3 inhibited apoptosis independently of its capacity to interact with μ1, we examined whether coexpression of σ3 with φ [construct pEGFP-C-M2(582-708)] would abrogate the capacity of φ to associate with intracellular membranes and/or induce apoptosis. We reasoned that since φ lacks the vast majority of residues in μ1 that interact with σ3 (37), σ3 and φ should not interact upon coexpression and therefore any other antiapoptotic effect of σ3 would be revealed. As a control, we coexpressed φ with the reovirus σ2 protein. Coexpression of σ3 or σ2 with φ neither altered the intracellular distribution of φ (Fig. 7C) nor reduced the induction of apoptosis (Fig. 7D, right panel).
In summary, we conclude that coexpression of σ3 with full-length μ1, as a function of relative levels of the two proteins, progressively abrogates μ1 association with intracellular membranes and induction of apoptosis, most likely because of μ1 sequestration into μ1:σ3 heterohexamers. Since the φ region lacks the vast majority of μ1:σ3 contacts apparent in the structure of the μ1:σ3 heterohexamer, we conclude that σ3 cannot sequester φ when those two proteins are coexpressed; thus, φ retains its activities at membrane association and apoptosis induction.
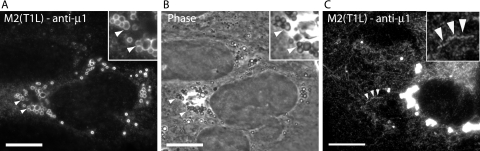
μ1 localizes to lipid droplets, ER, and mitochondria in infected cells.
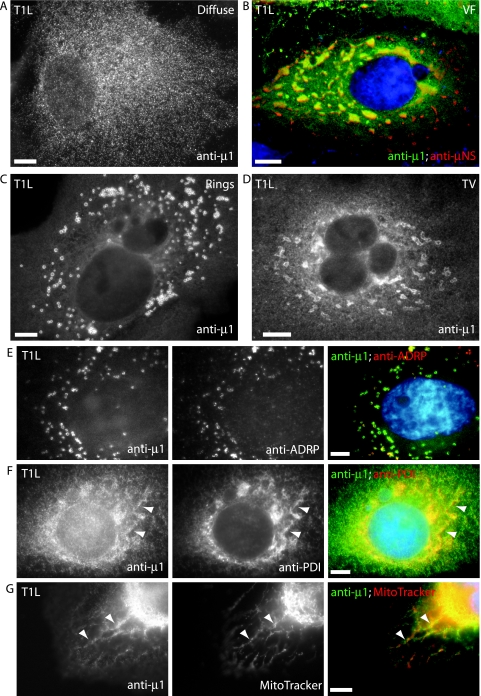
The μ1 protein is known to localize to viral factories in reovirus-infected cells (5, 53) but has previously not been localized to membranous organelles. Upon examining the distribution of μ1 in T1L-infected CV-1 cells by IF microscopy, we found that at 24 h postinfection (p.i.), in addition to localizing to viral factories, μ1 localized to ring-like and tubulovesicular structures in a subset of infected cells (see Fig. 9). We discerned four patterns of μ1 distribution at 24 h p.i.: (i) diffuse through the cytosol (Fig. 8A), (ii) colocalized with μNS in viral factories (Fig. 8B), (iii) localized to ring-like structures (Fig. 8C), and (iv) localized to tubulovesicular structures (Fig. 8D). Many cells displayed more than one of these patterns (e.g., in Fig. 8B, diffuse and localized to viral factories). As with our findings in transfected cells, μ1 colocalized with markers for lipid droplets (Fig. 8E), ER (Fig. 8F), and mitochondria (Fig. 8G) in infected cells. We found similar distributions of μ1 in infected HeLa, CHO, and L929 cells (data not shown, but see Fig. 9). We conclude that μ1 localizes to lipid droplets, ER, and mitochondria in T1L-infected cells in addition to viral factories; thus, its distribution partially mirrors that seen in transfected cells.
