Abstract
Hepatitis C virus (HCV) cell entry involves interaction between the viral envelope glycoprotein E2 and the cell surface receptor CD81. Knowledge of conserved E2 determinants important for successful binding will facilitate development of entry inhibitors designed to block this interaction. Previous studies have assigned the CD81 binding function to a number of discontinuous regions of E2. To better define specific residues involved in receptor binding, a panel of mutants of HCV envelope proteins was generated, where conserved residues within putative CD81 binding regions were sequentially mutated to alanine. Mutant proteins were tested for binding to a panel of monoclonal antibodies and CD81 and for their ability to form noncovalent heterodimers and confer infectivity in the retroviral pseudoparticle (HCVpp) assay. Detection by conformation-sensitive monoclonal antibodies indicated that the mutant proteins were correctly folded. Mutant proteins fell into three groups: those that bound CD81 and conferred HCVpp infectivity, those that abrogated both CD81 binding and HCVpp infectivity, and a final group containing mutants that were able to bind CD81 but were noninfectious in the HCVpp assay. Specific amino acids conserved across all genotypes that were critical for CD81 binding were W420, Y527, W529, G530, and D535. These data significantly increase our understanding of the CD81 receptor-E2 binding process.
Hepatitis C virus (HCV) is the sole member of the Hepacivirus genus within the family Flaviviridae. It is a major cause of community-acquired and posttransfusion hepatitis. More than 170 million people worldwide are seropositive for HCV, and only 20% of those infected are able to clear the virus. In the remaining 80% of individuals, the virus persists and can lead to chronic liver disease, including cirrhosis and hepatocellular carcinoma (2, 3). While combination therapies have increased treatment response rates, a significant proportion of patients fail to respond (25). Consequently there is a need to develop additional antiviral agents.
HCV has a positive-sense RNA genome that encodes a polyprotein of approximately 3,000 amino acids. This contains the structural proteins core, E1, and E2 (E2-p7) and several nonstructural proteins. E1 and E2, the putative viral envelope glycoproteins, are cleaved from the polyprotein precursor by host signal peptidases. Both are predicted to be transmembrane proteins with a large N-terminal ectodomain and a C-terminal hydrophobic region (18) and are heavily modified by N-linked glycosylation (22).
HCV entry into cells requires the interaction of the envelope glycoproteins with cell surface receptors. The ability of E2 to bind to CD81 has been widely documented; however, widespread distribution of CD81 on nonpermissive cells (30) led to the view that other liver-specific molecules must also be involved in facilitating entry (19). Several other cell surface molecules have been reported to bind to native HCV particles or recombinant E2, including the low-density lipoprotein receptor (1, 51), dendritic-cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN) and liver/lymph node-specific intercellular adhesion molecule 3-grabbing integrin (L-SIGN or DC-SIGNR) (20, 32, 42), glycosaminoglycans (4), and scavenger receptor class B type I (SR-B1) (6, 44). Despite early doubts about the function of CD81 in HCV entry, data using recently developed assays of retroviral pseudoparticles (HCVpp) and in vitro infectious clones support a central role for CD81 in mediating infection (6, 11, 29, 31).
CD81 is a membrane-bound protein that contains four transmembrane regions and two extracellular loops (30). CD81 colocalizes with a number of other cell surface molecules, and its function varies greatly across cell types (30). CD81 engagement may be associated with a number of manifestations including mixed cryoglobulinemia and B-cell malignancies (55), B-cell proliferation (36), and immunoglobulin gene hypermutation (33). The HCV E2 binding region maps to the large extracellular loop (LEL) of CD81 (41). The site is conformational, and a number of specific residues within the LEL, particularly Phe186, are critical for binding a soluble truncated form of E2 (13, 23), although this residue seems less important in retroviral pseudotype infectivity (54).
The CD81 binding region of E2 is less well characterized. The interaction requires correctly folded E2 (17), and antibody blocking experiments have identified a number of regions important for CD81 binding. The first of these lies immediately downstream of the second hypervariable region between positions 480 and 493 (8, 17, 38), the second spans residues 528 to 535 (8, 38), and a third region encompasses residues 544 to 551 (17, 38). In addition, antibodies targeting amino acids 412 to 423 and 432 to 447 are also capable of blocking CD81 binding, although antibodies targeting the latter region are able to block binding only by virus-like particles and not by soluble E2 or full-length E1E2 (38). Finally, structural modeling identified putative CD81 binding regions similar to those identified by antibody blocking experiments, as well as an additional region between residues 612 and 620 (52). Mutation of this region abolished CD81 binding (43). Although the E2-CD81 interaction is an important target for the development of entry inhibitors, individual residues within E2 that are critical for binding have not yet been identified.
HCV exhibits extensive genetic variability, particularly within the envelope genes. Six distinct genotypes have been identified, which differ by approximately 30% at the nucleotide level (45, 46). Even within a single infected individual HCV exists as a complex population of genetically distinct variants, termed a quasispecies (7, 34). To block diverse strains of HCV, effective entry inhibitors will need to target those conserved regions of the envelope proteins required for entry.
