Abstract
African swine fever virus (ASFV) produces a fatal acute hemorrhagic fever in domesticated pigs that potentially is a worldwide economic threat. Using an expressed sequence tag (EST) library-based antisense method of random gene inactivation and a phenotypic screen for limitation of ASFV replication in cultured human cells, we identified six host genes whose cellular functions are required by ASFV. These included three loci, BAT3 (HLA-B-associated transcript 3), C1qTNF (C1q and tumor necrosis factor-related protein 6), and TOM40 (translocase of outer mitochondrial membrane 40), for which antisense expression from a tetracycline-regulated promoter resulted in reversible inhibition of ASFV production by >99%. The effects of antisense transcription of the BAT3 EST and also of expression in the sense orientation of this EST, which encodes amino acid residues 450 to 518 of the mature BAT3 protein, were investigated more extensively. Sense expression of the BAT3 peptide, which appears to reversibly interfere with BAT3 function by a dominant negative mechanism, resulted in decreased synthesis of viral DNA and proteins early after ASFV infection, altered transcription of apoptosis-related genes as determined by cDNA microarray analysis, and increased cellular sensitivity to staurosporine-induced apoptosis. Antisense transcription of BAT3 reduced ASFV production without affecting abundance of the virus macromolecules we assayed. Our results, which demonstrate the utility of EST-based functional screens for the detection of host genes exploited by pathogenic viruses, reveal a novel collection of cellular genes previously not known to be required for ASFV infection.
Viruses and other infectious agents that reproduce but are not free-living require genetic and biochemical functions of host cells in order to propagate and produce disease (see reviews in references 20 and 51). Viral pathogenesis begins with the binding of virus to host proteins at the cell surface and ends with the release of infectious particles. The host-pathogen relationship is dynamic; even as the virus is exploiting host cell functions, various host genes are concurrently implementing surveillance mechanisms to detect the invading viral pathogen and initiate steps toward its eradication. Successful completion of the virus life cycle normally is dependent on the ability of the virus to interfere with host functions that actively limit viral replication, evade host immune response mechanisms, and effectively conscript the host gene products it requires (17, 46).
African swine fever (ASF) is a tick-borne disease associated with acute hemorrhagic fever in domesticated pigs and a mortality of nearly 100% (33). ASF outbreaks have decimated domestic pig populations where the disease is endemic, with devastating consequences to the economy of affected areas. As no vaccine or other preventative measure is available and slaughtering of the infected animals is the routine method of halting spread of the disease, ASF currently is included on list A of infectious diseases of the World Organization for Animal Health (66) and is considered to be a potentially important vector of bioterrorism (48).
The genome of African swine fever virus (ASFV), the causative agent of ASF, consists of double-stranded DNA 170 kb in length encoding 151 identified open reading frames (ORFs) (68). ASFV has similarities to the poxvirus and iridovirus families but is sufficiently different from these viruses that it has been assigned as the only member of the Asfarviridae family. As occurs for other viruses, ASFV recruits and subverts host cell functions to complete its life cycle (12). ASFV enters susceptible cells using host-encoded pathways of receptor-mediated endocytosis and uses host-encoded enzymes to express viral proteins that restrict host cell defenses and enable viral genome replication. Virus core particles are then transported by the microtubule/dynein motor complex to the perinuclear region of the cytoplasm of infected cells, where the virus is wrapped by the endoplasmic reticulum (52). Energy required for these processes is derived from host cell mitochondria, which migrate en masse to the site of virus assembly. Ultimately, the virus particle acquires a plasma membrane envelope, and the release of mature virus occurs at the edge of the cell via budding (30).
Studies of the reproductive cycles of specific mammalian viruses have identified some of the host gene functions that these viruses require (14, 22, 45). Recently, the direct discovery of cellular genes exploited by viruses and other pathogens (i.e., cellular genes exploited by pathogens, or CGEPs) has become practical through the development of methods that accomplish homozygous functional inactivation of chromosomal genes in mammalian cells, coupled with screens that identify cell clones that consequently acquire phenotypic properties of interest (8, 31, 35, 36, 43, 47, 64, 65). In general, these approaches employ RNA complementary to genomically encoded transcripts (antisense RNA [36] or small interfering RNA [32]) to silence expression from both copies of chromosomal genes in individual cell clones. We applied an antisense method that uses RNA encoded by a library of ∼40,000 expressed sequence tags (ESTs) (36) to functionally inactivate cellular genes in HeLa and HT144 cell populations and identified cell clones that limit the replication of ASFV. Here we report (i) that EST libraries can be used effectively for the discovery of genes whose inactivation interferes with viral replication in mammalian cells, (ii) the identification of several host genes previously unknown to be required for normal completion of the ASFV life cycle, and (iii) the important role of the HLA-B-associated transcript 3 gene, BAT3, in ASFV infection.
MATERIALS AND METHODS
Construction of the EST expression vector and preparation of the HeLatTA and HT144tTA pLentiEST libraries.
The EST expression construct we used has been described in detail previously (36). Briefly, a collection of ∼40,000 human sequence-verified ESTs (Invitrogen) was pooled and amplified by PCR using two directional universal primers flanking the EST DNA fragments. The resulting amplified EST products were then digested with NheI and cloned using a modified version of a self-inactivating lentiviral backbone plasmid derived from pRRLsinPPT.CMV.MCS.Wpre (a gift of L. Naldini, HSR-TIGET, Milan, Italy) (19), in which the constitutive cytomegalovirus (CMV) promoter of the original plasmid was replaced with a DNA fragment containing a neomycin resistance expression cassette and a CMV minimal promoter controlled by the tetracycline-regulated tetracycline responsive element (TRE) (24). The tetracycline derivative doxycycline (Dox) was used to regulate the promoter in the experiments reported here.
The host cells used in this work, a HeLa human cervical cancer cell line and HT144, a line of human metastatic melanoma cells (18, 25), were modified to overexpress tetracycline-dependent transcriptional activator (tTA) (24) from a pBabetTAPuro retrovirus. We then generated pLentiEST libraries in both HeLatTA and HT144tTA cells as described elsewhere (36). Briefly, 20 subconfluent 15-cm2 plates of HeLatTA cells (ca. 2 × 107 cells in total) were infected with 500 μl of pLentiEST virus supernatant derived from 293T packaging cells and pseudotyped with the G protein of vesicular stomatitis virus envelope gene in the presence of 4 μg/ml of Polybrene. Following G418 selection (1,000 μg/ml for HeLatTA and 600 μg/ml for HT144 tTa), the G418-resistant clones were pooled in several aliquots and expanded one generation to form the pLentiEST libraries of HeLatTA and HT144tTa, respectively.
Genomic DNA extraction and PCR.
Genomic DNA was isolated from 3 × 106 to 4 × 106 cultured cells from each clone of interest using the Gentra DNA extraction kit (Gentra Systems), and DNA was dissolved in 100 μl of the hydration buffer included in the kit. The EST DNA fragment in each cell clone was isolated by PCR amplification of the genomic DNA with the universal primers (36) flanking the EST insert in the viral construct. The amplified PCR products were purified, sequenced, and identified by a BLAST search of the NCBI database. The orientation of the EST in the cell clone was determined by PCR amplification using a Lenti 3′ primer derived from the viral vector and a universal EST forward or EST reverse primer.
Viruses, cell cultures, and infection.
The ASFV used in these experiments is the Malawi Lil-20/1 isolate (27), which is a virulent pathogenic African swine fever virus isolated from ticks of the Ornithodoros moubata complex (Ixodoidea: Argasidae) that were collected in the ASF enzootic area of Malawi. The virulent pathogenic virus isolated from swine was adapted to grow in Vero cell cultures by 25 serial passages prior to culture on HeLa or HT144 cells; it also retained virulence properties in swine in vivo (L. Zsak, unpublished data). The replicative cycle of this virus was longer in Vero and HeLa cells than in pig cells and yielded the p30 early ASFV protein in 30% of cells 4 to 8 h after infection (T. Burrage, unpublished data). Progression to late protein was observed at later time points, as indicated below, and death of the entire culture was observed. Cultures of HeLatTA and HT144tTA EST libraries, which did not show any detectable growth alterations compared to the parental cell lines, were infected with the adapted ASFV.