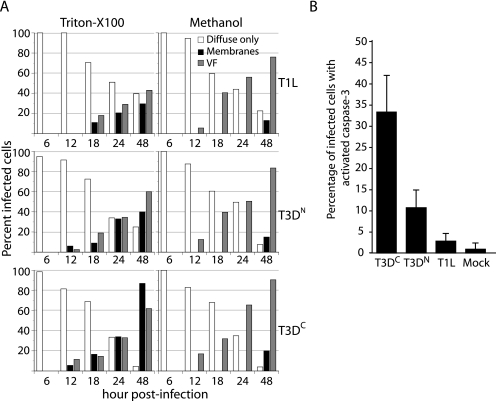
FIG. 9.
Distribution of μ1 and apoptosis induction in T1L-, T3DN-, and T3DC-infected L929 cells. (A) Reovirus T1L-, T3DN-, or T3DC-infected cells (MOI = 10) were fixed at the indicated times p.i., permeabilized with Triton X-100 (left panel) or methanol (right panel), and then immunostained with anti-μ1 (MAb 4A3) and anti-μNS serum followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. The patterns of μ1 distribution in individual infected cells were scored for each time point as diffuse only, associated with intracellular membranes (tubulovesicular and ring-like structures) (membranes), or associated with viral factories (VF). At least 200 cells were scored per replicate. Each data point represents the average of two replicates. (B) Cells were infected as above, fixed at 48 h p.i., permeabilized with Triton X-100, and then immunostained with anti-μ1 (MAb 4A3) and rabbit anti-activated caspase-3 polyclonal antibody followed by goat anti-mouse IgG conjugated to Alexa 488 and goat anti-rabbit IgG conjugated to Alexa 594. Cells were scored for caspase-3 activation as in Fig. 1. The means and standard deviations of three determinations are shown.
FIG. 8.
Distribution patterns and subcellular localizations of μ1 in infected cells examined by fluorescence microscopy. CV-1 cells infected with T1L reovirus were fixed at 24 h p.i., and the distribution patterns of μ1 were detected by immunostaining with anti-μ1 (MAb 4A3) followed by goat anti-mouse IgG conjugated to Alexa 488. Four patterns of μ1 staining were detected as follows. (A) Diffuse. (B) Associated with viral factories (VF). Factories were detected by immunostaining with a rabbit polyclonal serum to μNS followed by goat anti-rabbit IgG conjugated to Alexa 594. In this merged image, yellow indicates colocalization between μ1 (green) and μNS (red). (C) Associated with annular ring-like structures (Rings). (D) Associated with tubulovesicular structures (TV). (E, F, and G) To ascertain the subcellular localization of μ1, fixed cells were first immunostained with anti-ADRP to detect lipid droplets (E) and anti-PDI to detect ER (F) followed by goat anti-mouse IgG conjugated to Alexa 594. Cells were then fixed again and immunostained with anti-μ1 (MAb 4A3) conjugated to Cy2. Alternatively, cells were first stained with MitoTracker CMXros to detect mitochondria (G), after which they were fixed and immunostained with anti-μ1 followed by goat anti-mouse IgG conjugated to Alexa 488. Nuclei were stained with DAPI. Arrowheads indicate areas of colocalization between μ1 and ER (F) or mitochondria (G). Scale bars, 5 μm.
Given the findings at 24 h p.i., we interpreted the different μ1 distribution patterns as representing a continuum that may vary with time p.i. In preliminary experiments, we noted that the distribution of μ1 was affected by the method used to permeabilize cellular membranes: methanol reduced the staining of μ1 with ring-like and tubulovesicular structures but increased its staining within viral factories, whereas 0.1% Triton X-100 had the opposite effects (data not shown). We therefore compared the distribution of μ1 in T1L-, T3DN-, and T3DC-infected L929 cells at 6, 12, 18, 24, and 48 h p.i. in cells permeabilized with either 0.1% Triton X-100 or 100% methanol (Fig. 9A). With all three strains, we found that μ1 was predominantly diffuse in the cytosol at 6 and 12 h p.i. regardless of the permeabilization method. Thereafter, in methanol-permeabilized cells, μ1 increasingly stained with viral factories and was seen associating with intracellular membranes (ring-like structures) only at the latest time point (48 h). In contrast, in Triton X-100-permeabilized cells, μ1 staining of intracellular membranes (tubulovesicular and ring-like structures) became visible at 12 to 18 h p.i. and increased to being seen in ∼30% of T1L-, ∼40% of T3DN-, and ∼87% of T3DC-infected cells by 48 h p.i. We moreover noted that as μ1 became associated with viral factories and membrane structures, fewer cells had a diffuse distribution of μ1. From these results, we conclude that the pattern of μ1 distribution in infected cells changes as infection progresses, being initially diffuse and then becoming localized to viral factories and associated with ring-like and tubulovesicular structures from 12 to 18 h p.i. and beyond.