To identify conserved residues involved in CD81 interaction, we have performed site-directed mutagenesis of E2 residues conserved across a large panel of functional E1E2 clones and assessed the ability of these mutant proteins to bind to CD81 and to a variety of antibodies, as well as their ability to confer infectivity in the HCVpp assay. Using this approach we have identified a number of residues critical for CD81 binding. These data increase our understanding of HCV glycoprotein function, and this knowledge will aid the future development of small-molecule inhibitors of the CD81-E2 interaction.
MATERIALS AND METHODS
Antibodies.
The anti-E2 monoclonal antibodies (MAbs) AP33, ALP98, H48, H35, and H53 and a rabbit polyclonal antiserum R646 have been described previously (5, 10, 15, 38). MAbs were purified from hybridoma supernatants using a HiTrap protein G column according to the manufacturer's protocol (GE Healthcare). The purity of the antibodies was assessed using silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and their concentrations were determined using a commercially available protein quantification kit (Sigma, United Kingdom), according to the manufacturer's protocol.
Cell culture.
Human hepatoma Huh-7 cells (35) and human epithelial kidney (HEK) 293T cells (ATCC CRL-1573) were grown in Dulbecco's modified Eagle's medium (Invitrogen, United Kingdom) supplemented with 10% fetal calf serum, 5% nonessential amino acids, and 200 mM glutamine.
Plasmid expression constructs, generation of HCVpp, and infection assays.
The plasmids expressing the HCV genotype 1a strain H77c-derived full-length E1E2, murine leukemia virus (MLV) Gag-Pol, and the MLV transfer vector carrying the green fluorescent protein (GFP) coding sequence under the control of human cytomegalovirus promoter were described previously (5, 39). cDNA sequences encoding full-length E1E2 (representing amino acid residues 170 to 746, encoded by the HCV open reading frame referenced to strain H77c [53]) were generated and cloned into the expression vector pCR3.1 (Invitrogen) and their nucleotide sequences determined as described previously (29). HCVpps were produced essentially as described previously (5). Briefly, HEK293T cells were cotransfected with the MLV Gag-Pol packaging vector, the MLV-GFP transfer construct, and a plasmid expressing HCV E1E2 using a calcium phosphate transfection kit (Sigma, United Kingdom). In all experiments control particles devoid of envelope glycoproteins were used. Two days following transfection, the medium containing HCVpp was collected, clarified, filtered through a 0.45-μm-pore-size membrane, and used for infection of Huh-7 cells. Four days following infection, the cells were harvested and analyzed on a FACSCalibur (Becton Dickinson) using CellQuest software. The transduction efficiency was determined as the percentage of GFP-positive cells (following subtraction of the percentage of GFP-positive Huh-7 cells “infected” with the no-envelope control, which was typically 0.05%). The infectious titers, expressed as transducing units per ml, were calculated from the transduction efficiency.
To rule out the possible effects of differential incorporation of mutant E1E2 in HCVpp and/or nonincorporated glycoproteins in infection, the medium containing pseudoparticles was subjected to ultracentrifugation through a 20% sucrose cushion at 116,000 × g for 2 h at 4°C. The quantity of HCVpp present in the resulting pellet was then normalized with respect to their E2 content by deglycosylation with peptide N-glucosidase followed by Western blotting with the anti-E2 antiserum R646, and the bound antibodies were detected using enhanced chemiluminescence reagents (GE Healthcare). The E2 protein bands in the Western blot were quantitated using Quantity One volume analysis software (Bio-Rad). The HCVpp preparations with normalized E2 content were subsequently used in infection assays described above. Such normalized HCVpp preparations also contained comparable levels of MLV capsid protein as determined by Western blotting.
Site-directed mutagenesis.
An amino acid alignment of E1E2 coding regions corresponding to full-length E1E2 clones capable of conferring reproducible levels of infectivity in the HCVpp assay was generated using CLUSTAL_X (48) with manual adjustment. Residues located within regions previously implicated in CD81 binding (8, 17, 38, 43, 52) that were conserved across the functional E1E2 clones analyzed were subject to alanine replacement mutagenesis using the commercial QuikChange-II site-directed mutagenesis kit (Stratagene, The Netherlands), according to the manufacturer's protocol. Sequences of the primers used for mutagenesis are available upon request from the authors. Resulting mutant clones were sequenced across the entire E1E2 insert, using dye terminator sequencing methods (Roche, United Kingdom), to ensure that the clones possessed the desired mutation.
GNA capture and CD81 binding assay.
The enzyme-linked immunosorbent assay (ELISA) to detect MAb binding to E2 glycoprotein was performed essentially as described previously (40). Briefly, HEK293T cells were cotransfected with E1E2 coding sequence-containing plasmids, and the expressed glycoproteins present in clarified lysates of these cells were captured on to GNA (Galanthus nivalis) lectin-coated Immulon II enzyme immunoassay (EIA) plates (Thermolabsystems). Bound glycoproteins were detected using anti-E2 MAbs, followed by an anti-species immunoglobulin G-horseradish peroxidase (Sigma, United Kingdom) and TMB (3,3′,5,5′-tetramethylbenzidine; Sigma, United Kingdom) substrate. Absorbance values were determined at 450 nm.
