Abstract
Recovery from Lassa virus (LASV) infection usually precedes the appearance of neutralizing antibodies, indicating that cellular immunity plays a primary role in viral clearance. To date, the role of LASV-specific CD8+ T cells has not been evaluated in humans. To facilitate such studies, we utilized a predictive algorithm to identify candidate HLA-A2 supertype epitopes from the LASV nucleoprotein and glycoprotein precursor (GPC) genes. We identified three peptides (GPC42-50, GLVGLVTFL; GPC60-68, SLYKGVYEL; and GPC441-449, YLISIFLHL) that displayed high-affinity binding (≤98 nM) to HLA-A*0201, induced CD8+ T-cell responses of high functional avidity in HLA-A*0201 transgenic mice, and were naturally processed from native LASV GPC in human HLA-A*0201-positive target cells. HLA-A*0201 mice immunized with either GPC42-50 or GPC60-68 were protected against challenge with a recombinant vaccinia virus that expressed LASV GPC. The epitopes identified in this study represent potential diagnostic reagents and candidates for inclusion in epitope-based vaccine constructs. Our approach is applicable to any pathogen with existing sequence data, does not require manipulation of the actual pathogen or access to immune human donors, and should therefore be generally applicable to category A through C agents and other emerging pathogens.
Lassa virus (LASV), a member of the family Arenaviridae, is a rodent-borne pathogen that causes an estimated 200,000 cases of Lassa fever (LF) annually in the West African countries of Nigeria, Sierra Leone, Liberia, and the Republic of Guinea (20). Mortality due to LASV infection in hospitalized patients ranges from 16 to 70%, while an estimated 5% of all infections are thought to be fatal (20). LF is particularly lethal in pregnant women, especially during the third trimester, when mortality rates can reach 30% with fetal and neonatal losses of 88% (27). Deafness, a common complication of LASV infection, occurs in ∼30% of cases (20). There are no approved human vaccines for LASV; the only effective treatment against infection is ribavirin, provided it is administered within the first week after onset of symptoms (19, 21).
LASV has an RNA genome that is carried in an ambisense fashion and consists of two single-stranded RNA segments, the 3.4-kb small (S) and 7.2-kb large (L) segments. The L segment encodes the viral polymerase and zinc binding protein, while the S segment encodes the nucleoprotein (NP) and glycoprotein precursor (GPC), which is posttranslationally cleaved to yield GP1 and GP2 (36). Considerable genetic diversity exists among LASV isolates within the endemic region, and phylogenetically, four genetic lineages exist (7). Isolates from the western end of the endemic zone (Sierra Leone, Liberia, and the Republic of Guinea) are fairly conserved and compose a single lineage, while those from the far eastern end in Nigeria are more diverse, constituting the remaining three lineages.
One of the hallmarks of LASV infection is the absence of detectable neutralizing antibodies during acute infection (9, 14). Low-titer neutralizing antibodies, if formed, typically appear several weeks to months after the resolution of infection (9, 14, 20). Furthermore, the treatment of LF patients with immune plasma did not protect against disease (14, 19). These observations strongly suggest that cell-mediated immunity is critical for protection against LASV infection in humans. At present, our understanding of the T lymphocyte response to LASV infection is limited to empirical observations of LASV-specific CD4+ T cells present in convalescent patients (39, 40).
More detailed studies of the immune response against LASV infection have been conducted utilizing experimental animal infection models. Similar to human infection, LASV infection in either rodents or nonhuman primates failed to induce high-titer neutralizing antibodies prior to the resolution of acute infection (9, 25). The vaccination of animals with recombinant vaccinia viruses (rVV) that express native LASV proteins has been shown to be protective against lethal LASV challenge (3, 8, 13, 15, 22). While rVV constructs that expressed either NP or GPC were protective against challenge in a guinea pig model of infection, only GPC-immunized nonhuman primates were protected against fatal disease (3, 13, 22). In both models, protection was mediated in the absence of detectable neutralizing antibodies (3, 13, 22). Recently, Geisbert and colleagues demonstrated that an attenuated recombinant vesicular stomatitis virus engineered to express the LASV glycoprotein was able to confer protection against a lethal LASV infection in nonhuman primates (16). Considering the fact that both neutralizing antibodies and virus-specific T cells were detected following challenge, the basis for protection with this vaccine was likely due to a priming of a synergistic humoral and cell-mediated immune response. A parallel can be drawn with the work of Baldridge et al. in which a synergistic effect was observed between antibody and CD8+ T cells in recovery from infection by the closely related arenavirus lymphocytic choriomeningitis virus (LCMV) (4). Finally, in two separate studies, the transfer of immune splenocytes (from either guinea pigs or CBA mice that had been successfully vaccinated against LASV) to naïve recipients demonstrated that splenocytes but not serum were capable of providing protection against a subsequent lethal LASV challenge in recipient animals (18, 25). These observations from animal infection models help illustrate the importance of the antiviral T-cell response during LASV infection.
Considering the central role that cell-mediated immunity plays in providing protection against LASV infection, it is important that sensitive reagents for measuring this response in the context of human infection or in response to vaccine candidates are developed. The identification of HLA-restricted class I and class II epitopes from LASV is required to develop tetrameric staining reagents and enzyme-linked immunospot (ELISPOT) assays. In addition to being of diagnostic use, these epitopes would also serve to determine the quality of immune responses, define correlates of protection and immunopathology, and ultimately guide the selection of candidate vaccines (30). Several approaches can be taken to identify viral epitopes. One approach involves screening lymphocytes from LASV-immune individuals for reactivity against a series of overlapping peptides that correspond to selected viral proteins. While effective, studies such as these are difficult to implement for a number of reasons. First, the approach from the outset potentially requires LASV-infected material and, thus, biosafety level 4 (BSL-4) containment (41). Second, procuring lymphocytes from LASV-immune individuals in West Africa can be difficult due to a number of logistical and political constraints. Finally, identifying epitopes that provide broad population coverage can be challenging using this approach, because the HLA restrictions identified are limited to those randomly represented by the study cohort.
In the present study, we utilized an alternative approach to identify human CD8+ T-cell epitopes from LASV. Our goal was to identify epitopes that would provide broad population coverage and be conserved among diverse LASV strains. Accordingly, we screened the LASV NP and GPC amino acid sequences for CD8+ T-cell epitopes by using an HLA motif algorithm to predict potential epitopes corresponding to the HLA-A2 supertype family, which is represented in ∼50% of the worldwide population, irrespective of ethnicity. HLA-A*0201 binding peptides were screened for immunogenicity in HLA transgenic mice. Three peptides (GPC42-50, GLVGLVTFL; GPC60-68, SLYKGVYEL; and GPC441-449, YLISIFLHL) were able to be endogenously processed from native LASV GPC in HLA-restricted human target cells. Two of these epitopes (GPC42-50 and GPC60-68) protected HLA-A*0201 mice from challenge with an rVV that expressed LASV GPC.
MATERIALS AND METHODS
Bioinformatic analyses.
LASV open reading frames utilized in this study were strain Josiah NP (NCBI accession number NP_694869) and GPC (NCBI accession number NP_694870) and strain GA391 GPC (NCBI accession number CAA36645). Candidate HLA-A2 supertype epitopes were identified using a previously described algorithm (11, 17). Potential cleavage sites for the human proteasome were identified using NetChop 3.0 (http://www.cbs.dtu.dk/services/NetChop/) (23) and MHC Pathway (http://www.mhc-pathway.net) (38).