As we had found that coexpression of μ1 with σ3 caused μ1 to redistribute from intracellular membranes to a predominantly diffuse distribution in cells and abrogated μ1-induced apoptosis, we speculated that the association of μ1 with intracellular membranes in infected cells was related to apoptosis induction. In support of this hypothesis, we found that that the level of apoptosis induced by the T1L, T3DN, and T3DC strains at 48 h p.i. in L929 cells (Fig. 9B) correlated with the percentage of infected cells in which μ1 associated with intracellular membranes (Fig. 9A, left panels).
DISCUSSION
Consistent with genetic reassortant studies showing that the reovirus M2 gene is a determinant of virus strain differences in the capacity to induce apoptosis during infection (51, 60, 61), we found that expression of the M2-encoded μ1 protein induced apoptosis in uninfected, transfected cells. Both S1 and M2 viral genes were previously identified as genetic determinants of strain differences in reovirus-induced apoptosis (60); however, since these earlier studies, most attention has been paid to the roles of the S1 gene products σ1 and σ1s, particularly to receptor interactions by σ1 with JAM-A and α-sialic acid and their possible importance for proapoptotic signaling (reviewed in reference 26). Recently, Danthi et al. showed that in CHO cells expressing the Fc receptor, but not JAM-A or sialic acid, infection and reovirus-induced apoptosis can occur when virion-associated σ1 is prebound with MAbs such that the Fc portion of the antibody mediates virus attachment (23). These authors also found that under such conditions the sole genetic determinant of apoptosis induction was the M2 gene. The results presented here extend and support those conclusions, and we therefore propose that the μ1 protein plays a more primary role in reovirus-induced apoptosis than was previously appreciated.
If sufficient numbers (a high MOI) of reovirus particles are added to cells, viral transcription or genome replication is not required for induction of apoptosis (19, 20, 61). Nevertheless, an unidentified postattachment or disassembly step is required (20). In this study, we found that the φ region of μ1 is necessary and sufficient for inducing apoptosis in transfected cells. During infectious entry, proteolytic processing of virion-associated μ1 within endo/lysosomes produces partially uncoated particles, ISVPs, which are primed for membrane penetration (reviewed in reference 10). The φ fragment remains associated with ISVPs (41), and so it seems possible that particle-derived φ could be released into the cytosol after membrane penetration, where if present in sufficient concentrations, it could induce apoptosis. Our previous finding that the particle-derived δ fragment of μ1 is present in the cytosol and nucleus of the infected cell soon after penetration is consistent with this hypothesis (11). At lower MOI, it is possible that fragments of μ1 could be released into the cytosol in smaller amounts that do not directly induce apoptosis but instead prime the cells for apoptosis induction later in the infectious cycle.
In the current study, μ1 and all of its derived regions that induced apoptosis in transfected cells also associated with mitochondria and, to a lesser extent, with ER. It is tempting to speculate that association of these proteins with mitochondria and/or ER is important for proapoptotic activity, as these organelles are intricately involved in the intrinsic apoptotic pathway (22, 50). However, one protein, EGFP/φ(585-708), associated with both mitochondria and ER but induced only low levels of apoptosis, suggesting that association with one or both of these organelles is not sufficient for apoptosis induction, though perhaps it is still required. This construct reflects the φ fragment that is generated by trypsin digestion of μ1 during generation of ISVPs (41). The relative difference in the proapoptotic abilities of the 582-to-708 versus the 585-to-708 forms of φ is intriguing, as it raises the possibility that levels of apoptosis induction might be determined by cell-type-specific proteolytic processing of μ1, perhaps during cell entry.