For CD81 binding assays, a glutathione S-transferase (GST) fusion protein expressing the second extracellular region (EC2) of the LEL of human CD81 (GST-hLEL) (23) was used to coat Immulon II EIA plates (Thermolabsystems) at 0.5 μg/ml for 4 h at 37°C or overnight at 4°C. Binding of E1E2 protein, normalized by ELISA for H53-reactive E2 to specifically bind GST-hLEL, was performed as above using R646 antiserum to detect CD81-bound E2.
Radiolabeling of proteins and immunoprecipitation.
HEK293T cells were cotransfected with plasmids as described above. Eighteen hours following transfection, cells were washed with phosphate-buffered saline (PBS) and incubated in methionine/cysteine-free medium containing 25 μCi/ml of l-35S-Redivue Pro-Mix (Amersham Biosciences) for 48 h. The medium of transfected cells was harvested and clarified by centrifugation. The cells were washed with PBS, lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 20 mM iodoacetamide, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100), and the lysate was spun briefly to remove nuclei. The clarified cell lysates and the medium containing HCVpp (obtained following centrifugation through a sucrose cushion as described above) were incubated with a mixture of anti-E2 MAbs as described above for 2 h at 4°C and the immune complexes precipitated using protein A-Sepharose. Following washes of the protein A-Sepharose beads, the immune complexes were analyzed by 10% SDS-PAGE. The gels were dried and exposed to a phosphor screen and the radiolabeled proteins visualized with a Bio-Rad Personal FX phosphorimager.
RESULTS
Identification of conserved residues within putative CD81 binding regions.
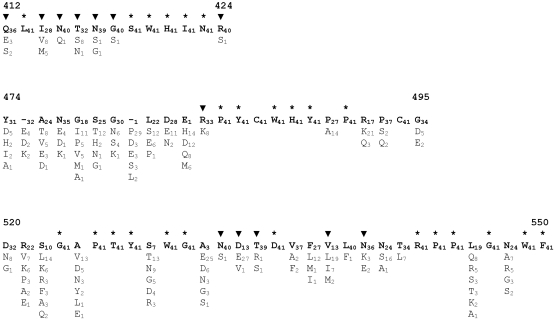
To identify E2 residues critical for binding to CD81, we adopted an approach previously used to map amino acids present in human immunodeficiency virus type 1 gp120 that are essential for CD4 binding (37). This relies on mutagenesis of residues showing absolute conservation across genetically diverse and functional envelope genes. While a large number of full-length E1E2 sequences are available through the Los Alamos HCV Sequence Database, only a limited number were known to be functional. Therefore, we isolated and tested a large number of E1E2 clones for their ability to confer infectivity to HCVpp. A total of 37 full-length E1E2 clones representative of genotypes 1 through to 6 were shown to be functional in the HCVpp entry assay (not shown), some of which have been reported previously (29, 39). Amino acid sequence alignments, corresponding to positions 412 to 424, 474 to 495, and 520 to 550 were generated for these E1E2 clones, together with E1E2 sequences of known infectious reference clones available through the Los Alamos HCV Sequence Database (http://hcv.lanl.gov/content/hcv-db/index). These regions were selected on the basis of previously published antibody blocking and protein modeling experiments that indicated their potential role in mediating E1E2 binding to CD81 (8, 17, 38, 52). A high degree of sequence conservation was observed between the functional E1E2 clones (Fig. 1). Of the 66 sites included in the analysis, 27 showed absolute conservation among functional E1E2 clones. The numbers of conserved sites within regions 412 to 424, 474 to 495, and 520 to 550 were 6, 8, and 13, respectively. As expected the second hypervariable region (HVR2), located between positions 474 and 482, contained the highest level of variability.
FIG. 1.
Identification of amino acids conserved in functional E1E2 clones located in regions previously shown to be involved in CD81 binding. E1E2 clones recovered ex vivo by PCR were analyzed for their ability to confer infectivity in a retroviral pseudoparticle (HCVpp) assay. HCVpp functional clones, together with E1E2 of infectious clones available from the Los Alamos HCV database, were aligned and the alignment divided into the three regions previously implicated in CD81 binding (26-29, 48). Amino acid numbering relative to the AUG start codon of HCV strain H (http://hcv.lanl.gov/content/hcv-db/LOCATE/locate.html) is also shown above each alignment. The sequence of HCV strain H77 is shown on the top line, while variant amino acids are shown on subsequent lines. The frequency that each residue appeared in the alignment of 41 full-length functional E1E2 clones is also shown as a subscript. *, conserved residues that were subjected to alanine replacement mutagenesis. A number of additional mutant E1E2 clones were available from other studies and were included in some of the analyses described here. ▾, sites of alanine substitution for these mutants.
Expression and antigenic analysis of mutated E1E2.