Peptides.
Peptides (≥90% pure) were obtained from Genemed Synthesis, Inc. (South San Franscisco, CA). Hepatitis B virus (HBV) ENV 378 (LLPIFFCLWV) was used as an irrelevant HLA-A*0201-restricted peptide; HRP274-82 (KVDDTFYYV) is an HLA-A*0201-restricted peptide derived from the VV host range protein 2 (35).
MHC-peptide binding assays.
Major histocompatibility complex (MHC) molecules were purified, and binding assays were performed as previously described (34). Briefly, 1 to 10 nM of radiolabeled peptide was coincubated with 1 μM to 1 nM of purified MHC in the presence of 1 to 3 μM human β2-microglubulin. After 2 days, binding of the radiolabeled peptide to the corresponding MHC class I molecule was determined by capturing MHC/peptide complexes on Greiner Lumitrac 600 microplates (Greiner Bio-One, Longwood, FL) coated with the W6/32 antibody, and measuring bound counts per minute using the TopCount microscintillation counter (Packard Instrument Co.).
Mice.
HLA-A*0201/Kb (referred to as HLA-A*0201 in the text) transgenic mice were bred at Pharmexa-Epimmune. These mice represent the F1 generation resulting from a cross between HLA-A*0201/Kb transgenic mice (expressing a chimeric gene consisting of the α1 and α2 domains of HLA-A*0201 and the α3 domain of H-2Kb) created on the C57Bl/6 background with BALB/c mice (The Jackson Laboratory) (43). All studies were conducted in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care and according to Institutional Animal Care and Use Committee-approved animal protocols.
Cells.
JA2.1 cells (human Jurkat cells that express the HLA-A*0201/Kb chimeric gene) (43) and lipopolysaccharide (LPS) blasts (prepared as previously described) (33) were grown as previously described (33). CV-1 cells (CCL-70) and BSC-40 cells (CRL-2761; both from American Type Culture Collection) were grown as directed from the supplier.
Viruses.
VLSN and VLSGPC (which express LASV Josiah NP and GPC, respectively) are rVV constucts that were generated on the Wyeth background as previously described (3, 22). rVV-NP and rVV-GPC (which express LASV Josiah NP and GPC, respectively) were generated on the Western Reserve background according to the guidelines established by Blasco and Moss (5). Briefly, the LASV NP and GPC genes were PCR amplified using primers that each contained a unique restriction site (see the underlined sequences below) (NheI, forward primers; HindIII, reverse primers). The NP gene was amplified using the forward primer 5′-CGCGCCGCTAGCATGAGTGCCTCAAAGG-3′ and the reverse primer 5′-GGGCCCAAGCTTTTACAGAACGACTCTAGGTG-3′; the GPC gene was amplified using the forward primer 5′-GGGCCGGCTAGCATGGGACAAATAGTGACATTC-3′ and the reverse primer 5′-GGGCCCAAGCTTTCATCTCTTCCATTTCACAG-3′. PCR products were cloned into the pRB21 transfer vector. CV-1 cells were infected with VV strain vRB12 (multiplicity of infection [MOI] of 2) and transfected with 10 μg of transfer vector containing either NP or GPC. Viruses that underwent homologous recombination with the transfer vector were selected on the basis of their ability to form plaques. Each rVV was screened for NP or GPC expression via Western blotting using rabbit polyclonal antibody 2165 or mouse monoclonal antibody (MAb) L52-161-6 (29) (provided by M. Guttieri), respectively (data not shown). vRB12 and pRB21 were provided by B. Moss.
ELISPOT assay.
ELISPOT assays were performed as previously described (37). Briefly, 4 × 105 splenic CD8+ T cells (isolated by anti-CD8 coated magnetic beads [Miltenyi Biotec, Auburn, CA]) were cultured with 1 × 105 peptide-pulsed or rVV-infected JA2.1 target cells. Target cells were pulsed by incubating them with peptide for at least 2 h at room temperature, followed by three washes to remove free peptide. For rVV infections, JA2.1 cells were infected (at an MOI of 10) 18 h prior to the assay. Effector and target cells were incubated in flat-bottom, 96-well nitrocellulose plates (Immobilon-P membrane; Millipore) precoated with 50 μl/well of anti-gamma interferon (IFN-γ) MAb (Mabtech AN18; 10 μg/ml). After 16 to 20 h, plates were washed and wells were incubated with 100 μl biotinylated anti-IFN-γ MAb (Mabtech R4-6A2; 1 μg/ml) for 2 h. After additional washing, spots were developed by sequential incubation with Vectastain ABC peroxidase (Vector Laboratories) and 3 amino-9-ethyl carbazole solution (Sigma-Aldrich) and counted by computer-assisted image analysis (Zeiss KS ELISPOT reader).
For endogenous processing assays, splenocytes were restimulated with peptide-pulsed LPS blasts for 6 days in vitro (as previously described [33]) prior to CD8+ T-cell isolation. The remainder of the assay was carried out as described above.
Each assay was performed in three replicate wells, and the experimental values were expressed as the mean spots/106 CD8+ T cells ± standard deviations (SD) for each peptide. Responses of CD8+ T cells derived from peptide-immunized mice against JA2.1 cells pulsed with irrelevant peptide (HBV ENV 378) or infected with irrelevant VV constructs were measured to establish background values. In addition, responses of CD8+ T cells derived from naïve mice against the same set of targets were also used to determine background values.
Immunizations and viral challenges.
HLA-A*0201 mice (8 to 12 weeks old) were inoculated subcutaneously at the base of the tail with a mixture of CD8+ T-cell peptides (50 μg of each peptide per mouse) and the helper T-cell peptides human lambda repressor 12 (YLEDARRLKAIYEKKK), chicken ovalbumin 323 (ISQAVHAAHAEINE), and HBV core 128 (TPPAYRPPNAPIL) (46.7 μg of each peptide per mouse) that had been emulsified 1:1 in incomplete Freund's adjuvant (IFA). For challenge studies, mice were immunized with peptide emulsions and challenged 14 days postimmunization via intraperitoneal (i.p.) inoculation with 1 × 107 PFU of rVV-NP or rVV-GPC. Five days postchallenge, spleens were harvested for CD8+ T-cell purification and ovaries were harvested for rVV titer determinations. To determine viral titer, ovaries were homogenized, freeze-thawed three times, and sonicated prior to plating serial 10-fold dilutions of this homogenate on BSC-40 cells. The minimal detectable level of virus was 10 PFU/ovary.
Statistical analyses.
To evaluate whether a CD8+ T-cell response to peptide-pulsed or rVV-infected target cells was statistically significant, we utilized the Student t test to compare two sets of variables: (i) mean IFN-γ spots generated by CD8+ T cells (from immunized mice) in response to JA2.1 cells pulsed with peptide (relevant versus irrelevant) or infected with an rVV construct (relevant versus irrelevant) or (ii) mean IFN-γ spots generated in response to peptide-pulsed or rVV-infected JA2.1 cells by CD8+ T cells from immunized versus naïve mice. We utilized 2-by-2 contingency tables to determine whether an association existed between peptides with binding affinities of less than or greater than 100 nM and peptides that were immunogenic in HLA-A*0201 mice. P values from the two-tailed Fisher exact test are reported. To determine whether peptide immunization led to viral titer reduction in the ovary following challenge, we used the Student t test to compare mean titers from immunized mice versus that of control mice.