Regions encompassing two amphipathic α-helices (residues 582 to 611 and 643 to 675) within the φ region of μ1 were important for its proapoptotic activity. We also found that the disordered region in the crystal structure of μ1:σ3 at the C terminus of μ1 (μ1 residues 676 to 708) was dispensable for this activity and may even serve to downregulate it. The first 30 residues of φ (residues 582 to 611 of μ1), consisting of a short hydrophobic loop followed by an amphipathic helix, was the minimal identified region sufficient for induction of apoptosis and was required for association with mitochondrial and ER membranes. Several other viral proteins associate with mitochondrial membranes and induce apoptosis. These include human immunodeficiency virus type 1 Vpr, influenza A virus PB1-F2, and the human T-cell leukemia virus type 1 p13II accessory protein (16, 21, 33). All of these proteins have predicted amphipathic α-helical regions that are required for interactions with mitochondrial membranes. However, the mechanism(s) of apoptosis induction is not completely understood for any of these proteins. The Vpr and PB1-F2 proteins are thought to promote apoptosis by directly interacting with components of the mitochondrial permeability transition pore, thereby causing loss of the transmembrane potential with a resultant increase in mitochondrial outer membrane permeability (33, 66). However, both of these proteins may induce and/or promote apoptosis in other ways. Vpr interacts with the antiapoptotic protein HAX-1 on the outer mitochondrial membrane and may promote apoptosis by counteracting the HAX-1 antiapoptotic effect (63), and PB1-F2 may directly permeabilize mitochondrial and/or other cellular membranes by forming lipidic or proteolipidic pores (13). Previous authors have shown that mitochondrial apoptotic pathways are activated in reovirus-infected cells and have suggested that these pathways involve activation of caspase-8 and subsequent cleavage of the Bcl-2 family member Bid (35). However, it is possible that μ1 or μ1 fragments directly activate mitochondrial apoptotic pathways, and we are currently investigating this possibility.
We found that steady-state levels of μ1 and its constructs that induced apoptosis were much lower than those of constructs that did not induce apoptosis. Moreover, levels of the proapoptotic constructs were markedly increased by incubation of transfected cells with the broad-spectrum caspase inhibitor z-VAD-fmk (Fig. 6B). As apoptosis is reported to inhibit translation generally (28), one explanation for this finding is that μ1 induction of apoptosis resulted in inhibition of its own translation. Apoptosis induction appeared to be downregulated by C-terminal residues 676 to 708 within either μ1 or φ. Interestingly, this polypeptide sequence contains a predicted PEST motif (PESTfind [http://emb1.bcc.univie.ac.at/content/view/21/45/]). PEST motifs are short regions of polypeptide sequence that are often found at the C terminus of proteins; are enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) residues; and are usually flanked by basic residues. PEST sequences are thought to act as signals for rapid protein degradation (49). If this were true of μ1, it would explain the enhancement of apoptosis seen in constructs lacking this region. Avian reoviruses (ARVs) induce higher levels of apoptosis in infected cells at 24 h p.i. than do mammalian reoviruses (36). We have found that expression of the ARV-176 μB protein (the homolog of μ1) also robustly induces apoptosis in transfected cells (C. M. Coffey and J. S. L. Parker, unpublished data). We note that ARV μB lacks the C-terminal extension (68), including the putative PEST motif found in μ1, and we are currently investigating the role of this region of μ1 in protein stability.