Titration of ASFV.
Parental HeLatTA cell, HeLatTA cell EST library pools, and individual ASFV-resistant HeLa cell clones were cultured in T75 flasks until they reached 90% confluence. Cells were trypsinized and counted for viability using trypan blue. Cell cultures for growth curve experiments were made in 24-well Primaria plastic plates; cells were plated at a density of 5 × 106 per well in 10% Dulbecco's modified Eagle's medium (DMEM). For the doxycycline-positive cultures, 5 μg per ml Dox was used in the culture medium.
EST libraries and control cells were infected after at least 24 h of growth in culture with ASFV Malawi isolate at the indicated multiplicities of infection (MOIs) and incubated for 2 h at 37°C. Following incubation, cultures were washed three times with prewarmed 10% DMEM, and 1 ml of 10% DMEM was added following the final wash. Samples were collected at various times postinoculation. Cells were scraped into the medium, and the mixture was transferred into an Eppendorf tube and centrifuged at 3,000 rpm for 5 min. Supernatant was removed, transferred into a new Eppendorf tube, and designated as extracellular virus. Cell pellets were resuspended in 1 ml of fresh medium, freeze-thawed, and sonicated to lyse the cells and disrupt viral aggregates. The “intracellular virus” titers determined from these samples closely paralleled those determined for extracellular virus by testing the supernatants from cultures containing unlysed cells. Two samples were taken at each time point and were stored at −70°C until titration.
Virus titers were determined in primary cultures of swine macrophages. Virus titers were calculated based on the hemadsorption of infected cells and calculated as described previously (50).
Dot blot DNA hybridization.
Genomic DNA was isolated from the indicated cell lines that had been plated at the same density and then infected at approximately the 80% confluent stage with ASFV (MOI, 5). Duplicate samples were taken at various times postinfection (16 to 72 h), and DNA was extracted from the cell pellet and adjusted to an identical concentration (1 μg per ml). For quantitative hybridization, serial twofold dilutions were made from an initial 100-ng sample and equal amounts of DNA dilutions were transferred onto nitrocellulose membranes. DNA was hybridized with 32P-labeled ASFV genomic DNA probes using standard procedures.
Quantitation of viral antigen-containing cells.
Expression of ASFV early and late genes in BAT3 sense and antisense clones was assayed by fluorescence microscopy of antigens/antibody staining following infection of HeLatTA and BAT3 clones with ASFV. At different times after infection, cell cultures were fixed and immunostained with either anti-p30 (ASFV early protein) or anti-p72 (ASFV late protein). The experiments were performed by counting the viral antigen-containing positive cells in cultures at the different time points. Several thousand cells were observed per time point for each sample.
Quantitative PCR.
Quantitative PCR experiments for evaluation of cellular BAT3 mRNA production were performed using the Bio-Rad iCycler real-time PCR detection system and the IQ SYBR Green Super Mix kit (catalog no. 170-8880). Oligonucleotide primers were synthesized by the Stanford PAN Facility. The sequences of the forward and reverse probes correspond to exons of the BAT3 sequence available from the National Center for Biotechnology Information. These were as follows: BAT3.1512F, GTGGAACCCGTGGTCATGATGCA; BAT31629.F(F2), GTCATGATGCACATGAACATTC; BAT3.1634VariantReverse (VR), GGTGGAGCCCAGGGTTTGG; BAT3.1743R(R), CCTGCTGTCCCAGGGTTTGG. Cell lines were grown until the exponential phase, and total RNA was isolated by using a QIAGEN RNAeasy Plant Mini kit. Five μg of total RNA was used in the synthesis of cDNA using avian myeloblastosis virus reverse transcriptase (Invitrogen) under conditions recommended by the vendor. PCR was performed in a 15-μl reaction mixture volume in the presence of 10 nM forward and reverse primers, 7.5 μl IQ SYBR Green Super mix, and 0.2 μl of cDNA. Each assay consisted of an initial denaturation period of 5 min at 95°C to activate the polymerase followed by 45 cycles of 95°C for 15 s, 55°C for 30 s, and 70°C for 45 s. Melting curve analysis was used to determine levels of BAT3 transcript produced in the antisense and sense orientations with respect to the control cell line, HeLatTA. The stably expressed β-glucuronidase (GUS) gene (1) was used to monitor the input RNA for the three cell lines. Three separate experiments were carried out; the values shown represent the averages from four repeats in each experiment and had a mean standard deviation of <0.5%.
Microarray analysis.
The cell clones HeLatTA (parental cell line), BAT3S (dominant negative clone), and BAT3A (BAT3 EST in antisense configuration) were grown to exponential phase, and poly(A) RNA was extracted (Invitrogen PolyA RNA extraction kit) for microarray analysis. The cDNA labeled with Cy3TTP or Cy5TTP was hybridized to human 40K cDNA arrays prepared by the Stanford Functional Genomics Facility. The results were analyzed by the GABRIEL software (44) using either pattern-based rules (44) or a modified t-score algorithm (63). Missing values in the microarray data set were estimated with KNNimpute using 14 neighbors (63).
MTT viability assay for apoptotic potential.
Cells were grown in 96-well plates to a density of 10,000 cells per well and treated with different dosages of staurosporine to induce apoptosis (10). After 24 h of treatment, cell viability was measured by the 3-(4,5-dimethylthiozol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (38). The percentage of surviving cells was determined by comparing these values with those obtained for untreated control cells.
RESULTS
Construction of ASFV-infected HeLa and HT144 cell libraries expressing ESTs and isolation of ASFV-resistant cell clones.
Whereas swine macrophages are the natural target cells for ASFV, laboratory strains of the virus have been adapted for replication in a variety of permanent cell lines derived from multiple sources, including primates, where extensive investigations of ASFV molecular biology have been carried out; these cell lines include Vero cells (4), monkey kidney MS cells (55), Jurkat cells (26), and the human myeloid leukemia cell line K562. The human cervical carcinoma (HeLa) and melanoma (HT144) cell lines, which we previously had shown to enable ASFV replication at a rate similar to that observed in Vero cells (L. Zsak, unpublished data) and which were also highly infectible by our lentivirus EST libraries, were chosen as targets for the isolation of cellular genes required for the propagation of ASFV. We infected these cell lines with an ASFV isolate that had been previously adapted to passage in Vero cells but which also remained virulent in swine (see Materials and Methods). The rate of virus replication we observed for this isolate in HeLa and HT144 cells was similar to that observed for Vero cells but was slower than replication in swine macrophages (Zsak, unpublished data).
A previously described pLenti EST library (36) that expresses a collection of ESTs from a promoter controlled by the TRE (24) was introduced into HeLa and HT144 cells that contain the tTA (24), which enables regulation of expression of inserted ESTs by the tetracycline derivative doxycycline.
Pools of library cells constructed as described in Materials and Methods were grown for one generation before being infected with ASFV at MOIs ranging from 1 to 10. Clones of surviving cells were recultured and challenged through three additional cycles of ASFV infection at a multiplicity of 10, at which time no surviving cells were detected in ASFV-infected cultures of HeLatTA or H144tTA cells that had not received the EST library. PCR amplification and sequence analysis of the EST inserts (see Materials and Methods) identified 18 ESTs corresponding to seven previously annotated genes, shown in Table 1. Importantly, in some instances multiple clones derived independently from different EST library pools and, in the case of MYOHD, ASFV-resistant cell clones derived from different kinds of host cell lines (i.e., both HeLa and HT44), contained the same EST insert.
TABLE 1.