A panel of H77c-derived E1E2 mutants was generated, where each of the conserved sites within the putative CD81 binding regions (Fig. 1) was mutated to alanine. Given their importance in maintaining protein structure, the conserved cysteine residues were not mutated. Each of the mutant proteins was named according to the identification and position of the wild-type (isolate H77c) amino acid and the substitution made; for example, clone L413A has leucine at position 413 mutated to alanine. Additional mutants, available from other ongoing studies, were also included in some of the subsequent analyses. Mutant E1E2 proteins were assessed for their reactivity to a panel of MAbs recognizing linear and conformational epitopes (Fig. 2). All of the constructs tested produced detectable amounts of E2 protein, as shown by reactivity to the ALP98 antibody, which recognizes an epitope located between positions 644 and 651. The majority of the substitutions were tolerated by MAb AP33, although binding to mutants L413A, N415A, G418A, and W420A was reduced by at least 50% compared to the wild-type protein. We have recently shown that these residues form part of the AP33 epitope (47). Recognition of the mutant proteins by three conformation-dependent antibodies was more variable. All of the proteins were recognized by at least one of the conformation-sensitive antibodies, highlighting that the mutations had minimal effect on the overall conformation of the E2 protein. Assessing the reactivity of MAbs H53, H48 and H35 to the mutant proteins highlights a number of residues critical for binding to these MAbs. Mutations introduced between residues 540 and 550 reduced recognition by H53. Even more striking was that the G523A and G530A mutations reduced the binding of H35 by greater than 90%, while G530A had a similar effect on H48 binding. Indeed all mutations introduced between position 523 and 530 resulted in greater than 50% reductions in binding by both MAbs. By contrast, these mutations had minimal effect on H53 binding, highlighting that the overall conformation of the E2 protein was intact (10). Therefore it seems likely that this region, and residue 530 in particular, contributes to the epitope recognized by these two antibodies.
FIG. 2.
Reactivity of mutated E1E2 glycoproteins to monoclonal antibodies. E1E2 clones transiently expressed in HEK293T cells were captured using GNA and probed with anti-E2 MAbs AP33 and ALP98 (A) and H53, H48, and H35 (B), which recognize linear and conformational epitopes, respectively. Binding for each mutant protein is expressed as a percentage of that seen for the wild-type H77c protein. Mutant proteins are annotated according to the amino acid occurring in the H77 sequence, the amino acid position relative to the start of the H77 polyprotein chain, and the substitution introduced. WT, wild-type E1E2. Experiments repeated on different days gave comparable results. Values shown are the means and standard errors of the means for at least two replicate assays.
Alanine replacement mutagenesis identifies conserved E2 residues critical for CD81 binding.
Having confirmed that mutations had minimal effect on the overall conformation of the E2 protein, we next assessed the ability of the mutant panel to bind to a recombinant form of CD81 (Fig. 3). To compensate for any minor differences in the relative amounts of correctly folded and misfolded proteins, lysate volumes were normalized according to the amount of MAb H53-reactive E2. Alanine substitution at positions 420, 527, 529, 530, and 535 resulted in a greater-than-90% (1 log10) reduction in CD81 binding compared to wild-type H77c E1E2. In addition, a number of other mutations, specifically H421A, I422A, S424A, G523A, T526A, and F550A, reduced CD81 binding to less than 50% of that observed for wild-type E1E2. By contrast, many of the mutations introduced, for example, L413A, I414A, T416, W487A, N523A, and T534A, resulted in moderately increased CD81 binding.
FIG. 3.
Alanine replacement mutagenesis identifies a number of residues critical for CD81 binding. E1E2 mutant proteins were expressed in HEK293T cells and recovered by cell lysis. The amount of correctly folded E2 protein was determined by GNA capture enzyme immunoassay using MAb H53, and then aliquots containing H53-normalized amounts of E2 protein were allowed to bind to immobilized GST-CD81 LEL. CD81 binding for each mutant protein is expressed as a percentage of the wild-type (WT) H77c protein binding. Mutant protein annotations are as described in the legend for Fig. 2. Values shown are the means and standard errors of the means for at least two replicate assays.
Alanine replacements within the CD81 binding regions result in loss of infectivity conferred by E1E2 in the HCVpp assay.
As a number of mutations affected CD81 binding, we next assessed whether altered CD81 binding translated into changed infectivity, as determined by the HCVpp assay (Fig. 4). Few clones were capable of conferring levels of infectivity comparable to wild-type H77c E1E2. The only mutants capable of conferring infectivity levels greater than 50% of those observed for H77c were S419A, S424A, P484A, P491A, P525A, T526A, V538A, and P545A. Of the remaining mutants only N417A, N532A, D533A, T534A, N540A, and P544A were able to confer infectivity levels that were greater than 10% of those observed for the wild-type H77c E1E2.
FIG. 4.
Alanine replacement mutagenesis affects infectivity conferred by E1E2 in the HCVpp assay. HEK293T cells were cotransfected with the MLV packaging/transfer vector together with the different HCV E1E2 mutants and resulting HCVpp purified using sucrose cushion ultracentrifugation. HCVpp, normalized with respect to their E2 and gag protein contents, were used to infect Huh-7 cells. Infectivity was measured using a GFP reporter assay. The infectivity conferred by each mutant protein is expressed as a percentage of the infectivity conferred by the wild-type (WT) H77 E1E2. Mutant protein annotations are as described in the legend for Fig. 2. Values shown are the means and standard errors of the means for at least two replicate assays.