EGFP fusions of the φ region tended to localize to mitochondrial membranes, weakly to ER membranes, and occasionally to lipid droplets in transfected cells (Fig. 8), contrasting with the primary localization of full-length μ1 to lipid droplets and less so to mitochondria and ER in both transfected and infected cells. Although both μ1 and φ induced apoptosis in transfected cells, their differential localization patterns suggest that they may differ in their proapoptotic functions during reovirus infection. Caspase-3 and caspase-8 activation in cells infected with reovirus T3 Abney is biphasic, supporting the concept that two sequential proapoptotic signals may be present in infected cells (34). Kominsky et al. suggested that this biphasic pattern of caspase activation results from an initial tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-dependent activation of caspase-8 that leads to low-level activation of caspase-3 and cleavage of Bid, followed by more sustained activation of caspase-3 and caspase-8 that results from cleaved Bid-mediated activation of the intrinsic apoptotic pathway and release of cytochrome c and Smac/DIABLO from mitochondria (34, 35). This model proposes that during reovirus infection, TRAIL-dependent apoptotic pathways are activated before mitochondrial apoptotic pathways. Alternatively, it is possible that mitochondria become sensitized to TRAIL-dependent apoptosis before TRAIL secretion. The influenza A virus PB1-F2 protein, which has a localization pattern similar to that of φ, is believed to sensitize transfected cells to tumor necrosis factor alpha-induced apoptosis by modulating mitochondrial membrane permeability (66). We speculate that φ and/or a related fragment of μ1 released into the cytosol of infected cells during membrane penetration sensitizes cells to apoptosis induction by TRAIL, perhaps by modulating mitochondrial membrane permeability.
As noted above, full-length μ1 localized primarily to lipid droplets in both transfected cells and infected cells. The hepatitis C virus (HCV) core protein is similarly localized (2, 6, 54). Moreover, expression of the HCV core protein causes apoptosis induction in some, but not all, transfected cells (6, 25, 29). The determinants of lipid droplet localization of the HCV core protein have been mapped to amphipathic α-helices whose primary sequence is homologous to plant oleosins, which associate with lipid droplets (30). In addition, a 10-residue sequence at the C terminus of the HCV core protein mediates its localization to mitochondria (54). Our findings suggest that regions encompassing two amphipathic helices (residues 582 to 611 and 644 to 675) near the C terminus of μ1 are required for association with lipid droplets but that only the first of these regions is strictly required for mitochondrial localization (Fig. 6). The biological significance of associations by the HCV core protein and reovirus μ1 protein with lipid droplets remains uncertain. However, lipid droplets have recently been implicated as potential intracellular signaling platforms that might function analogously to lipid rafts on the plasma membrane (39). If this is the case, then it is possible that association of μ1 with lipid droplets may be important for activation of certain signaling pathways. In support of this idea, it is known that the M2 gene is the genetic determinant of strain differences in JNK activation during reovirus infection (18). Association of μ1 with lipid droplets may modulate its capacity to induce apoptosis. In support of this idea, we found that proteins EGFP/φ(582-675) and EGFP/φ(582-675ΔH2), which associated with mitochondria, ER, and lipid droplets, appeared to induce substantially higher levels of apoptosis than did EGFP/φ(582-643) and EGFP/φ(582-611), which associated only with mitochondria and ER.
It has been previously reported that coexpression of μ1 with the reovirus σ3 protein modulates the distribution of σ3 in transfected cells (58, 65). We have confirmed these findings and shown that coexpression of σ3 with μ1 abrogates both the membrane association of μ1 and its capacity to induce apoptosis. We hypothesize that this is a result of coassembly of μ1:σ3 heterohexamers. In our model, coexpression of μ1 with σ3 leads to sequestration of μ1 from membranes as assembled μ1:σ3 heterohexamers. Our finding that coexpression of σ3 with the φ domain of μ1 does not abrogate the capacity of φ to induce apoptosis supports this hypothesis. Late in infection, μ1 is more often associated with intracellular membranes in cells infected with T3 viruses such as T3D than in those infected with T1 viruses such as T1L (Fig. 9A). This observation correlates with the increased capacity of the T3 viruses to induce apoptosis (60; also Fig. 9B). Schmechel et al. have proposed that differences in the affinity of σ3 for μ1 regulate the subcellular distribution of σ3, which in turn determines its capacity to bind double-stranded RNA and prevent activation of PKR (53). Similarly, we speculate that strain-dependent differences in the affinity of μ1 for σ3 or in the kinetics of μ1:σ3 heterohexamer assembly may in turn determine the levels of “free” μ1 and thus μ1-determined strain differences in proapoptotic activity. The capacity of σ3 to interact with μ1 and to abrogate its proapoptotic activity in transfected cells is reminiscent of the control of proapoptotic Bcl-2 family members such as Bax and Bak by hetero-oligomerization with antiapoptotic members such as Bcl-XL and Bcl-2 (reviewed in reference 22). Relative levels of σ3-bound versus free μ1 may thus be an important determinant of phenotypes and strain differences relating to apoptosis induction, as well as to inhibition of host translation, by reovirus.