RESULTS
CD8+ T-cell epitope prediction and HLA binding studies.
To identify candidate CD8+ T-cell epitopes from LASV, the NP and GPC amino acid sequences of LASV strain Josiah were screened for potential HLA-A2 supertype epitopes (11, 17). The highest probability peptides (n = 83; 31 NP and 52 GPC) predicted by the algorithm were screened for binding affinity to purified HLA-A*0201 molecules, the prototypic member of the HLA-A2 supertype family. Previous studies demonstrated that a correlation exists between peptides that possess in vitro binding affinity values of ≤500 nM and are immunogenic in vivo (in HLA transgenic mice and human recall responses) (33). A total of 32 peptides (9 NP; 23 GPC) bound purified HLA-A*0201 with affinity values of ≤500 nM (Table 1).
TABLE 1.
LASV peptide screening: binding affinity, immunogenicity, functional avidity, and endogenous processing from native antigen
| LASV peptidea | LASV peptide sequence | HLA-A*0201 binding affinity (nM) | Immunogenicb | Functional avidity (M)c | Processing from human APCd |
|---|---|---|---|---|---|
| NP530-539 | ALMDCIMFDA | 2 | + | 6 × 10−11 | − |
| GPC258-266 | LLGTFTWTL | 5 | + | 6 × 10−11 | − |
| GPC434-442 | FVFSTSFYL | 5 | + | ≤3 × 10−12 | − |
| GPC127-136 | NLYDHALMSI | 8 | + | 6 × 10−11 | − |
| GPC257-266 | RLLGTFTWTL | 10 | + | 6 × 10−11 | − |
| GPC441-450 | YLISIFLHLV | 12 | + | ≤3 × 10−12 | − |
| GPC441-449 | YLISIFLHL | 14 | + | 6 × 10−11 | + |
| GPC34-43 | GLYNFATCGL | 31 | + | ≤3 × 10−12 | − |
| NP531-539 | LMDCIMFDA | 36 | + | 1 × 10−9 | − |
| GPC42-50 | GLVGLVTFL | 41 | + | 6 × 10−11 | + |
| GPC60-68 | SLYKGVYEL | 47 | + | 6 × 10−11 | + |
| GPC213-221 | IMTSYQYLI | 67 | + | ≤3 × 10−12 | − |
| GPC257-265 | RLLGTFTWT | 67 | + | 6 × 10−11 | − |
| GPC478-487 | GLYKQPGVPV | 89 | + | 1 × 10−9 | − |
| NP101-109 | ILAADLEKL | 92 | − | NAe | NA |
| GPC314-322 | RLFDFNKQA | 94 | + | 1 × 10−5 | − |
| GPC442-450 | LISIFLHLV | 95 | − | NA | NA |
| GPC194-202 | MAWGGSYIA | 97 | − | NA | NA |
| GPC23-31 | LIALSVLAV | 102 | − | NA | NA |
| GPC133-142 | LMSIISTFHL | 113 | − | NA | NA |
| NP38-47 | LLHGLDFSEV | 132 | − | NA | NA |
| NP240-248 | NISGYNFSL | 138 | + | 6 × 10−11 | − |
| GPC343-351 | ALINDQLIM | 191 | + | 1 × 10−9 | − |
| NP444-452 | GLTSAVIDA | 198 | − | NA | NA |
| NP66-74 | RLRDLNQAV | 259 | − | NA | NA |
| GPC193-201 | RMAWGGSYI | 296 | − | NA | NA |
| GPC22-30 | VLIALSVLA | 324 | − | NA | NA |
| GPC136-144 | IISTFHLSI | 330 | − | NA | NA |
| NP376-384 | AMLQLDPNA | 383 | − | NA | NA |
| NP365-373 | LTYSQLMTL | 454 | + | 1 × 10−5 | − |
| GPC437-445 | STSFYLISI | 474 | − | NA | NA |
| GPC22-31 | VLIALSVLAV | 485 | − | NA | NA |
Peptide position within LASV Josiah NP or GPC. Each peptide that induced a significant IFN-γ response is set in bold type.
CD8+ T cells were isolated from peptide-immunized HLA-A*0201 transgenic mice 11 to 14 days postimmunization and tested for their abilities to generate IFN-γ in response to JA2.1 target cells that had been pulsed with 1 × 105 M of peptide; +, statistically significant response; −, response not statistically significant.
CD8+ T cells were isolated from peptide-immunized HLA-A*0201 transgenic mice 11 to 14 days after immunization and tested for their abilities to generate IFN-γ in response to peptide-pulsed JA2.1 target cells that received decreasing gradient doses of peptide to identify endpoint reactivity. Each value represents the endpoint quantity of peptide required to generate a statistically significant IFN-γ response.
Each immunogenic peptide was screened for its ability to be naturally processed and presented by HLA-A*0201 molecules following endogenous expression of either LASV NP or GPC in JA2.1 human target cells as a result of infection with recombinant vaccinia virus constructs expressing either LASV NP or GPC. +, processed; −, not processed.
NA, not applicable. A value of NA in the avidity or endogenous processing column indicates that the parameter was not tested for a given peptide because it was not found to be immunogenic.
Peptide immunogenicity in HLA transgenic mice.
To determine whether the 32 peptides identified above were immunogenic in vivo, HLA-A*0201 transgenic mice were immunized with pools of three to five nonoverlapping LASV peptides (Fig. 1). CD8+ T cells (effector cells) isolated from the spleens of peptide-immunized mice (11 to 14 days postimmunization) or naïve mice were incubated with peptide-pulsed HLA-A*0201/Kb-restricted human Jurkat (JA2.1) cells directly ex vivo. We screened each LASV peptide for its ability to induce IFN-γ secretion from effector cells via ELISPOT assay. Peptides were considered immunogenic if they induced IFN-γ spot formation from immunized CD8+ T cells that was significant relative to IFN-γ spot formation from naïve CD8+ T cells exposed to identical peptide-pulsed target cells and immunized CD8+ T cells exposed to JA2.1 target cells that had been pulsed with an irrelevant HLA-A*0201-restricted peptide.
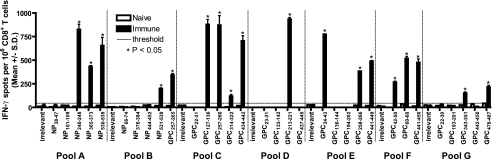
FIG. 1.
Identification of immunogenic peptides in HLA-A*0201 mice following immunization with pools of candidate LASV peptides. HLA-A*0201 mice were immunized with pools (A through G) of nonoverlapping LASV peptides (range, 3 to 5 peptides). Splenic CD8+ T cells were isolated 11 to 14 days postimmunization and exposed to JA2.1 target cells that had been pulsed with 1 × 10−5 M of each of the immunizing peptides in an ex vivo IFN-γ ELISPOT assay. Peptides that induced significant IFN-γ spot formation are denoted by an asterisk; the dotted line indicates the maximum number of spots produced by naïve CD8+ T cells exposed to peptide or immunized CD8+ T cells exposed to the irrelevant peptide. Error bars indicate standard deviations.