Having determined the effects of mutagenesis on HCVpp infectivity, we next assessed if this was related to CD81 binding (Table 1). Those mutations that led to a greater-than-90% reduction in CD81 binding had a concomitant effect on HCVpp infectivity. However, binding to GST-CD81 LEL was not predictive of a clone's ability to confer HCVpp infectivity. A large number of clones that exhibited good CD81 binding (Fig. 3) failed to facilitate HCVpp infection of Huh7 cells (Fig. 4). For example, replacement at positions 485, 487, 488, and 489 moderately enhanced CD81 binding yet reduced HCVpp infectivity by more than 90%. There was no correlation between CD81 binding and infectivity (P > 0.1, Spearman's rank correlation test). Therefore, CD81 binding appeared necessary, but was not sufficient, for HCVpp infectivity.
TABLE 1.
CD81 binding of mutant E1E2 clones does not predict their infectivity in the HCVpp assaya
| CD81 binding | Mutants for which HCVpp infectivity was:
|
|
|---|---|---|
| Positive | Negative | |
| Positive | N417A, S419A, P484A, P491A, P525A, T526A, N532A, D533A, T534A, V538A, N540A, P544A, P545A | Q412, L413A, I414A, N415A, T416A, G418A, H421A, I422A, N423A, R483A, Y485A, W487A, H488A, Y489A, G523A, R543A, G547A, W549A, F550A |
| Negative | W420A, Y527A, W529A, G530A, D535A | |
A panel of alanine replacement E1E2 mutants were generated, expressed in HEK 293T cells, and tested for their ability to bind to MAb H53 and GST-CD81 LEL, as well as their ability to confer infectivity in the HCVpp assay. Binding and infectivity were expressed as percentages of that observed for wild-type H77c E1E2 protein. CD81 binding and HCVpp infectivity were normalized relative to the amount of H53-reactive E2 protein. CD81 binding and HCVpp infectivity levels that were at least 10% of that observed for wild-type H77 E1E2 were considered positive.
Mutant E2 proteins maintain their ability to form noncovalent E1E2 heterodimers.
Introduction of mutations within E2, while not affecting the overall conformation of the E2 protein, might still abrogate E1E2 noncovalent heterodimer formation. To assess whether this might explain the low level of HCVpp infectivity conferred by many of the E1E2 mutants, radiolabeled E1E2s present in cell lysates were precipitated using either MAb ALP98 or a mixture of MAbs AP33 and ALP8 and then resolved by nonreducing SDS-PAGE (Fig. 5). Both E1 and E2 were detected in all of the mutant and wild-type samples, indicating that the proteins were capable of forming noncovalent heterodimers of E1 and E2. While E1E2 was detectable, differences in the amounts of precipitated proteins were evident; for example, W487A, R543A, and F550A resulted in lower yields of precipitated E1E2 protein. Finally, mutations that abolished potential NXT/S N-linked glycosylation sites (N417A and S419A, N423A, N532A and T534A, and N540A) produced proteins that had faster migration rates, indicating that these asparagine residues were modified by the addition of glycans.
FIG. 5.
E1E2 mutants are capable of noncovalent heterodimer formation. E1E2 mutant proteins, expressed in HEK293T cells in the presence of radiolabeled methionine and cysteine, were immunoprecipitated with E2-specific MAb ALP98 alone (A) or an equal mix of anti-E2 MAbs AP33 and ALP98 (B) and then resolved using nonreducing polyacrylamide gel electrophoresis. Bands corresponding to fully glycosylated E1 and E2 and partially glycosylated E2 are indicated by E1, E2, and E2*, respectively. Clones are labeled according to the identity and location of the mutated amino acid in the H77c polyprotein.
E1E2 proteins conferring different levels of HCVpp infectivity are incorporated into HCVpp.
Having established that the mutations had little effect on E1E2 heterodimer formation, we assessed whether differences in HCVpp infectivity could be explained by differential incorporation of the mutant E1E2s into the pseudoparticles. Radiolabeled HCVpp present in the cell culture medium were harvested and pelleted by centrifugation through a 20% sucrose cushion and then immunoprecipitated with MAb H53. Immunoprecipitates of the cushion-purified HCVpp were visualized by PAGE or by gag-specific Western blotting (Fig. 6). Seven clones exhibiting different HCVpp infectivities and CD81-binding affinities were analyzed together with the wild-type H77c. The levels of E1E2 and gag in the HCVpp preparations generated using the mutated E1E2s were comparable to those observed for the HCVpp incorporating wild-type E1E2, indicating that differential incorporation was unlikely to be the reason underlying altered infectivity. E1E2 heterodimers could be precipitated from all HCVpp-producing supernatants, further supporting successful incorporation of the glycoproteins into HCVpp (data not shown).
FIG. 6.