Annotated functions of ESTs identified in surviving cell clones after three challenges with ASFV
| Group | Cell clone | GenBank reference | Annotated designation | Length of EST (bp) | Clone ID | ORF length (aa) | Chromosome location | Annotated biological role |
|---|---|---|---|---|---|---|---|---|
| 1 | HeLa 15 | NM_004639.21 | Homo sapiens BAT3 | 447 | 770887 | 1,126 | 6p21.3 | Apoptosis |
| HeLa 19 | ||||||||
| HeLa 20 | ||||||||
| HeLa 24 | ||||||||
| 2 | HeLa 25 | NM_031910.2 | Homo sapiens C1QTNF | 519 | 454445 | 278 | 22q13.1 | Apoptosis |
| HeLa 33 | ||||||||
| HeLa 37 | ||||||||
| 3 | HeLa 28 | NM_020185 | Homo sapiens MKPX | 1,000 | 240748 | 184 | 6q25.3 | Signal transduction |
| HeLa 35 | ||||||||
| HeLa 36 | ||||||||
| HeLa 49 | ||||||||
| 4 | HT 08 | NM_025109 | Homo sapiens MYOHD | 517 | 364800 | 317 | 17q12 | Myosin associated |
| HT 07 | ||||||||
| HeLa 12 | ||||||||
| 5 | HeLa 21 | gi:29125369 | Human DNA sequence | 779 | 780964 | ??? | 13q34 | Unknown |
| HeLa 39 | gi:34364894 | that contains GAS6 and six novel genes of unknown function | ||||||
| 6 | HeLa 16 | NM_006114.1 | Homo sapiens TOM40 | 592 | 809466 | 361 | 19q13.1 | Apoptosis |
| 7 | HeLa 40 | NM_017607.1 | Homo sapiens PPP1R12C | 473 | 810389 | 792 | 19q13.42 | Actin cytoskeleton |
Two of the ESTs identified in ASFV-resistant HeLa clones contained coding sequences of peptides annotated as being involved in pathways related to apoptosis: BAT3 (6) and C1q complement, tumor necrosis factor-related protein 6 (C1qTNFR6) (54). The BAT3 gene, which encodes a nuclear protein, previously was mapped to the major histocompatibility complex class III (MHC III) region of human chromosome 6 (6), where it is located between the MHC class I and class II regions. C1qTNFR6, a protein of 278 amino acids, contains a small globular N-terminal domain, a collagen-like Gly/Pro-rich central region, and a conserved C-terminal region, the C1q domain, and is part of the C1 enzyme complex. C1q complement protein is involved in binding of virus (60) and recognition of microbial surfaces (56).
An EST identified in another one of the clones we isolated (Table 1) encodes the channel-forming subunit of TOM40, a translocase present in the outer membrane of mitochondria (21, 49). TOM40 is essential for processing of apocytochrome c to cytochrome c and for protein import into mitochondria (16). Two other ESTs, mitogen-activated protein kinase phosphatase (MKPX) and protein phosphatase 1 regulatory subunit 12C (PP1R12C), are related to known signal transduction pathways (5, 59).
cDNAs representing five of the seven ESTs we identified by this screen were present also in a porcine EST library (NCBI accession number CB483054) (2) prepared from the primary cultures of porcine macrophages, and all of these human ESTs had high nucleotide sequence homologies (indicated in parentheses) with their swine counterparts: C1QTNFR6 (88%), MKPX (92%), TOM40 (93%), PP1R12C (93%), and BAT3 (93%).
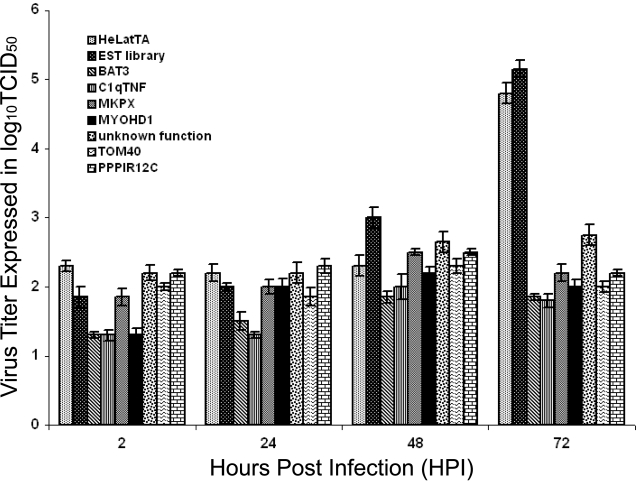
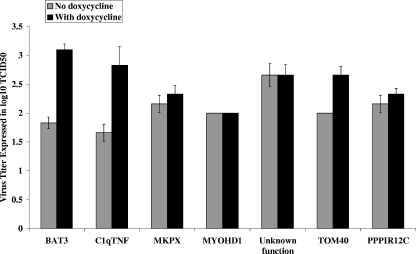
Experiments using the identified ESTs to reconstitute the ASFV resistance phenotype in naïve cells were carried out in order to confirm that the observed survival to repeated challenges of ASFV infection resulted from the EST sequences. Lentiviral constructs expressing ESTs of each of the seven identified genes from a promoter regulated by the TRE were introduced by transfection into 293T cells and harvested as pLentiEST virus stock mixtures. These were then used to infect the naïve HeLatTA parental cell line, and 10 randomly selected reconstituted clones containing pLentiEST insertions at different chromosomal locations were isolated for each of the seven genes and tested individually for ASFV production following infection by ASFV. A dramatic decrease in virus production (2 to 4 logs) was observed relative to control cultures of parental HeLatTA cells and the HeLatTA EST library pool for each of the tested clones (Fig. 1), and the decrease was partially reversed by the addition of doxycycline to the medium used to culture cells containing the BAT3, C1qTNFR6, and TOM40 EST constructs (Fig. 2). While the ASFV resistance phenotype was reconstituted in naïve cells for the other genes identified by our initial screen, phenotypic reversal by doxycycline was not observed.
FIG. 1.
Extracellular ASFV virus titers of the reconstituted EST clones. The patterned bars represent the extracellular virus titer assayed in culture media used to grow reconstituted EST-expressing clones at the indicated hpi at an MOI of 10. The virus titer is expressed as the log10 of the 50% tissue culture infectious dose (TCID50). The first two bars are controls and showed the same pattern when infected with ASFV: HeLatTA, a parental cell line, and an EST library that represents HeLatTA cells containing the collection of EST inserts in the parental cell line. Bars three through nine represent the HeLa cell clones, each of which contained a different EST insert and showed a reduced titer when infected with ASFV virus: BAT3S, C1qTNF6, MKPX, MYOHD1, an EST cell clone of unknown gene function, TOM40, and PPPIR12C.
FIG. 2.
Virus titers in reconstituted EST clones taken at 72 hpi. The ASFV virus titer of reconstituted clones was grown in the absence and presence of doxycycline, and the sample was taken at 72 hpi. The names of the EST clones are shown below the bar. The y axis denotes the virus titer expressed as the log10 of the 50% tissue culture infective dose (TCID50).
The human BAT3 gene, which encodes the BAT3 EST identified here, is shown diagrammatically in Fig. 3. The gene is estimated to specify a protein of 120 kDa that contains several structural domains of possible relevance to the phenotypic properties we observed. The N-terminal portion of the BAT3 protein (amino acids 120 to 190) contains a ubiquitin-like region, a central segment that includes a polyproline-rich stretch, and a C-terminal region that includes both a nuclear localization signal (37) and a caspase 3 cleavage site that triggers apoptosis when released from the BAT3 protein (67).
FIG. 3.
Structural similarity between BAT3 domains and domains of other proteins, as determined by NCBI conservative domain homology analysis. The numbers on top denote the amino acid residues of the BAT3 peptide. The location of the BAT3 dominant negative peptide (BATdn) is shown as a box at the top of the BAT3 full-length protein. UBQ represents the ubiquitin-like peptide on BAT3; the region on the BAT3 peptide that is responsible for apoptotic activity contains a DEAD box, indicated as apoptosis. CAP and BAG1 indicate regions of homology to the adenylyl cyclase protein domain and BAG1 peptide, respectively. The box labeled ppro denotes the polyproline regions of BAT3, and the nuclear localization signal (37) is shown in the box labeled N.
Effects of BAT sense and antisense constructs on ASFV DNA production.
During the reconstitution experiments described above, we isolated a HeLatTA cell clone containing a construct (here designated as BAT3 1.3-3s) that contains the BAT3 EST in the sense direction relative to the Dox-controlled promoter (see below) and which showed a Dox-reversible decrease in ASFV titer that was quantitatively similar to the decrease observed for several cell clones expressing the BAT3 EST in the antisense direction (1.3-4as, 1.3-9as, and 1.2-14as) (Table 2). Further investigation of the role of BAT3 in ASFV production is the principal focus of the remainder of this report.