The results of our immunogenicity screening are summarized in Fig. 1 and Table 1. Eighteen of the 32 peptides were immunogenic in HLA-A*0201 mice. Four of these peptides were derived from NP, while the remaining 14 peptides corresponded to GPC. Of the seven peptide pools (A through G) utilized to immunize mice, each contained at least one immunogenic peptide (range of one to four peptides) (Fig. 1). The magnitude of peptide-induced IFN-γ spot formation (mean spots ± SD per 106 CD8+ T cells) ranged from 118 ± 18 for GPC314-322 to 933 ± 18 for GPC213-221. In vitro binding affinity values of ≤100 nM were highly predictive of in vivo peptide immunogenicity (P was 0.0009 via the Fisher exact test). Of the 18 peptides that bound HLA-A*0201 with affinities of 100 nM or less, 15 were immunogenic. In contrast, only 3 of the 14 peptides with affinities greater than 100 nM were immunogenic.
Functional avidity of CD8+ T-cell responses against LASV peptides.
To determine whether our immunogenic LASV peptides could induce CD8+ T-cell responses with functional avidity values representative of those seen in natural infections, in vitro peptide titration experiments were performed next. For these experiments, HLA-A*0201 mice were immunized with each of the 18 immunogenic LASV peptides. CD8+ T cells were isolated 11 to 14 days postimmunization and exposed to JA2.1 target cells that had been pulsed with serial 20-fold doses (range, 1 × 10−5 M to 3 × 10−12 M) of each peptide. A given dose was considered antigenic if it induced IFN-γ spot formation that was significantly higher than those of naïve CD8+ T cells exposed to identical peptide-pulsed target cells and immunized CD8+ T cells exposed to JA2.1 target cells pulsed with an irrelevant HLA-A*0201-restricted peptide.
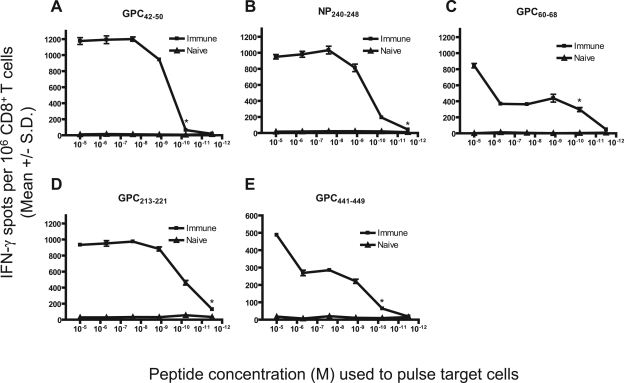
While a relatively large range (1 × 10−5 M to ≤3 × 10−12 M) of functional avidity values was observed, the majority of our peptides (n = 16) were able to generate highly avid (≤1 × 10−9 M) CD8+ T-cell responses that are representative of those seen following a natural infection with an intracellular pathogen (Table 1). Figure 2 displays representative functional avidity data.
FIG. 2.
Functional avidity of CD8+ T-cell responses against immunogenic LASV peptides in HLA-A*0201 mice following peptide immunization. HLA-A*0201 mice were immunized with (A) GPC42-50, (B) NP240-248, (C) GPC60-68, (D) GPC213-221, or (E) GPC441-449. Splenic CD8+ T cells were isolated 11 to 14 days postimmunization and exposed to JA2.1 cells that had been pulsed with gradient doses of the immunizing LASV peptide in an ex vivo IFN-γ ELISPOT assay. An asterisk denotes the lowest concentration of peptide that yielded a significant response. Error bars indicate standard deviations.
Natural processing of immunogenic peptides from native antigens in HLA-restricted human APCs.
Next, we wished to address whether the peptides could be endogenously processed from native viral antigen by human antigen-presenting cells (APCs). To express LASV NP or GPC within JA2.1 cells, rVV constructs were designed to express full-length LASV (strain Josiah) NP (VLSN) or GPC (VLSGPC) (3, 22). These constructs were engineered using the Wyeth strain of VV, and in each case, LASV target gene expression was under the control of the VV P7.5K early/late promoter (12). To test for optimal LASV protein expression, JA2.1 cells were infected with each construct at several MOIs (1, 3, and 10). At several time points (7, 19, 32, and 42 h) postinfection (p.i.), cellular protein lysates were collected and screened for LASV NP or GPC expression via Western blotting. Peak LASV protein expression was reproducibly detectable from each construct as early as 32 h p.i. following infection at an MOI of 10 (data not shown). No significant difference in cell viability was observed between infected cells and uninfected control cells at any of the time points tested (data not shown).
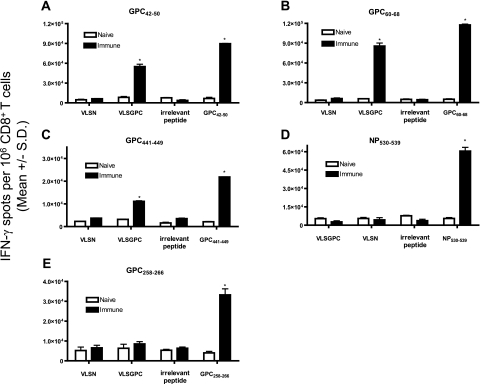
To assess processing from intact LASV antigens, HLA-A*0201 mice were immunized with individual LASV peptides. Splenocytes from immunized (days 11 to 14 postimmunization) or naïve mice were collected. Following one in vitro restimulation with peptide, CD8+ T cells were isolated and screened for IFN-γ secretion in response to JA2.1 cells that had been either pulsed with peptide or infected with rVV. It was found that three (n = 18) peptides (GPC42-50, GPC60-68, and GPC441-449) were endogenously processed from native LASV antigen by human APCs and recognized by LASV peptide-primed CD8+ T cells (Fig. 3; Table 1). The three positive peptides were derived from LASV GPC, whereas none of the four LASV NP peptides tested displayed the capacity to be naturally processed. In all cases, a significant IFN-γ response to target cells pulsed with the immunizing peptide was observed, indicating that the failure to recognize target cells expressing native LASV antigen was not due to a paucity of epitope-specific effector cells (Fig. 3 and additional data not shown).
FIG. 3.
Natural processing and recognition of immunogenic LASV peptides in HLA-A*0201/Kb-restricted human target cells that endogenously express native LASV antigens. HLA-A*0201 mice were immunized with (A) GPC42-50, (B) GPC60-68, (C) GPC441-449, (D) NP530-539, or (E) GPC258-266. Splenocytes from immunized or naïve mice were restimulated once in vitro with peptide-pulsed syngeneic LPS blasts. Following restimulation, CD8+ T cells were isolated and screened by ELISPOT assay for IFN-γ secretion in response to JA2.1 cells that had been either pulsed with peptide (LASV peptide or irrelevant peptide) or infected with rVV (irrelevant or relevant rVV construct). Peptides or rVV were considered immunogenic if they induced IFN-γ spot formation from immunized CD8+ T cells that was significant compared to IFN-γ spot formation from naïve CD8+ T cells exposed to identical peptide-pulsed or rVV-infected target cells and immunized CD8+ T cells exposed to JA2.1 target cells that had been pulsed with an irrelevant HLA-A*0201-restricted peptide or infected with an irrelevant rVV construct. Immunogenic responses are denoted by an asterisk. Error bars indicate standard deviations.