E1E2 mutants conferring different levels of infectivity are incorporated into HCVpp in comparable amounts. HEK-293T cells were cotransfected with the MLV packaging/transfer vector together with the HCV E1E2 mutants shown and resulting HCVpp purified using sucrose cushion ultracentrifugation. Sucrose cushion-purified protein pellets were immunoprecipitated with MAb H53 and analyzed by polyacrylamide gel electrophoresis for the presence of E1 and E2 glycoproteins. In addition, Western blotting with a gag-specific monoclonal antibody was used to detect gag proteins in the sucrose cushion-purified HCVpp preparations. CD81 binding of the E1E2 proteins together with the HCVpp infectivity are given for reference. CD81 binding and HCVpp infectivity levels were scored as follows: <10%, −; 10 to 49%, +; 50 to 124%, ++.
DISCUSSION
There is overwhelming evidence that CD81 facilitates entry of hepatitis C virus into hepatocytes via interaction with the viral E2 glycoprotein (6, 24, 29, 31, 41). Definition of specific residues critical for this interaction will greatly assist future development of E2-CD81 targeted entry inhibitors. Antibody blocking experiments, together with molecular modeling, identified several relatively large and discontinuous regions of E2 that were potentially involved in CD81 binding (8, 17, 38, 43, 52). However, it was unclear whether these regions formed part of the CD81 binding site or were simply in close proximity to the binding site. Specific CD81 residues involved in E2 binding have been defined (13, 23), whereas reciprocal identification of residues on E2 interacting with CD81 has not previously been attempted. One probable reason for this was the relatively poorly defined CD81 binding regions; hence a large number of residues would need to be mutated to identity those involved in binding. Successful glycoprotein-receptor interaction will exert significant purifying selection on those specific residues involved in binding. Therefore, identification of residues conserved across diverse strains of virus effectively identifies those residues most likely involved in binding, and these residues can then be targeted in mutagenesis experiments. This approach successfully identified human immunodeficiency virus type 1 gp120 residues critical for CD4 binding (37), residues whose involvement was later confirmed when the crystal structure of gp120 was obtained (28). However, in the case of HCV, sequence databases contain E1E2 sequences from both functional and nonfunctional viruses; therefore, sites potentially involved in CD81 binding may exhibit amino acid heterogeneity that is contributed by nonfunctional E1E2 clones. To overcome this problem, we first generated a panel of E1E2 clones representing genotypes 1 through 6 and screened these for functionality in the HCVpp assay. We then combined these sequences with those of known infectious clones to generate an alignment of 41 functional and diverse E1E2s. Using this approach we were able to identify a number of residues, located within the putative CD81 binding regions, for alanine replacement mutagenesis. Not only does this approach help define receptor binding at a molecular level, it has the added advantage of identifying those antiviral targets that are conserved across diverse strains of HCV.
Mutations were introduced in the context of full-length E1E2 because the E1E2 heterodimer exhibits superior CD81 binding compared to soluble monomeric E2 (9). To recover E1E2, the cells have to be lysed with a mixture of detergents, which precludes their use in cell binding assays. However, previous work has shown good correlation between plate-based CD81-LEL binding data and cell-associated CD81-binding data (41, 43), therefore negating the need to perform cell-based assays.
Before testing the ability of the mutants to bind CD81, we first assessed the effect of each mutation on expression and overall conformation of E2 by probing each mutant protein with a panel of murine monoclonal antibodies. MAb H53, which selectively recognizes correctly folded E2 (10), showed similar reactivity to the mutant proteins except for some of the mutations introduced between positions 540 and 550, which reduced binding by more than 50%. Similar reductions in binding to these mutants by MAbs H48 and H35 were seen, indicating that the E2 conformation may be sensitive to mutations within this region. Alanine substitution at positions 523 and 530 resulted in a greater-than-90% reduction in binding by MAb H35, while substitution at position 530 had a similar effect on H48 binding. Indeed many of the mutations introduced between positions 523 and 530 diminished the binding of these MAbs. These findings indicate that this region forms part of the epitope recognized by these two MAbs, with residues 523 and 530 and 523 alone as probable contact residues for MAb H35 and MAb H48, respectively. The observed overlap between residues important for CD81, MAb H35, and MAb H48 binding indicates that these MAbs target the CD81 binding site. Importantly, these MAbs have recently been shown to inhibit CD81 binding and neutralize HCVpp infectivity (12). Similarly, W420A mutation abolished both MAb AP33 (itself a broadly neutralizing antibody [39]) and CD81 binding, indicating overlap between the CD81 binding site and the AP33 epitope. Finally, mutations at L413, N415, and G418 also reduced the binding of MAb AP33, and we have recently shown that these, together with W420, are probable contact residues (47).
Having established that the mutant proteins were correctly folded, we compared their abilities to bind to CD81. We used MAb H53 normalization to ensure that cell lysates contained equivalent amounts of correctly folded E2. In addition, we used subsaturating amounts of E2 so that we could identify mutations that enhanced CD81 binding, as well as those that reduced binding. Alanine substitution at a number of sites resulted in increased CD81 binding. Enhanced CD81 and MAb binding might be due to increased exposure of the receptor and/or antigenic sites. For example, replacement of the threonine at position 534 would lead to the loss of an N-linked glycosylation site, as would the asparagine substitution upstream at position 532. Both of these resulted in moderately increased CD81 binding (Fig. 3). It is feasible that removal of a large glycan moiety led to better exposure of the CD81-binding site.