TABLE 2.
Growth characteristics of ASFV in cell cultures expressing BAT3 sense and antisense transcripts
| Cell clone | Titer at hpi:
|
||
|---|---|---|---|
| 24 | 48 | 72-120 | |
| HeLatTa (parental control) | 3.5 | 5 | 5.5 |
| 1.3-3 sense | 3.8 | 4 | 2.5 |
| 1.3-4 sense | 4.1 | 4.2 | 2.8 |
| 1.3a-4 antisense | 4 | 4.1 | 3.5 |
| 1.3a-9 antisense | 3.8 | 3.3 | 3.1 |
| 1.3a-14 antisense | 3.6 | 4.2 | 3.5 |
The basis for defective ASFV virus production in HeLa cell clones when transfected with BAT3 1.3-3s or BAT3 1.3-4as constructs was investigated in dot blot hybridization experiments using labeled ASFV DNA as a probe. In these experiments, the extent of viral DNA replication in different HeLa cells was inferred by quantitating viral DNA at different time points after ASFV infection. The hybridization signal on the dot blot shown in Fig. 4 indicates that the ASFV DNA in infected parental HeLa cells and in the BAT3 1.3-4as clone increased steadily from 16 h postinfection (hpi) to 48 hpi and leveled off at 72 hpi. In contrast, cells containing the BAT3 1.3-3s construct showed a decrease in the hybridization signal between 16 and 48 hpi, with a further decrease at 72 hpi. These results argue that despite the similar ability of the BAT3 EST inserted into pLentiEST in the sense or antisense direction to limit ASFV virus production, the sense and antisense constructs had disparate effects on ASFV DNA replication.
FIG. 4.
Dot blot hybridization of genomic DNA isolated from ASFV-infected EST-expressing clones with ASFV DNA, as a measure of ASFV replication. Genomic DNA was isolated from BAT3 sense and antisense clones. One hundred nanograms of total DNA was diluted serially at 1:2, and 100-μl samples of the increasing dilutions were transferred to wells from the left to right orientation (columns 1 to 6). One hundred nanograms of DNA was spotted in wells marked 1, half of that amount was spotted in wells numbered 2, etc. Malawai (Mal) DNA is the ASFV viral DNA that served as a positive control; genomic DNA from a parental HeLa cell line was used as a negative control.
Expression of ASFV early and late genes in BAT3 EST sense and antisense clones.
Production of the ASFV early and late proteins p30 and p72, respectively (7, 11, 23, 70) was assayed by fluorescence microscopy following ASFV infection of HeLatTA cells containing the BAT3 1.3-3s or BAT3 1.3-4as constructs. At different times after infection, cells were fixed and the presence of these early or late proteins was determined using fluorescein-labeled antibodies generated against the ASFV p30 or p72 proteins. As seen in Table 3, the fraction of cells showing staining for the p72 late protein was decreased dramatically at all time points in the BAT3 1.3-3s cells versus controls; the fraction of cells containing detectable p30 early protein was not affected 24 h after infection but, like the fraction containing p72, was sharply decreased at later times. The BAT3 1.3-4as clone showed a very different pattern staining for ASFV early and late proteins: the cell fraction showing detectable p30 was unaffected by the BAT3 EST at any time, whereas cells that stained for p72 were increased by two- to three fold compared with the parental cell line. This result, together with the ASFV DNA dot blot data shown in Fig. 4, suggests that in the BAT3 sense clone, blockage of ASFV replication was affected by BAT3 1.3-3s at an early stage of the virus life cycle, whereas the effects of antisense-mediated interference with BAT3 expression did not occur until a much later stage.
TABLE 3.
Expression of ASFV early (p30) and late (p72) genes in cells expressing BAT3 transcripts
| Time after infection and protein | Cell line | No. of cells/field | No. positive/field | % Positive | Total no. of cells |
|---|---|---|---|---|---|
| 24 hpi | |||||
| p30 | HeLatTa | 375 ± 21.0 | 40 ± 3.0 | 11 | 3,750 |
| 1.3-3s | 166 ± 6.0 | 58 ± 4.0 | 35 | 1,660 | |
| 1.3-4as | 183 ± 9.0 | 43 ± 4.0 | 23 | 1,830 | |
| p72 | HeLatTa | 118 ± 15.0 | 4.3 ± 1.3 | 4 | 1,180 |
| 1.3-3s | 179 ± 10.0 | 0.1 ± 0.1 | 0.05 | 1,790 | |
| 1.3-4as | 135 ± 12.0 | 8.4 ± 1.2 | 6 | 1,350 | |
| 48 hpi | |||||
| p30 | HeLatTa | 92 ± 8.0 | 22.2 ± 1.8 | 24 | 915 |
| 1.3-3s | 148 ± 8.0 | 7.3 ± 1.0 | 5 | 1,480 | |
| 1.3-4as | 95 ± 9.0 | 23 ± 3.1 | 24 | 950 | |
| p72 | HeLatTa | 150 ± 14 | 4.8 ± 1.2 | 3 | 1,500 |
| 1.3-3s | 178 ± 10 | 1.3 ± 0.5 | 0.7 | 1,780 | |
| 1.3-4as | 135 ± 0.4 | 15.6 ± 2.0 | 12 | 1,350 | |
| 72 hpi | |||||
| p30 | HeLatTa | 98 ± 6.0 | 43 ± 4.0 | 44 | 980 |
| 1.3-3s | 108 ± 6.0 | 8.0 ± 0.0 | 7 | 1,080 | |
| 1.3-4as | 50 ± 3.0 | 24 ± 3.0 | 48 | 500 | |
| p72 | HeLatTa | 96 ± 4.0 | 4.2 ± 0.1 | 4 | 960 |
| 1.3-3s | 116 ± 7.0 | 1.3 ± 0.4 | 1 | 1,160 | |
| 1.3-4as | 62 ± 9.0 | 13 ± 2.0 | 20 | 620 |
Analysis of ESTs present in the BAT3 1.3-3s and 1.3-4as constructs and their effects on cellular gene expression.
The BAT3 ESTs we cloned are contained within an NheI fragment introduced into the pLentivirus vector. The orientation of the BAT3 EST was determined by PCR amplification using a nested 3′ primer derived from the pLentiviral vector and one of the universal forward or reverse primers that flanks the EST fragment. Analysis of the sequence of the amplified PCR DNA product indicated that it encodes a predicted fusion protein of 299 amino acids consisting of an ORF starting in the BAT3 1.3-3s clone, at an ATG translation start codon (Fig. 5A) and extending into a vector-derived woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (Fig. 5B) (19). The BAT3-encoded segment of this predicted fusion peptide corresponds to amino acids 450 through 518 of the human native BAT3 as identified by accession number CAI18506.
FIG. 5.
cDNA sequence corresponding to BAT3 EST DNA and to the ORF of the fused BAT3 peptide. (A) The sequence of the BAT3 EST in the sense clone is bracketed by a pair of universal primers: the AEK reverse primer in the lined box is located at the 5′ side of the BAT3 sense sequence, and the AEK forward primer in the dotted box is positioned at the 3′ side of the BAT3 EST. The translational initiation codon of the BAT3 dominant peptide is underlined. (B) Putative ORF of the BAT3 peptide fused to a vector-encoded peptide corresponding to a segment of a woodchuck hepatitis virus DNA polymerase. The dotted box encompassing 71 amino acids is derived from the BAT3S EST, and the dashed box is the sequence of woodchuck hepatitis virus DNA polymerase encoded by the pLenti vector. The box formed by solid lines shows the NheI site where the EST was introduced into the vector, and the underlined sequence is the universal primer AEK forward, which is located on the 3′ side of the BAT3 EST sequence. The numbering at the left and right sides of the figure corresponds to amino acid and nucleotide sequence numbers of the ORF. The putative peptide contains 298 amino acids.