The in vitro restimulation step was utilized to avoid missing naturally processed peptides due to insufficient epitope-specific effector cells. Additional experiments utilized effector cells directly ex vivo. Each of the three GPC peptides was recognized in this format as well (data not shown).
LASV peptides are protective against rVV challenge in HLA-A*0201 mice.
We next wished to address whether the GPC-derived epitopes could confer protection against viral challenge in HLA-A*0201 mice. Because challenge with LASV would require BSL-4 containment, we utilized an alternative viral challenge model. Peptide-immunized HLA-A*0201 mice were challenged with an rVV construct that expressed either LASV GPC or LASV NP. The Wyeth-based rVV constructs (VLSN and VLSGPC) utilized in the endogenous processing assays could not be employed in these challenge studies because of their highly attenuated growth properties in mice. We therefore constructed a second set of rVV constructs on the Western Reserve background, which is permissive for murine infection. These new constructs expressed either LASV GPC (rVV-GPC) or NP (rVV-NP) from strain Josiah (see Materials and Methods). Intraperitoneal inoculation of HLA-A*0201 mice with 106 or 107 PFU of these constructs reliably led to relatively high virus titers (range, 105 to 107 PFU) in the ovaries (data not shown).
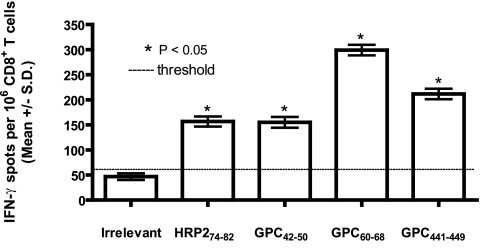
To verify that murine APCs could naturally process our GPC-derived epitopes from endogenously expressed LASV GPC, we infected HLA-A*0201 mice with 107 PFU of rVV-GPC via i.p. inoculation. Five days later, we isolated splenic CD8+ T cells and exposed them to JA2.1 cells pulsed with LASV GPC42-50, GPC60-68, or GPC441-449 in an ex vivo ELISPOT assay. As a control, we also included JA2.1 cells pulsed with HRP274-82, an HLA-A*0201-restricted peptide (derived from the VV host range protein 2) that has been shown to protect HLA-A*0201 mice from lethal VV challenge (35). As shown in Fig. 4, each of the three LASV GPC epitopes induced significant IFN-γ responses comparable to that of HRP274-82. Therefore, because our LASV peptides were processed by murine APCs, they were acceptable for inclusion in an HLA-A*0201 mouse challenge model.
FIG. 4.
Murine processing and recognition of immunogenic LASV peptides. HLA-A*0201 mice were inoculated i.p. with 107 PFU of rVV-GPC. On day 5 p.i., splenic CD8+ T cells were isolated and screened in an ex vivo ELISPOT assay for IFN-γ secretion in response to peptide-pulsed JA2.1 cells. Immunogenic peptides are denoted by an asterisk; the dotted line indicates the number of IFN-γ spots produced in response to irrelevant peptide. Error bars indicate standard deviations.
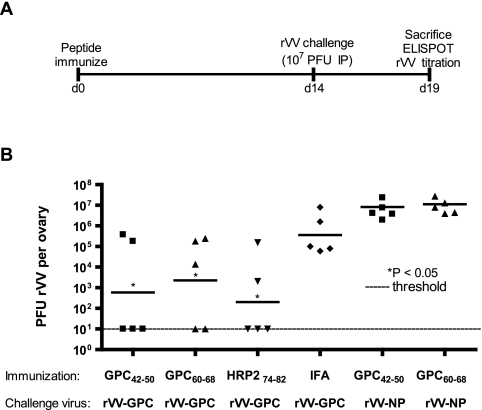
Figure 5A outlines the logistics of the challenge experiment. Groups of HLA-A*0201 mice were initially immunized with the following CD8+ T-cell peptides: GPC42-50, GPC60-68, HRP274-82, or no CD8+ T-cell peptide (IFA plus helper peptides). On day 14 postimmunization, mice were challenged with 107 PFU of rVV-GPC via i.p. inoculation. Ovaries were harvested from each mouse on day 5 postchallenge and screened for rVV-GPC titer. Each of the IFA control animals displayed evidence of rVV replication following challenge (Fig. 5). In contrast, compared to that in IFA control mice, we observed a significant reduction in mean viral titer following immunization with GPC42-50 (2.79 log10 reduction; P = 0.030), GPC60-68 (2.2 log10 reduction; P = 0.045), or HRP274-82 (3.26 log10 reduction; P = 0.007).
FIG. 5.
Peptide immunization protects HLA-A*0201 mice from rVV-GPC challenge. The logistics of the challenge experiment are outlined in panel A. Female HLA-A*0201 mice were immunized with GPC42-50, GPC60-68, HRP274-82, or IFA (plus helper T lymphocyte peptides alone). On day 14 postimmunization, mice were inoculated i.p. with 1 × 107 PFU of rVV-GPC or rVV-NP. Ovaries were harvested on day 5 postchallenge, and viral titers were determined via plaque assay. Individual viral titers are shown in log10 values; the solid line represents the geometric mean titer for each group. The limit of detection (10 PFU/ovary) in the plaque assay is indicated by the dotted line. Mean titers from each group were compared to that of the IFA control group using the Student t test to determine if differences were significant. Significant reductions in viral titer are indicated by an asterisk.
Protection is epitope specific.
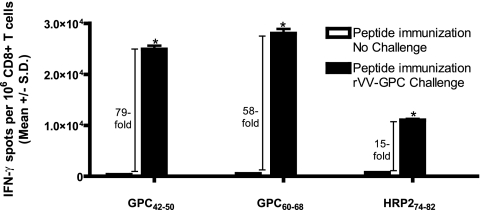
To rule out the possibility that immunization with GPC42-50 or GPC60-68 led to generalized immunity against rVV constructs, groups of mice immunized with GPC42-50 or GPC60-68 were subsequently challenged on day 14 postimmunization with 107 PFU of rVV-NP. Immunization with GPC42-50 or GPC60-68 did not result in protection against challenge with the irrelevant rVV-NP construct, confirming that the protection observed was epitope specific (Fig. 5). To provide further evidence of epitope-specific protection, we measured the frequency of epitope-specific (GPC42-50, GPC60-68, or HRP274-82) CD8+ T cells in mice that were immunized with peptide and then either challenged or not challenged with rVV-GPC. We reasoned that if protection was epitope-specific, we would see a substantial expansion of epitope-specific CD8+ T cells following challenge with rVV-GPC (relative to unchallenged mice). Figure 6 summarizes the results from this experiment. Peptide-immunized mice that did not receive a challenge displayed significant numbers of epitope-specific T cells on days 19 postimmunization (range, 312 ± 11.46 to 711 ± 17.34 mean spots ± SD per 106 CD8+ T cells). However, peptide-immunized mice that were challenged with rVV-GPC showed a clear expansion of CD8+ T cells specific for the immunizing peptide (Fig. 6). The increase in peptide-specific effector cells ranged from 15-fold for HRP274-82 to 79-fold and 58-fold, respectively, for GPC42-50 and GPC60-68.
FIG. 6.