Acknowledgments
We are grateful to Roy Duncan for the generous gift of reagents, to Hollis Erb for advice on statistical analyses, and to Melina Agosto, Tijana Ivanovic, and Leslie Schiff for helpful comments on the manuscript.
This work was supported by a Burroughs Welcome Fund Investigators of Pathogenesis of Infectious Disease award (to J.S.L.P.) and by NIH grants K08 AI052209 (to J.S.L.P.), R01 AI063036 (to J.S.L.P.), R01 AI46440 (to M.L.N.), and T32 AI07618 (to C.M.C.).
REFERENCES
- 1.Attoui, H., Q. Fang, F. M. Jaafar, J. F. Cantaloube, P. Biagini, P. de Micco, and X. de Lamballerie. 2002. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol. 83:1941-1951. [DOI] [PubMed] [Google Scholar]
- 2.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, E. S., J. L. Connolly, J. C. Forrest, J. D. Chappell, and T. S. Dermody. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 276:2200-2211. [DOI] [PubMed] [Google Scholar]
- 4.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 5.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σ NS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benali-Furet, N. L., M. Chami, L. Houel, F. De Giorgi, F. Vernejoul, D. Lagorce, L. Buscail, R. Bartenschlager, F. Ichas, R. Rizzuto, and P. Paterlini-Brechot. 2005. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24:4921-4933. [DOI] [PubMed] [Google Scholar]
- 7.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 8.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran, K., and M. L. Nibert. 2003. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 11:374-382. [DOI] [PubMed] [Google Scholar]
- 11.Chandran, K., J. S. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanturiya, A. N., G. Basanez, U. Schubert, P. Henklein, J. W. Yewdell, and J. Zimmerberg. 2004. PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J. Virol. 78:6304-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chappell, J. D., V. L. Gunn, J. D. Wetzel, G. S. Baer, and T. S. Dermody. 1997. Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein σ1. J. Virol. 71:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chappell, J. D., A. E. Prota, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 17.Clarke, P., R. L. Debiasi, R. Goody, C. C. Hoyt, S. Richardson-Burns, and K. L. Tyler. 2005. Mechanisms of reovirus-induced cell death and tissue injury: role of apoptosis and virus-induced perturbation of host-cell signaling and transcription factor activation. Viral Immunol. 18:89-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke, P., S. M. Meintzer, C. Widmann, G. L. Johnson, and K. L. Tyler. 2001. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J. Virol. 75:11275-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly, J. L., E. S. Barton, and T. S. Dermody. 2001. Reovirus binding to cell surface sialic acid potentiates virus-induced apoptosis. J. Virol. 75:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connolly, J. L., and T. S. Dermody. 2002. Virion disassembly is required for apoptosis induced by reovirus. J. Virol. 76:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Agostino, D. M., L. Ranzato, G. Arrigoni, I. Cavallari, F. Belleudi, M. R. Torrisi, M. Silic-Benussi, T. Ferro, V. Petronilli, O. Marin, L. Chieco-Bianchi, P. Bernardi, and V. Ciminale. 2002. Mitochondrial alterations induced by the p13II protein of human T-cell leukemia virus type 1. Critical role of arginine residues. J. Biol. Chem. 277:34424-34433. [DOI] [PubMed] [Google Scholar]
- 22.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116:205-219. [DOI] [PubMed] [Google Scholar]
- 23.Danthi, P., M. W. Hansberger, J. A. Campbell, J. C. Forrest, and T. S. Dermody. 2006. JAM-A-independent, antibody-mediated uptake of reovirus into cells leads to apoptosis. J. Virol. 80:1261-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denzler, K. L., and B. L. Jacobs. 1994. Site-directed mutagenic analysis of reovirus σ3 protein binding to dsRNA. Virology 204:190-199. [DOI] [PubMed] [Google Scholar]