Three alternatively spliced transcript variants have been reported for human BAT3. Quantitative PCR analysis using oligonucleotide primers designed to detect cellular transcripts that include the segment of BAT3 present in the fusion protein showed an approximately 40% decrease in the abundance of these transcripts in three separate experiments (Fig. 6) in the antisense clone BAT3 1.3-4as. However, we were unable to detect a corresponding change in the overall abundance of BAT3 proteins by Western blotting using a polyclonal antibody generated against a synthetic peptide corresponding to the segment of BAT3 present as inserts in the BAT3 1.3 clones (data not shown), possibly because BAT3 protein epitopes in the peptide used to generate the antibody are predicted from sequence analysis to be present also in BAT3 protein isoforms that may not be affected by the transcript reduction we have observed. As expected, there was no detectable effect on the abundance of cellular BAT3 transcripts as a result of expression of the BAT3 insert in the sense direction, as shown in Fig. 6.
FIG. 6.
Expression of BAT3 transcripts as measured by reverse transcription-PCR in HeLatTA, BAT3 antisense, and BAT3 sense cells. Three sets of primers derived from different exons were used to quantify BAT3 mRNA. The oligonucleotide sequence for each primer is shown in Materials and Methods. The y axis represents the percentage of expression relative to the GUS β-glucuronidase positive control.
To learn whether the vector-encoded peptide sequence fused to the C-terminal end of the BAT3-encoding EST that we cloned is required for antiviral effects observed for BAT3 1.3-3s, two additional constructs were tested for possible effects on ASFV production. In the first, the leucine codon TTA that specifies the last amino acid of the BAT3 peptide segment (Fig. 5B) was mutated to a TGA translation termination codon. In the second construct, the BAT3 EST was cloned into a plasmid vector that yielded a BAT3-encoded peptide lacking any vector-encoded component. Neither manipulation affected the ability of the BAT3 EST to reduce the ASFV titer, indicating that the observed effects on ASFV production are independent of the vector-encoded segment of the fusion peptide, as well as that of the lentivector-derived WTRE sequences.
Previous work has shown that Scythe, a Xenopus laevis homolog of BAT3, can modulate apoptosis through its interaction with a protein called Reaper (61, 62), and recent experiments have implicated BAT3 itself as a regulator of apoptosis in mammalian cells (15). Microarray analysis of mRNA abundance in host cells carrying the BAT3 EST inserted in either the sense or antisense orientation showed that perturbation of host gene expression was observed in both types of cells. In cells containing the BAT3 EST expressed in the sense direction, 247 ESTs representing 63 previously annotated genes (false discovery rate score of 0.049; see also Tables SA and SB in the supplemental material) were upregulated, and 133 ESTs representing 62 previously annotated genes were downregulated (false discovery rate score of 0.091; see Table SB). Genetic ontology analysis indicated that half the number of genes whose expression was altered by the BAT3 1.3-3s construct, as shown in Table 4, included those associated with cell cycle events, ubiquitination, or apoptotic processes, as well as those implicated in cell adhesion, signal transduction, and transcription, suggesting that expression of the BAT3 1.3-3s EST has broad cellular effects. As shown in Table 4, four proapoptotic genes were upregulated. In contrast with our results for BAT3 1.3-3s, the BAT3 antisense clone BAT3 1.3-4as showed upregulation for only 11 mRNA species, including two genes involved in cell cycle regulation: cyclin-dependent kinase 6 (Cdk6) and Mdm2 (see Table SC in the supplemental material). The results from the same set of microarrays showed 81 genes that were downregulated; 37 of the 53 unique genes submitted to the GeneOntology software were annotated (see Table SC). Whereas uninfected cells expressing BAT3 1.3-3s and BAT3 1.3-3as grew normally in the absence of apoptotic agents, they showed increased sensitivity to the apoptosis-inducing agent staurosporine (Fig. 7), consistent with earlier evidence implicating BAT3 and its homologs in the modulation of apoptosis (15, 61, 62). Transcripts downregulated by the BAT3 1.3-3as clone included those encoded by genes related to proteins that have been localized to mitochondria, as shown in Table 5. In this context, it is interesting that the Scythe protein of Xenopus laevis (61) has been found to reversibly inhibit the actions of the heat shock chaperone protein Hsp70 (62), which has a well-established key role in apoptosis (41, 57).
TABLE 4.
Upregulated and downregulated genes in the BAT3 sense clone versus HeLatTA
| Regulation, function, and accession no. | Gene name |
|---|---|
| Upregulated genes | |
| Response to stimulus | |
| AI268394 | Killer cell lectin-like receptor subfamily C, member 2 |
| R93590 | Prolactin receptor |
| AI021894 | Mitogen-activated protein kinase kinase kinase kinase 3 |
| W92134 | Serine protease inhibitor, Kazal type 5 |
| R94660 | Major histocompatibility complex class I, E |
| AA875933 | EGF-containing fibulin-like extracellular matrix protein 1 |
| N39161 | CD36 antigen (collagen type I receptor, thrombospondin receptor) |
| Cell adhesion | |
| AI348559 | Protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| N39161 | CD36 antigen (collagen type I receptor, thrombospondin receptor) |
| AA156802 | Laminin, beta 2 (laminin S) |
| Intracellular signaling cascade | |
| AI091460 | Son of sevenless homolog 1 (Drosophila) |
| AA286902 | Serine/threonine kinase 17a (apoptosis inducing) |
| R93590 | Prolactin receptor |
| AI021894 | Mitogen-activated protein kinase kinase kinase kinase 3 |
| AA704240 | Phospholipase C, beta 1 (phosphoinositide specific) |
| Transcription | |
| H87271 | Neuronal PAS domain protein 3 |
| AA633957 | Proline-rich nuclear receptor coactivator 2 |
| AI668604 | Zinc finger protein 198 |
| H80335 | Suppressor of Ty 3 homolog (Saccharomyces cerevisiae) |
| AA987815 | Putative homeodomain transcription factor 1 |
| H51992 | Nuclear receptor coactivator 3 |
| AA676636 | Tripartite motif-containing 33 |
| N90781 | Mesenchymal stem cell protein DSC92 |
| R80235 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) |
| AI017342 | Interferon regulatory factor 6 |
| R26855 | Hypothetical protein DKFZp686L21136 |
| Cell cycle | |
| H80335 | Suppressor of Ty 3 homolog (S. cerevisiae) |
| AA704240 | Phospholipase C, beta 1 (phosphoinositide specific) |
| R80235 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) |
| AI346635 | Kinesin family member 11 |
| N23941 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| Ubiquitin | |
| H92965 | F-box protein 3 |
| AA676636 | Tripartite motif-containing 33 |
| R80235 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) |
| R36614 | F-box and leucine-rich repeat protein 7 |
| AA857859 | F-box protein 32 |
| R08866 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b |
| AA977010 | Tripartite motif-containing 9 |
| Ubiquitin ligase complex | |
| AA676636 | Tripartite motif-containing 33 |
| R80235 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) |
| R36614 | F-box and leucine-rich repeat protein 7 |
| R08866 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b |
| AA977010 | Tripartite motif-containing 9 |
| Apoptosis | |
| AA286902 | Serine/threonine kinase 17a (apoptosis inducing) |
| R93590 | Prolactin receptor |
| AA194983 | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| N23941 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| Downregulated genes | |
| Response to stimulus | |
| AA083671 | CD83 antigen (activated B lymphocytes, immunoglobulin superfamily) |
| AA449133 | CTP synthase |
| AA214574 | Mitogen-activated protein kinase kinase kinase kinase 5 |
| AA620519 | Thioredoxin domain containing 4 (endoplasmic reticulum) |
| AA910836 | Ankylosis, progressive homolog (mouse) |
| AI457797 | Chemokine (C-C motif) ligand 22 |
| AA486082 | Serum/glucocorticoid regulated kinase |
| H84871 | Serine threonine kinase 39 (STE20/SPS1 homolog, yeast) |
| R26732 | Peripheral myelin protein 22 |
| AA486738 | Bardet-Biedl syndrome 2 |
| R09561 | Decay accelerating factor for complement (CD55, Cromer blood group system) |
| H84871 | Serine threonine kinase 39 (STE20/SPS1 homolog, yeast) |
| Signal transduction | |
| AI457797 | Chemokine (C-C motif) ligand 22 |
| AA083671 | CD83 antigen (activated B lymphocytes, immunoglobulin superfamily) |
| AA488324 | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) |
| AA857563 | Ankyrin repeat and SOCS box-containing 3 |
| H98072 | Ran GTPase activating protein 1 |
| AA676404 | Peptidylprolyl isomerase C (cyclophilin C) |
| AA214574 | Mitogen-activated protein kinase kinase kinase kinase 5 |
| Cell cycle-related genes | |
| AI337108 | P300/CBP-associated factor |
| R59212 | Meningioma (disrupted in balanced translocation) 1 |
| AA488324 | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) |
| AA448799 | Anaphase promoting complex subunit 7 |
| AA434135 | CDKN1A interacting zinc finger protein 1 |
| AA608583 | NHP2 nonhistone chromosome protein 2-like 1 (S. cerevisiae) |
| AA488757 | Microspherule protein 1 |
| R26732 | Peripheral myelin protein 22 |
| Ubiquitination | |
| AI970881 | Tripartite motif-containing 32 |
| AA448799 | Anaphase promoting complex subunit 7 |
| R85212 | Ubiquitin protein ligase E3A (human papilloma virus E6-associated protein, Angelman syndrome) |
| AA670215 | Tumor susceptibility gene 101 |
| AA293847 | Tripartite motif-containing 36 |
| R27585 | Proteasome (prosome, macropain) subunit, alpha type, 1 |
FIG. 7.