Removal of other N-linked glycosylation sites had variable effects on CD81 binding and infectivity. Increased migration rates of E2 in immunoprecipitation assays confirmed that all of the glycosylation sites were modified by the addition of glycans. Loss of the glycosylation site at 423 resulted in an E2 protein that was capable of noncovalent heterodimer formation and of CD81 binding. However, this mutant was unable to confer infectivity in the HCVpp assay. By contrast, loss of the glycosylation sites at 417, 532, and 540 did not abolish HCVpp infectivity, although infectivity conferred by E2 mutated at amino acids 417 and 532 was less than 50% of that conferred by wild-type E1E2. While these findings are in general agreement with a recently published analysis of the effect of N glycosylation on HCVpp infectivity (21), we observed a more marked reduction in HCVpp infectivity associated with loss of the glycosylation sites at 417 and 423. This is most likely due to differences in the nature of the amino acid replacement; we used alanine substitution whereas Gofford et al. used a more conservative glutamine residue.
A number of residues in the first putative CD81 binding region, which is located immediately downstream of the first hypervariable region (HVR1), reduced CD81 binding by greater than 50%. In particular, substitution W420A reduced binding by greater than 1 log10, while replacement at residues 421 and 422 both reduced binding by more than 50%. This finding is at odds with previously published data where MAb 3/11, whose epitope maps to this region of E2, only partially blocked CD81 binding (17). Based on this observation the authors argued that antibodies to this region did not directly block the CD81 binding site, but rather exerted their inhibitory effect through steric hindrance. However, an equally plausible argument to explain their findings is that CD81 may have a much greater affinity for E2 than the 3/11 MAb used in their analysis (17).
Surprisingly, none of the mutations introduced into the second putative CD81 binding region (residues 474 to 495) reduced CD81 binding. This region had been implicated in CD81 binding, as MAb 6/41a, which recognizes a linear epitope between residues 480 to 493, was apparently capable of blocking E2 binding to CD81-LEL and vice versa (17). However, in our previous work we did not observe any CD81-E2 blocking effect for this MAb (38). In addition, MAb 6/41a was also unable to block the infectivity of HCVpp carrying autologous H77 E1E2 proteins (24). These observations, together with our current mutagenesis experiments, suggest that this region is not directly involved in CD81 binding. However, it is still possible that more variable residues within this and the other regions under analysis, while not directly mediating binding, may influence CD81 binding affinity. This notion is supported by domain-swapping experiments that showed that the second hypervariable region (HVR2), encompassing residues 474 to 478, is capable of modulating CD81-E2 interaction (43).
The third putative binding region analyzed in our studies contained a number of residues that were pivotal in CD81 binding. In particular, substitutions Y527A, W529A, G530A, and D535A resulted in greater-than-90% reductions in CD81 binding, without affecting the overall conformation of the E2 protein. This region contains other sites that had indirectly been implicated in CD81 binding. For example, insertional mutations at P545 inhibit E1E2-mediated cell fusion, and this was proposed to be due to loss of CD81 binding (27). However, our data show that this residue per se is not directly involved in CD81 interaction.
Residues in CD81 LEL shown to be critical for binding to E2 include a phenylalanine residue at position 186 (13, 23). This residue is located on the head subdomain of CD81 and forms part of a low-polarity patch with other additional solvent-exposed aliphatic hydrophobic residues, for example, isoleucine at positions 181 and 182 and leucine at position 185 (26). These data, together with our mutational analyses, suggest that E2-CD81 binding is mediated by interaction of predominantly hydrophobic residues present on both molecules.
The effects of mutagenesis on HCVpp infectivity were interesting. Firstly, some mutations that reduced (but did not abrogate) CD81 binding had minimal impact on HCVpp infectivity. This suggests that suboptimal binding conferred by each E1E2 complex at a cell surface does not adversely affect entry. This is in agreement with previous studies that showed that E2 molecules that had different CD81 binding affinities were all capable of conferring comparable HCVpp infectivity (54). Secondly, while there was good concordance between loss of CD81 binding and abrogation of HCVpp infectivity, many mutations that had minimal, if any, effect on CD81 binding resulted in a large reduction in HCVpp infectivity. We showed that loss of infectivity was unlikely due to unsuccessful formation of the E1E2 noncovalent heterodimer, the supposed functional form of the viral glycoproteins (14, 16), or inefficient HCVpp incorporation. Abrogation of HCVpp infectivity conferred by some of the mutations has previously been shown. HCV entry is a multifactorial process (6, 49); therefore, it is possible that some of the mutations interfere with other steps in the viral entry process, for example with SR-BI binding or the fusion process. It will be important to confirm these findings in alternative infection systems. Introduction of these mutations into the recently described in vitro infectious clone JFH-1 (31, 50) will help resolve this issue.