Challenge with rVV-GPC significantly increases the frequency of epitope-specific CD8+ T cells in peptide immunized HLA-A*0201 mice. HLA-A*0201 mice were immunized with GPC42-50, GPC60-68, or HRP274-82. On day 14 postimmunization, a subset of mice in each immunization group were inoculated i.p. with 1 × 107 PFU of rVV-GPC. On day 19 postimmunization, splenic CD8+ T cells were isolated from mice that were immunized without an accompanying viral challenge (open boxes) or immunized and challenged with rVV-GPC (black boxes). CD8+ T cells were exposed to JA2.1 target cells that had been pulsed with 1 × 10−5 M of the immunizing peptide in an ex vivo IFN-γ ELISPOT assay. We compared challenged versus unchallenged CD8+ T cells for their abilities to produce IFN-γ in response to peptide-pulsed JA2.1 cells using the Student t test. Significant increases in epitope-specific CD8+ T-cell frequencies are indicated by an asterisk. The increase (n-fold) of epitope-specific CD8+ T-cell frequencies in challenged mice compared to unchallenged mice are indicated. Error bars indicate standard deviations.
Relevance of LASV epitopes for vaccine design and diagnostics.
Considerable genetic diversity exists among LASV isolates within the endemic region (7). To investigate this issue, we compared the primary amino acid sequence of each immunogenic peptide (derived from LASV strain Josiah) to the corresponding region in LASV strain GA391. Strain GA391 was chosen for this comparison because it is both genetically and geographically distinct from strain Josiah. Genetically, the NP and GPC amino acid sequences of GA391 differ from Josiah by 10 and 7%, respectively, while geographically, each strain was isolated at either the far eastern (GA391) or western (Josiah) border of the endemic region. Of our three LASV epitopes, GPC441-449 was the only one that was identical to strain GA391 in primary amino acid sequence. The amino acid sequence differences observed between viral strains from the remaining two epitopes are listed in Table 2. Differences in primary sequence were restricted to a conservative amino acid substitution at a nonanchor residue in GPC42-50, whereas GPC60-68 differed from GA391 with a conservative substitution occurring at the B pocket anchor residue, a nonconservative substitution at position 1 and a semiconserved substitution at position 6.
TABLE 2.
LASV epitopes that differ in primary sequence among isolates are cross-reactive
| Peptidea | LASV strain | Peptide sequenceb | Functional avidity (M)c | Binding affinity (nM) to HLA allotype
|
||||
|---|---|---|---|---|---|---|---|---|
| A*0201 | A*0202 | A*0203 | A*0206 | A*6802 | ||||
| GPC42-50 | Josiah | GLVGLVTFL | 6 × 10−11 | 41 | 88 | 7 | 118 | >10,000 |
| GPC42-50 | GA391 | --I------ | 6 × 10−11 | NDd | ND | ND | ND | ND |
| GPC60-68 | Josiah | SLYKGVYEL | 6 × 10−11 | 47 | 115 | 12 | 395 | >10,000 |
| GPC59-67 | GA391 | LI---T--- | (−) | ND | ND | ND | ND | ND |
| GPC441-449 | Josiah | YLISIFLHL | 6 × 10−11 | 14 | 173 | 11 | 10 | 6,657 |
| GPC440-448 | GA391 | --------- | ||||||
Peptide position within LASV (strain Josiah or GA391) NP or GPC.
Boldface letters indicate amino acid differences in LASV strain GA391 compared to LASV strain Josiah.
CD8+ T cells were isolated from peptide (LASV strain Josiah peptide)-immunized HLA-A*0201 transgenic mice 11 to 14 days after immunization and tested for their abilities to generate IFN-γ in response to peptide-pulsed JA2.1 target cells that received decreasing gradient doses of LASV strain Josiah or GA391 peptide to identify endpoint reactivity. Each value represents the endpoint quantity of peptide required to generate a statistically significant IFN-γ response. (−), not immunogenic.
ND, not tested.
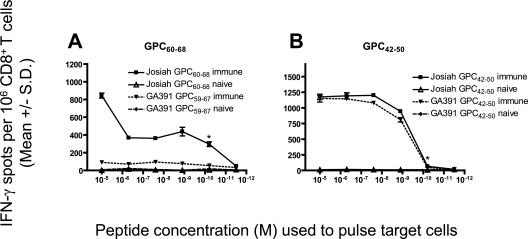
To determine whether the variant GA391 peptides would be cross-reactively recognized by CD8+ T cells specific for LASV Josiah, HLA-A*0201 mice were immunized with each of the two LASV Josiah peptides (GPC42-50 and GPC60-68). On days 11 to 14 postimmunization, splenic CD8+ T cells were isolated and exposed to JA2.1 cells that had been pulsed with a dose range of either the immunizing LASV Josiah epitope or the corresponding GA391 variant peptide. One of the two GA391 peptides, GPC42-50, was antigenic in this setting and retained a high functional avidity value that was equal to that obtained with the corresponding Josiah epitope (Fig. 7; Table 2). The other variant peptide, GA391 GPC59-67, was not cross-reactive with CD8+ T cells induced by immunization with the corresponding Josiah epitope.
FIG. 7.
Functional avidity of CD8+ T-cell responses against immunogenic LASV Josiah peptides and the corresponding LASV GA391 peptides in HLA-A*0201 mice following peptide immunization. HLA-A*0201 mice were immunized with (A) GPC60-68 or (B) GPC42-50 (from LASV strain Josiah). Splenic CD8+ T cells were isolated 11 to 14 days postimmunization and exposed to JA2.1 target cells that had been pulsed with gradient doses of the immunizing LASV strain Josiah peptide or the corresponding LASV strain GA391 peptide in an ex vivo IFN-γ ELISPOT assay. An asterisk denotes the lowest concentration of peptide that yielded a significant response. Error bars indicate standard deviations.
As a final criterion of clinical relevance, we examined each epitope for its binding affinity against a panel of the five most commonly occurring HLA-A2 supertype alleles (HLA-A*0201, -A*0202, -A*0203, -A*0206, and -A*6802) (Table 2). Each of the three peptides displayed promiscuous binding capacity to the majority of HLA-A2 supertype alleles tested. More specifically, GPC42-50, GPC60-68, and GPC441-449 were each able to bind four out of five alleles with a high affinity of 500 nM or less.
DISCUSSION
The CD8+ T-cell response is critical for protective immunity against intracellular pathogens. The humoral and cell-mediated immune responses act synergistically to provide protection appropriate to the pathogen. LASV infections are atypical in that they generally resolve prior to the appearance of neutralizing antibodies (9, 14). Based largely upon indirect evidence, it is currently thought that the CD8+ T-cell response is the main determinant responsible for providing protection against LASV infection (14). In order to resolve the critical elements of the antiviral T-cell response to LASV and guide vaccine development, we have identified three HLA-A2 supertype-restricted epitopes from the LASV GPC gene. Because the HLA-A2 supertype family is present in about 50% of the general population (regardless of ethnicity), these epitopes should prove to be useful diagnostic reagents for evaluating CD8+ T-cell responses in the context of natural infection (active infection or recall response) and/or vaccine trials. These studies should allow for a better understanding of the natural history of infection with LASV and help to determine how CD8+ T-cell responses correlate with the outcome of LASV infection.
Previous studies have demonstrated that a correlation exists between strength of peptide binding to MHC and immunogenicity (33). On the basis of this observation, we limited our immunogenicity screening to the predicted peptides that were able to bind purified HLA-A*0201 molecules with affinities of 500 nM or less (n = 32) (Table 1). We identified 18 immunogenic peptides from these candidates and found that there was a significant association between peptides with binding affinities of ≤100 nM and those that were immunogenic; we propose that this more stringent binding affinity threshold be employed in future epitope identification studies.