Viability of BAT3 EST-expressing cell lines in the presence of staurosporine. Approximately 104 cells were grown for 24 h in each 96-well dish in different concentrations of staurosporine. The cells were then assayed for viability by the MTT method. Each point is the mean of three measurements, and the mean of the standard deviation for each point is shown as a bar in the figure. Three independent experiments were performed for this assay. The x axis shows the concentration of staurosporine used in the assay, and the y axis shows the percentage of cells that survived compared to cells without staurosporine treatment.
TABLE 5.
Downregulated genes in BAT3 antisense clone versus HeLatTa
| Function and accession no. | Gene name |
|---|---|
| Regulation of cell proliferation | |
| AA114957 | MAX interactor 1 |
| AA292054 | Growth arrest-specific 1 |
| Chromatin | |
| AA431321 | High-mobility group nucleosomal binding domain 3 |
| T66815 | Histone 1, H1c |
| W69399 | H1 histone family, member 0 |
| Mitochondrion | |
| AA670351 | Mitochondrial carrier homolog 1 (Caenorhabditis elegans) |
| N95462 | ATP-binding cassette, sub-family F (GCN20), member 2 |
| AA188563 | SH3-domain binding protein 5 (BTK associated) |
| AA677499 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19 kDa |
| AA976089 | Enoyl coenzyme A hydratase 1, peroxisomal |
| Generation of precursor metabolites and energy | |
| H99843 | Quinolinate phosphoribosyltransferase (nicotinate-nucleotide pyrophosphorylase [carboxylating]) |
| AA044326 | Kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| AA677499 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19 kDa |
| AA976089 | Enoyl coenzyme A hydratase 1, peroxisomal |
| Electron transporter activity | |
| AA044326 | Kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| AA171606 | Dehydrogenase/reductase (SDR family) member 3 |
| AA916325 | Aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) |
| AA677499 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19 kDa |
| AI924357 | Aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) |
| R93124 | Aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) |
| Apoptosis | |
| AA670351 | Mitochondrial carrier homolog 1 (C. elegans) |
| R63543 | Nerve growth factor receptor (TNFRSF16) associated protein 1 |
| AA281152 | Caspase 9, apoptosis-related cysteine protease |
| W72310 | FAST kinase |
DISCUSSION
In this report we have described the use of an EST-based genome-wide inactivation procedure (36) to functionally identify host cell genes implicated in ASFV replication. Among the genes identified are loci involved in the host cell immune response, signal transduction, mitochondrial stability, and functions related to actin cytoskeleton reorganization. The role of these genes in ASFV production was confirmed by reconstitution of the defective production phenotype in naïve cells, by the independent discovery of CGEPs in separate screens (some involving different types of host cells), and by our ability to reverse genetic inhibition of ASFV production by downregulation of the tetracycline-controlled promoter used for expression of these ESTs.
We have focused here on the effects of an EST from a gene that encodes a segment of BAT3, a member of the BAG1 protein family (9, 58) found in the MHC III region of human chromosome 6 (6) and which has been reported to modulate apoptosis and cell proliferation during mammalian development (15). The observed effects of BAT3, which produces 25 different transcripts and putatively a large number of protein isoforms as a result of alternative splicing (39), on ASFV infection are of special interest, as there is considerable evidence that host cell apoptotic processes are perturbed during such infection (42; for review, see reference 69). However, this perturbation appears to be complex: proapoptotic proteins of host cells are increased during ASFV infection (26) and a virus-encoded protein(s) has been reported to induce host cell apoptosis soon after viral entry (13, 28, 29). Conversely, ASFV proteins also have been reported to interfere with apoptosis and specifically to limit synthesis of the stress-induced proapoptotic transcription factor CHOP/Gadd153 (40).
Whereas altered expression of BAT3 or its Xenopus homolog Scythe can modulate apoptosis, the nature of this modulation also appears to be multifaceted. The apoptosis-related binding partners of Scythe include the apoptotic regulator Reaper (61), the cytoprotective stress sensor protein Hsp70 (62), and the regulatory proteins Grim, Hid (61), and X-1 (34). Our data indicate that dysfunction of BAT3 in mammalian cells results in the upregulation of apoptotic genes and that such dysfunction in ASFV-infected cells is associated with impairment of ASFV replication. In uninfected cells, BAT3 dysfunction did not affect cell growth per se but was observed to enhance staurosporine-induced apoptosis. Taken together, our findings suggest that perturbation of apoptosis-related signaling may account for the observed effects of BAT3 dysfunction on ASFV replication.
Whereas we found that expression of the BAT3 EST in either the sense or antisense orientation resulted in a reduction of ASFV titer, the mechanism underlying this reduction appears to be substantively different for the sense and antisense constructs. Dot blot DNA hybridization results indicated that the replication of ASFV DNA was not affected by BAT3 antisense transcripts; however, an increase of the late viral protein p72 was observed, accompanied by a decrease in the production or release of infectious virus. In the BAT3 1.3-3s sense clone, virus functions were severely inhibited starting at an early stage of infection, as evidenced by delayed expression of the early viral protein p30 and reduced viral DNA replication. Moreover, the overall effects of the sense construct on the expression of host cell genes were more extensive than those of the antisense construct, suggesting more complete interference with BAT3 functioning, or possibly differential effects of the sense and antisense constructs on the actions of different BAT3 isoforms.
Many of the transcripts affected by BAT3 1.3-3s were host response genes that react to external stimulation, while other transcripts affected by this construct are implicated in cell cycle regulation. Induction of the cyclin-dependent kinase inhibitor (p21), which also has been observed independently in cells infected with ASFV (26), was especially prominent. The ability of a short EST encoding 71 amino acids of BAT3 to extensively perturb cellular gene expression may occur by several mechanisms. These include possible dominant negative effects of competition between the peptide and native protein for a site on a BAT3 binding partner or reduced activity of native BAT3 by partial heterodimer formation with the peptide. While short peptide domains of larger proteins also can mimic the positive actions of a native protein (3, 20, 51), our finding that the BAT3 sense and antisense constructs both can interfere with ASFV production argues that the sense construct we have studied is affecting ASFV replication through a dominant negative mechanism.
Among the CGEPs identified by our screen was C1qTNFR6, which encodes a protein of the tumor necrosis factor superfamily (56). We note that members of the C1q superfamily have been implicated in the recognition of viral and microbial surfaces and in TNF-mediated cell death (56). Induction of the C1q complement component protein in conjunction with cytokines (e.g., TNF-α and interleukin-1α) has been observed in thymocytes of pigs infected with classical swine fever virus (54). C1q also has been shown to have a role in apoptosis of mononuclear blood cells, contributing to lymphopenia—one of the characteristic effects of ASFV infection (53).