- 25.Dumoulin, F. L., A. van dem Bussche, J. Sohne, T. Sauerbruch, and U. Spengler. 1999. Hepatitis C virus core protein does not inhibit apoptosis in human hepatoma cells. Eur. J. Clin. Investig. 29:940-946. [DOI] [PubMed] [Google Scholar]
- 26.Forrest, J. C., and T. S. Dermody. 2003. Reovirus receptors and pathogenesis. J. Virol. 77:9109-9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goral, M. I., M. Mochow-Grundy, and T. S. Dermody. 1996. Sequence diversity within the reovirus S3 gene: reoviruses evolve independently of host species, geographic locale, and date of isolation. Virology 216:265-271. [DOI] [PubMed] [Google Scholar]
- 28.Holcik, M., N. Sonenberg, and R. G. Korneluk. 2000. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16:469-473. [DOI] [PubMed] [Google Scholar]
- 29.Honda, M., S. Kaneko, T. Shimazaki, E. Matsushita, K. Kobayashi, L. H. Ping, H. C. Zhang, and S. M. Lemon. 2000. Hepatitis C virus core protein induces apoptosis and impairs cell-cycle regulation in stably transformed Chinese hamster ovary cells. Hepatology 31:1351-1359. [DOI] [PubMed] [Google Scholar]
- 30.Hope, R. G., D. J. Murphy, and J. McLauchlan. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261-4270. [DOI] [PubMed] [Google Scholar]
- 31.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 32.Hoyt, C. C., S. M. Richardson-Burns, R. J. Goody, B. A. Robinson, R. L. Debiasi, and K. L. Tyler. 2005. Nonstructural protein σ1s is a determinant of reovirus virulence and influences the kinetics and severity of apoptosis induction in the heart and central nervous system. J. Virol. 79:2743-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacotot, E., L. Ravagnan, M. Loeffler, K. F. Ferri, H. L. Vieira, N. Zamzami, P. Costantini, S. Druillennec, J. Hoebeke, J. P. Briand, T. Irinopoulou, E. Daugas, S. A. Susin, D. Cointe, Z. H. Xie, J. C. Reed, B. P. Roques, and G. Kroemer. 2000. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 191:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kominsky, D. J., R. J. Bickel, and K. L. Tyler. 2002. Reovirus-induced apoptosis requires both death receptor- and mitochondrial-mediated caspase-dependent pathways of cell death. Cell Death Differ. 9:926-933. [DOI] [PubMed] [Google Scholar]
- 35.Kominsky, D. J., R. J. Bickel, and K. L. Tyler. 2002. Reovirus-induced apoptosis requires mitochondrial release of Smac/DIABLO and involves reduction of cellular inhibitor of apoptosis protein levels. J. Virol. 76:11414-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labrada, L., G. Bodelon, J. Vinuela, and J. Benavente. 2002. Avian reoviruses cause apoptosis in cultured cells: viral uncoating, but not viral gene expression, is required for apoptosis induction. J. Virol. 76:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Londos, C., D. L. Brasaemle, C. J. Schultz, J. P. Segrest, and A. R. Kimmel. 1999. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10:51-58. [DOI] [PubMed] [Google Scholar]
- 39.Martin, S., and R. G. Parton. 2005. Caveolin, cholesterol, and lipid bodies. Semin. Cell Dev. Biol. 16:163-174. [DOI] [PubMed] [Google Scholar]
- 40.Nibert, M. L. 1998. Structure of mammalian orthoreovirus particles. Curr. Top. Microbiol. Immunol. 233:1-30. [DOI] [PubMed] [Google Scholar]
- 41.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noad, L., J. Shou, K. M. Coombs, and R. Duncan. 2006. Sequences of avian reovirus M1, M2 and M3 genes and predicted structure/function of the encoded μ proteins. Virus Res. 116:45-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odegard, A. L., K. Chandran, X. Zhang, J. S. Parker, T. S. Baker, and M. L. Nibert. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78:8732-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul, R. W., A. H. Choi, and P. W. K. Lee. 1989. The α-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology 172:382-385. [DOI] [PubMed] [Google Scholar]