Following identification of immunogenic peptides in HLA transgenic mice (via peptide immunization), it is important to address whether each peptide can be correctly processed from the native antigen and subsequently displayed on the surface of an APC. Of the 18 immunogenic peptides identified in this study, three were generated by natural processing in human APCs (Table 1; Fig. 3). No correlation was found between natural processing and either in vitro binding affinity or functional avidity. To address whether it would have been possible to predict the naturally processed peptides, we utilized NetChop 3.0 (http://www.cbs.dtu.dk/services/NetChop/), a neural network-based prediction method for cleavage sites of the human proteasome (23), and MHC Pathway (http://www.mhc-pathway.net), which combines matrix-based predictions of proteasomal cleavage and transporter associated with antigen processing transport (38), to predict which peptides would be naturally processed. While these algorithms correctly identified a subset of nonprocessed peptides (n = 4), they were not specific enough to eliminate false-positive predictions (n = 11) (data not shown). These observations illustrate the importance of experimentally evaluating whether immunogenic peptides are also generated by cellular processing.
Recently, Boesen and colleagues took a similar approach to identifying HLA-A*0201-restricted epitopes from LASV NP and GPC (6). The authors concluded that they had identified three novel HLA-A*0201-restricted epitopes (GPC60-68, GPC258-266, and GPC441-449) from LASV (on the basis of immunogenicity following peptide immunization) but did not evaluate these peptides for their abilities to be naturally processed in human APCs. Our studies show that GPC258-266 was not naturally processed in human APCs (Fig. 3) and further emphasizes the importance of experimentally determining cellular processing in the context of HLA transgenic mouse studies.
Two of the epitopes (GPC42-50 and GPC60-68) identified in this study were protective against rVV-GPC challenge in HLA-A*0201 mice and are thus potentially relevant for vaccine design. The exact immunological basis for protection from LASV infection has not been formally determined. While protection generally occurs independently of neutralizing antibodies in both animal infection models and human infection, what component(s) of the cell-mediated immune response is required for protective immunity has not been determined (3, 13, 18, 22, 25). Our group recently demonstrated that the immunization of mice with a single H-2d-restricted CD8+ T-cell epitope derived from LASV NP was sufficient to protect against a heterologous challenge with LCMV (28). We built upon this initial observation by demonstrating that CD8+ T cells primed against individual HLA-A*0201-restricted LASV GPC epitopes were protective against a heterologous challenge with an rVV (that expressed the native LASV antigen that each epitope was derived from) in HLA-A*0201 transgenic mice. This observation demonstrates that the CD8+ T-cell response is capable of providing protection against challenge with a virus that expresses an LASV antigen during the course of infection.
Epitope-based vaccines make excellent candidates for pathogens such as LASV for several reasons. First, it can be established that each epitope incorporated into the vaccine is able to mediate HLA-restricted protection. This is an important caveat considering that the majority of today's experimental vaccines are necessarily tested first in small animal models and, if successful in that format, then in nonhuman primate models. The inherent risk of this approach is making the assumption that the determinants required for protection in the experimental animal models are equivalent to those needed for humans (due to the differences in the genetic backgrounds represented in each format). A second advantage is that broad population coverage (greater than 99%) can be established, providing that epitopes corresponding to multiple HLA supertype families are incorporated into the vaccine (31, 32). Third, epitope-based vaccines avoid the potential for “immunosuppressive epitopes” to inadvertently drive the immune response from a Th1 to Th2 response (26, 44). In addition, there is great flexibility in the epitopes utilized in a given vaccine considering the fact that subdominant epitopes can equal the effectiveness of immunodominant epitopes as vaccine determinants (42). Last, monitoring immunogenicity following vaccination is easily done due to the fact that the epitopes used in the vaccine also serve as the required diagnostic reagents needed to follow T-cell responses in recipients.
Epitope-based vaccines can also be tailored to provide protection against multiple pathogens (or strains of a particular pathogen), providing that conserved or cross-reactive epitopes exist (30). ter Meulen et al. have already identified LASV-specific CD4+ T-cell epitopes that are conserved with other arenavirus family members (40). Similarly, in murine models, cross-reactive CD8+ T-cell epitopes have been identified among several arenaviruses, including LASV, LCMV, and Pichinde virus (24, 28) (J. Botten, unpublished data). In our case, GPC441-449 is highly conserved with other pathogenic arenaviruses. Following immunization of HLA-A*0201 mice with this epitope, peptides representing the corresponding regions of Mopeia, LCM, Junin, Machupo, Guanarito, Whitewater Arroyo, and Pichinde viruses are recognized by CD8+ T cells in a cross-reactive manner (J. Botten, unpublished data). The inclusion of conserved or cross-reactive peptides could potentially lead to the production of vaccines that are capable of providing protection against pathogens that are able to rapidly mutate in response to immune pressure (hepatitis C virus [HCV], human immunodeficiency virus [HIV], influenza, etc). Alternatively, epitopes corresponding to several pathogens can be incorporated into a single vaccine.
While our approach to epitope identification in this study was tailored specifically for LASV, it represents a model system that could be applied to any virus (or other intracellular pathogen). The minimum requirements needed to utilize this approach are (i) existing sequence data for the pathogen in question, (ii) access to the genome (DNA or RNA) of the pathogen for the purpose of generating rVV vectors, (iii) HLA transgenic mice, and (iv) access to BSL-2 facilities for in vitro and in vivo studies. Therefore, our approach to epitope identification as well as testing epitopes for their ability to protect against infection should be nearly universal regardless of the pathogen studied.
In the case of LASV, our approach allowed us to overcome several obstacles. First, we were able to avoid the logistical challenges associated with procuring lymphocytes from LASV-immune individuals. Because we carried out predictions to identify peptides that corresponded to an HLA supertype family, we were able to guarantee broad population coverage at the outset of the study. Second, the use of rVV vectors allowed us to circumvent the need for manipulating live LASV, which would have required BSL-4 containment (41). We instead were able to use rVV to drive the expression of native LASV antigen in HLA-restricted target cells for endogenous processing studies. We also utilized rVV constructs as challenge viruses in our protection experiments. This type of heterologous viral challenge has been used in several studies, including those involving immunizations with epitopes derived from viruses such as HIV, HCV, and others (1, 2, 10, 35). In a subset of these studies, an rVV (expressing a foreign viral open reading frame) was utilized as a challenge agent because the wild-type virus (HIV or HCV) was not permissive for infection in mice (or other relevant animal models) (1, 2, 10). Therefore, the use of an rVV in a challenge setting not only removes the need for high containment with certain pathogens but also makes it possible to evaluate epitope-based protection for pathogens that would otherwise not be permissive for a given animal infection model. Because a number of select agents/category A to C pathogens, particularly the hemorrhagic viruses (arenaviruses, bunyaviruses, flaviviruses, and filoviruses), possess part or all of the restrictions outlined above, our approach is also applicable in the biodefense setting.