In conclusion, these studies have identified a number of conserved E2 residues that are critical for CD81 binding and thus provide significant new insights into this important step in the virus life cycle. In addition, these data will prove important in the future design and targeting of small-molecule inhibitors to block CD81 binding, particularly if the detailed structure of the E2 molecule can be resolved.
Acknowledgments
We thank Duncan McGeoch for critically reading the manuscript and Jens Bukh, Francois-Loic Cosset, and Jean Dubuisson for provision of the H77c clone; the retroviral pseudotype assay; and MAbs H35, H48, and H53, respectively.
This work was supported by the Medical Research Council, European Union FP5 contract QLK2-CT-2001-01120, and the University of Nottingham Biomedical Research Committee. J.M.T. was supported by a Clinical Training Fellowship from the Wellcome Trust.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, H. J. 2000. Recovery, persistence and sequelae in hepatitis C infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1999. Global surveillance and control of hepatitis C. J. Viral Hepatitis 6:35-47. [PubMed] [Google Scholar]
- 4.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F.-L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 8.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., C. C. Kuo, J. Dubuisson, and S. Levy. 2003. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J. Virol. 77:10677-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreux, M., T. Pietschmann, C. Granier, C. Voisset, S. Ricard-Blum, P.-E. Mangeot, Z. Keck, S. Foung, N. Vu-Dac, J. Dubuisson, R. Bartenschlager, D. Lavillette, and F.-L. Cosset. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285-18295. [DOI] [PubMed]
- 13.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, M., C. Logvinoff, C. M. Rice, and J. A. McKeating. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J. Virol. 78:6875-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint, M., and J. A. McKeating. 2000. The role of the hepatitis C virus glycoproteins in infection. Rev. Med. Virol. 10:101-117. [DOI] [PubMed] [Google Scholar]
- 19.Flint, M., E. R. Quinn, and S. Levy. 2001. In search of hepatitis C virus receptors. Clin. Liver Dis. 5:873-893. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goffard, A., N. Callens, B. Bartosch, C. Wychowski, F.-L. Cosset, C. Montpellier, and J. Dubuisson. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goffard, A., and J. Dubuisson. 2003. Glycosylation of hepatitis C virus envelope proteins. Biochimie 85:295-301. [DOI] [PubMed] [Google Scholar]
- 23.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson, I., M. W. Russo, R. S. Brown, E. Lebovics, A. Min, S. Esposito, H. Tobias, F. Klion, D. Rovner, and C. Brass. 2002. Pegylated interferon alfa-2b plus ribavirin in patients with chronic hepatitis C: a trial in prior nonresponders to interferon monotherapy or combination therapy and in combination therapy relapsers. Hepatology 36:358a. [Google Scholar]
- 26.Kitadokoro, K., M. Ponassi, G. Galli, R. Petracca, F. Falugi, G. Grandi, and M. Bolognesi. 2002. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 383:1447-1452. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, M., M. C. Bennett, T. Bercot, and I. R. Singh. 2006. Functional analysis of hepatitis C virus envelope proteins, using a cell-cell fusion assay. J. Virol. 80:1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 30.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 32.Lozach, P. Y., H. Lortat-Jacob, A. De Lacroix De Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high-affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 33.Machida, K., K. T.-H. Cheng, N. Pavio, V. M.-H. Sung, and M. M. C. Lai. 2005. Hepatitis C virus E2-CD81 interaction induces hypermutation of the immunoglobulin gene in B cells. J. Virol. 79:8079-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martell, M., J. I. Esteban, J. Quer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 36.Ni, J. H., E. Hembrador, A. M. Di Bisceglie, I. M. Jacobson, A. H. Talal, D. Butera, C. M. Rice, T. J. Chambers, and L. B. Dustin. 2003. Accumulation of B lymphocytes with a naive, resting phenotype in a subset of hepatitis C patients. J. Immunol. 170:3429-3439. [DOI] [PubMed] [Google Scholar]
- 37.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 39.Owsianka, A., A. W. Tarr, V. S. Juttla, D. Lavillette, B. Bartosch, F.-L. Cosset, J. K. Ball, and A. H. Patel. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095-11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 41.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 42.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus - 15 years on. J. General Virology 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 46.Simmonds, P., J. Buch, C. Combet, G. Deléage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspé, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J.-M. Pawlotsky, F. Penin, E. Sablon, T. Shin-I, L. J. Stuyver, H.-J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 47.Tarr, A. W., A. Owsianka, J. M. Timms, C. P. McClure, R. J. P. Brown, T. P. Hickling, T. Pietschmann, R. Bartenschlager, A. H. Patel, and J. K. Ball. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592-601. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 50.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. J. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins Struct. Funct. Genet. 40:355-366. [DOI] [PubMed] [Google Scholar]
- 53.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zignego, A. L., L. Gragnani, E. Di Pietro, V. Solazzo, S. Puliti, G. Laffi, and P. Gentilini. 2003. HCV infection, malignancy, and liver transplantation. Transplant. Proc. 35:1032-1033. [DOI] [PubMed] [Google Scholar]