Another EST found to affect virus production when expressed in the antisense direction encodes a mitochondrial outer membrane translocase that is part of the TOM complex, which specifically mediates the transport of preproteins, e.g., apocytochrome c, a precursor of cytochrome c, and other precursor proteins into the mitochondrial intermembrane (16). Cytochrome c functions as an electron carrier between complex III and complex IV of the electron transport chain and is released into the cytoplasm in response to apoptotic stimuli. Potentially, reduction of TOM40 may limit the uptake of cytochrome c into the mitochondria and interfere with virus replication by affecting the electron transport chain and, consequently, apoptosis.
The gene silencing procedure described here has proven useful in identifying genes and proteins whose perturbed function limits ASFV replication. We suggest that such CGEPs may be targets for pharmacological or immunological interventions that treat ASF. Agents that mimic the effects of the BAT3 dominant negative peptide may prove to be of particular value in this regard.
Supplementary Material
Acknowledgments
We thank Limin Li for helpful discussions. We also thank Chih-Jian Lih for his advice with the MTT and staurosporine assays, Kevin Pan and Richard Lin for their help in the analysis of DNA microarray data, and Roberta Peterson for help with preparation of the manuscript.
These investigations were supported by grant N66001-01-1-8947 from the Defense Advanced Research Products Agency.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aerts, J. L., M. I. Gonzales, and S. L. Topalian. 2004. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. BioTechniques 36:84-91. [DOI] [PubMed] [Google Scholar]
- 2.Afonso, C. L., M. E. Piccone, K. M. Zaffuto, J. Neilan, G. F. Kutish, Z. Lu, C. A. Balinsky, T. R. Gibb, T. J. Bean, L. Zsak, and D. L. Rock. 2004. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J. Virol. 78:1858-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agou, F., G. Courtois, J. Chiaravalli, F. Baleux, Y. M. Coic, F. Traincard, A. Israel, and M. Veron. 2004. Inhibition of NF-kappa B activation by peptides targeting NF-kappa B essential modulator (nemo) oligomerization. J. Biol. Chem. 279:54248-54257. [DOI] [PubMed] [Google Scholar]
- 4.Alfonso, P., J. Rivera, B. Hernaez, C. Alonso, and J. M. Escribano. 2004. Identification of cellular proteins modified in response to African swine fever virus infection by proteomics. Proteomics 4:2037-2046. [DOI] [PubMed] [Google Scholar]
- 5.Alonso, A., J. J. Merlo, S. Na, N. Kholod, L. Jaroszewski, A. Kharitonenkov, S. Williams, A. Godzik, J. D. Posada, and T. Mustelin. 2002. Inhibition of T cell antigen receptor signaling by VHR-related MKPX (VHX), a new dual specificity phosphatase related to VH1 related (VHR). J. Biol. Chem. 277:5524-5528. [DOI] [PubMed] [Google Scholar]
- 6.Banerji, J., J. Sands, J. L. Strominger, and T. Spies. 1990. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc. Natl. Acad. Sci. USA 87:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barderas, M. G., F. Rodriguez, P. Gomez-Puertas, M. Aviles, F. Beitia, C. Alonso, and J. M. Escribano. 2001. Antigenic and immunogenic properties of a chimera of two immunodominant African swine fever virus proteins. Arch. Virol. 146:1681-1691. [DOI] [PubMed] [Google Scholar]
- 8.Berns, K., E. M. Hijmans, J. Mullenders, T. R. Brummelkamp, A. Velds, M. Heimerikx, R. M. Kerkhoven, M. Madiredjo, W. Nijkamp, B. Weigelt, R. Agami, W. Ge, G. Cavet, P. S. Linsley, R. L. Beijersbergen, and R. Bernards. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428:431-437. [DOI] [PubMed] [Google Scholar]
- 9.Bimston, D., J. Song, D. Winchester, S. Takayama, J. C. Reed, and R. I. Morimoto. 1998. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 17:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boix, J., N. Llecha, V. J. Yuste, and J. X. Comella. 1997. Characterization of the cell death process induced by staurosporine in human neuroblastoma cell lines. Neuropharmacology 36:811-821. [DOI] [PubMed] [Google Scholar]
- 11.Borca, M. V., P. Irusta, C. Carrillo, C. L. Afonso, T. Burrage, and D. L. Rock. 1994. African swine fever virus structural protein p72 contains a conformational neutralizing epitope. Virology 201:413-418. [DOI] [PubMed] [Google Scholar]
- 12.Brookes, S. M., A. D. Hyatt, T. Wise, and R. M. Parkhouse. 1998. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology 249:175-188. [DOI] [PubMed] [Google Scholar]
- 13.Carrascosa, A. L., M. J. Bustos, M. L. Nogal, G. Gonzalez de Buitrago, and Y. Revilla. 2002. Apoptosis induced in an early step of African swine fever virus entry into vero cells does not require virus replication. Virology 294:372-382. [DOI] [PubMed] [Google Scholar]
- 14.Clague, M. J., and S. Urbe. 2003. Hrs function: viruses provide the clue. Trends Cell Biol. 13:603-606. [DOI] [PubMed] [Google Scholar]
- 15.Desmots, F., H. R. Russell, Y. Lee, K. Boyd, and P. J. McKinnon. 2005. The reaper-binding protein Scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 25:10329-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diekert, K., A. I. de Kroon, U. Ahting, B. Niggemeyer, W. Neupert, B. de Kruijff, and R. Lill. 2001. Apocytochrome c requires the TOM complex for translocation across the mitochondrial outer membrane. EMBO J. 20:5626-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon, L. K., C. C. Abrams, G. Bowick, L. C. Goatley, P. C. Kay-Jackson, D. Chapman, E. Liverani, R. Nix, R. Silk, and F. Zhang. 2004. African swine fever virus proteins involved in evading host defence systems. Vet. Immunol. Immunopathol. 100:117-134. [DOI] [PubMed] [Google Scholar]
- 18.Fogh, J., J. M. Fogh, and T. Orfeo. 1977. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J. Natl. Cancer Inst. 59:221-226. [DOI] [PubMed] [Google Scholar]
- 19.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 20.Freed, E. O. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel, K., B. Egan, and T. Lithgow. 2003. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goff, A., L. S. Ehrlich, S. N. Cohen, and C. A. Carter. 2003. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 77:9173-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Puertas, P., F. Rodriguez, J. M. Oviedo, F. Ramiro-Ibanez, F. Ruiz-Gonzalvo, C. Alonso, and J. M. Escribano. 1996. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J. Virol. 70:5689-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouon, V., G. C. Tucker, L. Kraus-Berthier, G. Atassi, and N. Kieffer. 1996. Up-regulated expression of the beta3 integrin and the 92-kDa gelatinase in human HT-144 melanoma cell tumors grown in nude mice. Int. J. Cancer 68:650-662. [DOI] [PubMed] [Google Scholar]
- 26.Granja, A. G., M. L. Nogal, C. Hurtado, C. Del Aguila, A. L. Carrascosa, M. L. Salas, M. Fresno, and Y. Revilla. 2006. The viral protein A238L inhibits TNF-alpha expression through a CBP/p300 transcriptional coactivators pathway. J. Immunol. 176:451-462. [DOI] [PubMed] [Google Scholar]
- 27.Haresnape, J. M., P. J. Wilkinson, and P. S. Mellor. 1988. Isolation of African swine fever virus from ticks of the Ornithodoros moubata complex (Ixodoidea: Argasidae) collected within the African swine fever enzootic area of Malawi. Epidemiol. Infect. 101:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernaez, B., G. Diaz-Gil, M. Garcia-Gallo, J. Ignacio Quetglas, I. Rodriguez-Crespo, L. Dixon, J. M. Escribano, and C. Alonso. 2004. The African swine fever virus dynein-binding protein p54 induces infected cell apoptosis. FEBS Lett. 569:224-228. [DOI] [PubMed] [Google Scholar]
- 29.Hurtado, C., A. G. Granja, M. J. Bustos, M. L. Nogal, G. Gonzalez de Buitrago, V. G. de Yebenes, M. L. Salas, Y. Revilla, and A. L. Carrascosa. 2004. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology 326:160-170. [DOI] [PubMed] [Google Scholar]
- 30.Jouvenet, N., P. Monaghan, M. Way, and T. Wileman. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J. Virol. 78:7990-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimchi, A. 2003. Antisense libraries to isolate tumor suppressor genes. Methods Mol. Biol. 222:399-412. [DOI] [PubMed] [Google Scholar]
- 32.Kittler, R., G. Putz, L. Pelletier, I. Poser, A. K. Heninger, D. Drechsel, S. Fischer, I. Konstantinova, B. Habermann, H. Grabner, M. L. Yaspo, H. Himmelbauer, B. Korn, K. Neugebauer, M. T. Pisabarro, and F. Buchholz. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432:1036-1040. [DOI] [PubMed] [Google Scholar]
- 33.Kleiboeker, S. B., and G. A. Scoles. 2001. Pathogenesis of African swine fever virus in Ornithodoros ticks. Anim. Health Res. Rev. 2:121-128. [PubMed] [Google Scholar]
- 34.Kumar, R., W. Lutz, E. Frank, and H. J. Im. 2004. Immediate early gene X-1 interacts with proteins that modulate apoptosis. Biochem. Biophys. Res. Commun. 323:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, L., and S. N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319-329. [DOI] [PubMed] [Google Scholar]
- 36.Lu, Q., W. Wei, P. E. Kowalski, A. C. Chang, and S. N. Cohen. 2004. EST-based genome-wide gene inactivation identifies ARAP3 as a host protein affecting cellular susceptibility to anthrax toxin. Proc. Natl. Acad. Sci. USA 101:17246-17251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manchen, S. T., and A. V. Hubberstey. 2001. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem. Biophys. Res. Commun. 287:1075-1082. [DOI] [PubMed] [Google Scholar]
- 38.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 39.National Center for Biotechnology Information. 2005. AceView: identification and functional annotation of cDNA-supported genes in higher organisms. [Online.] http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly.