- 47.Poggioli, G. J., C. Keefer, J. L. Connolly, T. S. Dermody, and K. L. Tyler. 2000. Reovirus-induced G2/M cell cycle arrest requires σ1s and occurs in the absence of apoptosis. J. Virol. 74:9562-9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porter, A. G., and R. U. Janicke. 1999. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 6:99-104. [DOI] [PubMed] [Google Scholar]
- 49.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 50.Rizzuto, R., M. R. Duchen, and T. Pozzan. 2004. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE 215:1-9. [DOI] [PubMed] [Google Scholar]
- 51.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a σ1s-null mutant. J. Virol. 72:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmechel, S., M. Chute, P. Skinner, R. Anderson, and L. Schiff. 1997. Preferential translation of reovirus mRNA by a σ3-dependent mechanism. Virology 232:62-73. [DOI] [PubMed] [Google Scholar]
- 54.Schwer, B., S. Ren, T. Pietschmann, J. Kartenbeck, K. Kaehlcke, R. Bartenschlager, T. S. Yen, and M. Ott. 2004. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J. Virol. 78:7958-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198-210. [DOI] [PubMed] [Google Scholar]
- 56.Strong, J. E., G. Leone, R. Duncan, R. K. Sharma, and P. W. K. Lee. 1991. Biochemical and biophysical characterization of the reovirus cell attachment protein σ1: evidence that it is a homotrimer. Virology 184:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tauchi-Sato, K., S. Ozeki, T. Houjou, R. Taguchi, and T. Fujimoto. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277:44507-44512. [DOI] [PubMed] [Google Scholar]
- 58.Tillotson, L., and A. J. Shatkin. 1992. Reovirus polypeptide σ3 and N-terminal myristoylation of polypeptide μ1 are required for site-specific cleavage to μ1C in transfected cells. J. Virol. 66:2180-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyler, K. L., M. A. Mann, B. N. Fields, and H. W. Virgin IV. 1993. Protective anti-reovirus monoclonal antibodies and their effects on viral pathogenesis. J. Virol. 67:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyler, K. L., M. K. Squier, A. L. Brown, B. Pike, D. Willis, S. M. Oberhaus, T. S. Dermody, and J. J. Cohen. 1996. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J. Virol. 70:7984-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyler, K. L., M. K. Squier, S. E. Rodgers, B. E. Schneider, S. M. Oberhaus, T. A. Grdina, J. J. Cohen, and T. S. Dermody. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein σ1. J. Virol. 69:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yedavalli, V. S., H. M. Shih, Y. P. Chiang, C. Y. Lu, L. Y. Chang, M. Y. Chen, C. Y. Chuang, A. I. Dayton, K. T. Jeang, and L. M. Huang. 2005. Human immunodeficiency virus type 1 Vpr interacts with antiapoptotic mitochondrial protein HAX-1. J. Virol. 79:13735-13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yue, Z., and A. J. Shatkin. 1997. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234:364-371. [DOI] [PubMed] [Google Scholar]
- 65.Yue, Z., and A. J. Shatkin. 1996. Regulated, stable expression and nuclear presence of retrovirus double-stranded RNA-binding protein σ3 in HeLa cells. J. Virol. 70:3497-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zamarin, D., A. Garcia-Sastre, X. Xiao, R. Wang, and P. Palese. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, J., R. E. Campbell, A. Y. Ting, and R. Y. Tsien. 2002. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell. Biol. 3:906-918. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, X., J. Tang, S. B. Walker, D. O'Hara, M. L. Nibert, R. Duncan, and T. S. Baker. 2005. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 343:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]