- 40.Netherton, C. L., J. C. Parsley, and T. Wileman. 2004. African swine fever virus inhibits induction of the stress-induced proapoptotic transcription factor CHOP/GADD153. J. Virol. 78:10825-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nollen, E. A., J. F. Brunsting, J. Song, H. H. Kampinga, and R. I. Morimoto. 2000. Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol. Cell. Biol. 20:1083-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oura, C. A., P. P. Powell, and R. M. Parkhouse. 1998. African swine fever: a disease characterized by apoptosis. J. Gen. Virol. 79:1427-1438. [DOI] [PubMed] [Google Scholar]
- 43.Paddison, P. J., M. Cleary, J. M. Silva, K. Chang, N. Sheth, R. Sachidanandam, and G. J. Hannon. 2004. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods 1:163-167. [DOI] [PubMed] [Google Scholar]
- 44.Pan, K. H., C. J. Lih, and S. N. Cohen. 2002. Analysis of DNA microarrays using algorithms that employ rule-based expert knowledge. Proc. Natl. Acad. Sci. USA 99:2118-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panchal, R. G., G. Ruthel, T. A. Kenny, G. H. Kallstrom, D. Lane, S. S. Badie, L. Li, S. Bavari, and M. J. Aman. 2003. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. USA 100:15936-15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science. 280:248-253. [DOI] [PubMed] [Google Scholar]
- 47.Primiano, T., M. Baig, A. Maliyekkel, B. D. Chang, S. Fellars, J. Sadhu, S. A. Axenovich, T. A. Holzmayer, and I. B. Roninson. 2003. Identification of potential anticancer drug targets through the selection of growth-inhibitory genetic suppressor elements. Cancer Cell 4:41-53. [DOI] [PubMed] [Google Scholar]
- 48.Public Health Security and Bioterrorism Preparedness and Response Act of 2002. Public law 107-188. U.S. Statutes at Large 116(2002):594.
- 49.Rapaport, D. 2002. Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 27:191-197. [DOI] [PubMed] [Google Scholar]
- 50.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 51.Reeves, J. D., and A. J. Piefer. 2005. Emerging drug targets for antiretroviral therapy. Drugs 65:1747-1766. [DOI] [PubMed] [Google Scholar]
- 52.Rouiller, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez-Cordon, P. J., A. Nunez, F. J. Salguero, M. Pedrera, M. Fernandez de Marco, and J. C. Gomez-Villamandos. 2005. Lymphocyte apoptosis and thrombocytopenia in spleen during classical swine fever: role of macrophages and cytokines. Vet. Pathol. 42:477-488. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Cordon, P. J., S. Romanini, F. J. Salguero, A. Nunez, M. J. Bautista, A. Jover, and J. C. Gomez-Villamos. 2002. Apoptosis of thymocytes related to cytokine expression in experimental classical swine fever. J. Comp. Pathol. 127:239-248. [DOI] [PubMed] [Google Scholar]
- 55.Santurde, G., F. Ruiz Gonzalvo, M. E. Carnero, and E. Tabares. 1988. Genetic stability of African swine fever virus grown in monkey kidney cells. Brief report. Arch. Virol. 98:117-122. [DOI] [PubMed] [Google Scholar]
- 56.Shapiro, L., and P. E. Scherer. 1998. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8:335-338. [DOI] [PubMed] [Google Scholar]
- 57.Takayama, S., D. N. Bimston, S. Matsuzawa, B. C. Freeman, C. Aime-Sempe, Z. Xie, R. I. Morimoto, and J. C. Reed. 1997. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 16:4887-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takayama, S., Z. Xie, and J. C. Reed. 1999. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274:781-786. [DOI] [PubMed] [Google Scholar]
- 59.Tan, I., C. H. Ng, L. Lim, and T. Leung. 2001. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J. Biol. Chem. 276:21209-21216. [DOI] [PubMed] [Google Scholar]
- 60.Thielens, N. M., P. Tacnet-Delorme, and G. J. Arlaud. 2002. Interaction of C1q and mannan-binding lectin with viruses. Immunobiology 205:563-574. [DOI] [PubMed] [Google Scholar]
- 61.Thress, K., W. Henzel, W. Shillinglaw, and S. Kornbluth. 1998. Scythe: a novel reaper-binding apoptotic regulator. EMBO J. 17:6135-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thress, K., J. Song, R. I. Morimoto, and S. Kornbluth. 2001. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 20:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Troyanskaya, O., M. Cantor, G. Sherlock, P. Brown, T. Hastie, R. Tibshirani, D. Botstein, and R. B. Altman. 2001. Missing value estimation methods for DNA microarrays. Bioinformatics 17:520-525. [DOI] [PubMed] [Google Scholar]
- 64.Wei, W., Q. Lu, G. J. Chaudry, S. H. Leppla, and S. N. Cohen. 2006. The LDL receptor-related protein 6 (LRP6) mediates internalization and lethality of anthrax toxin. Cell 124:1141-1154. [DOI] [PubMed] [Google Scholar]
- 65.Westbrook, T. F., E. S. Martin, M. R. Schlabach, Y. Leng, A. C. Liang, B. Feng, J. J. Zhao, T. M. Roberts, G. Mandel, G. J. Hannon, R. A. Depinho, L. Chin, and S. J. Elledge. 2005. A genetic screen for candidate tumor suppressors identifies REST. Cell 121:837-848. [DOI] [PubMed] [Google Scholar]
- 66.World Organization for Animal Health. January. 2006, posting date. Diseases notifiable to the OIE. [Online.] http://www.oie.int/eng/maladies/en_classification.htm.
- 67.Wu, Y. H., S. F. Shih, and J. Y. Lin. 2004. Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J. Biol. Chem. 279:19264-19275. [DOI] [PubMed] [Google Scholar]
- 68.Yanez, R. J., J. M. Rodriguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodriguez, and E. Vinuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]
- 69.Zsak, L., and J. G. Neilan. 2002. Regulation of apoptosis in African swine fever virus-infected macrophages. Sci. World J. 2:1186-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zsak, L., D. V. Onisk, C. L. Afonso, and D. L. Rock. 1993. Virulent African swine fever virus isolates are neutralized by swine immune serum and by monoclonal antibodies recognizing a 72-kDa viral protein. Virology 196:596-602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.