Acknowledgments
We thank Jennifer Abma, Nicola Benning, Renaud Burrer, Claire Crimi, Rielle Giannino, Howard Grey, Marie-France del Guercio, Daniel Hassett, Carla Oseroff and David Seiber for helpful comments and/or technical assistance. We are grateful to Mary Guttieri for providing the anti-LASV GP1 mouse MAb, Bernard Moss for providing the reagents needed to generate rVV, and David Auperin for the Wyeth-based rVV.
This work was supported by National Institutes of Health grants AI50840 (to M.J.B.), AI27028 (to J.L.W.), AI065359 (to M.J.B.), T32 AI07354 (to J.B.), and F32 AI056827 (to J.B.) and contract HHSN266200400023C (to A.S.).
This is TSRI manuscript 18086-MIND.
REFERENCES
- 1.Ahlers, J. D., I. M. Belyakov, S. Matsui, and J. A. Berzofsky. 2001. Mechanisms of cytokine synergy essential for vaccine protection against viral challenge. Int. Immunol. 13:897-908. [DOI] [PubMed] [Google Scholar]
- 2.Arichi, T., T. Saito, M. E. Major, I. M. Belyakov, M. Shirai, V. H. Engelhard, S. M. Feinstone, and J. A. Berzofsky. 2000. Prophylactic DNA vaccine for hepatitis C virus (HCV) infection: HCV-specific cytotoxic T lymphocyte induction and protection from HCV-recombinant vaccinia infection in an HLA-A2.1 transgenic mouse model. Proc. Natl. Acad. Sci. USA 97:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Auperin, D. D., J. J. Esposito, J. V. Lange, S. P. Bauer, J. Knight, D. R. Sasso, and J. B. McCormick. 1988. Construction of a recombinant vaccinia virus expressing the Lassa virus glycoprotein gene and protection of guinea pigs from a lethal Lassa virus infection. Virus Res. 9:233-248. [DOI] [PubMed] [Google Scholar]
- 4.Baldridge, J. R., T. S. McGraw, A. Paoletti, and M. J. Buchmeier. 1997. Antibody prevents the establishment of persistent arenavirus infection in synergy with endogenous T cells. J. Virol. 71:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco, R., and B. Moss. 1995. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene 158:157-162. [DOI] [PubMed] [Google Scholar]
- 6.Boesen, A., K. Sundar, and R. Coico. 2005. Lassa fever virus peptides predicted by computational analysis induce epitope-specific cytotoxic-T-lymphocyte responses in HLA-A2.1 transgenic mice. Clin. Diagn. Lab. Immunol. 12:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen, M. D., P. E. Rollin, T. G. Ksiazek, H. L. Hustad, D. G. Bausch, A. H. Demby, M. D. Bajani, C. J. Peters, and S. T. Nichol. 2000. Genetic diversity among Lassa virus strains. J. Virol. 74:6992-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clegg, J. C., and G. Lloyd. 1987. Vaccinia recombinant expressing Lassa-virus internal nucleocapsid protein protects guinea pigs against Lassa fever. Lancet ii:186-188. [DOI] [PubMed] [Google Scholar]
- 9.Clegg, J. C. S. 1992. Current progress towards vaccines for arenavirus-caused diseases. Vaccine 10:89-95. [DOI] [PubMed] [Google Scholar]
- 10.Daftarian, P., S. Ali, R. Sharan, S. F. Lacey, C. La Rosa, J. Longmate, C. Buck, R. F. Siliciano, and D. J. Diamond. 2003. Immunization with Th-CTL fusion peptide and cytosine-phosphate-guanine DNA in transgenic HLA-A2 mice induces recognition of HIV-infected T cells and clears vaccinia virus challenge. J. Immunol. 171:4028-4039. [DOI] [PubMed] [Google Scholar]
- 11.Doolan, D. L., S. L. Hoffman, S. Southwood, P. A. Wentworth, J. Sidney, R. W. Chesnut, E. Keogh, E. Appella, T. B. Nutman, A. A. Lai, D. M. Gordon, A. Oloo, and A. Sette. 1997. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity 7:97-112. [DOI] [PubMed] [Google Scholar]
- 12.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 1998. Generation of recombinant vaccinia viruses, p. 16.17.1-16.17. 19. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 13.Fisher-Hoch, S. P., L. Hutwagner, B. Brown, and J. B. McCormick. 2000. Effective vaccine for lassa fever. J. Virol. 74:6777-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher-Hoch, S. P., and J. B. McCormick. 2001. Towards a human Lassa fever vaccine. Rev. Med. Virol. 11:331-341. [DOI] [PubMed] [Google Scholar]
- 15.Fisher-Hoch, S. P., J. B. McCormick, D. Auperin, B. G. Brown, M. Castor, G. Perez, S. Ruo, A. Conaty, L. Brammer, and S. Bauer. 1989. Protection of rhesus monkeys from fatal Lassa fever by vaccination with a recombinant vaccinia virus containing the Lassa virus glycoprotein gene. Proc. Natl. Acad. Sci. USA 86:317-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisbert, T. W., S. Jones, E. A. Fritz, A. C. Shurtleff, J. B. Geisbert, R. Liebscher, A. Grolla, U. Stroher, L. Fernando, K. M. Daddario, M. C. Guttieri, B. R. Mothe, T. Larsen, L. E. Hensley, P. B. Jahrling, and H. Feldmann. 2005. Development of a new vaccine for the prevention of Lassa fever. PLOS Med. 2:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulukota, K., J. Sidney, A. Sette, and C. DeLisi. 1997. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 267:1258-1267. [DOI] [PubMed] [Google Scholar]
- 18.Lukashevich, I. S., J. Patterson, R. Carrion, D. Moshkoff, A. Ticer, J. Zapata, K. Brasky, R. Geiger, G. B. Hubbard, J. Bryant, and M. S. Salvato. 2005. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J. Virol. 79:13934-13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormick, J. B. 1986. Clinical, epidemiologic, and therapeutic aspects of Lassa fever. Med. Microbiol. Immunol. 175:153-155. [DOI] [PubMed] [Google Scholar]
- 20.McCormick, J. B., and S. P. Fisher-Hoch. 2002. Lassa fever. Curr. Top. Microbiol. Immunol. 262:75-109. [DOI] [PubMed] [Google Scholar]
- 21.McCormick, J. B., I. J. King, P. A. Webb, C. L. Scribner, R. B. Craven, K. M. Johnson, L. H. Elliott, and R. Belmont-Williams. 1986. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 314:20-26. [DOI] [PubMed] [Google Scholar]
- 22.Morrison, H. G., S. P. Bauer, J. V. Lange, J. J. Esposito, J. B. McCormick, and D. D. Auperin. 1989. Protection of guinea pigs from Lassa fever by vaccinia virus recombinants expressing the nucleoprotein or the envelope glycoproteins of Lassa virus. Virology 171:179-188. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen, M., C. Lundegaard, O. Lund, and C. Kesmir. 2005. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33-41. [DOI] [PubMed] [Google Scholar]
- 24.Oldstone, M. B. A., H. Lewicki, D. Homann, C. Nguyen, S. Julien, and J. E. Gairin. 2001. Common antiviral cytotoxic T-lymphocyte epitope for diverse arenaviruses. J. Virol. 75:6273-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, C. J., P. B. Jahrling, C. T. Liu, R. H. Kenyon, K. T. McKee, Jr., and J. G. Barrera Oro. 1987. Experimental studies of arenaviral hemorrhagic fevers. Curr. Top. Microbiol. Immunol. 134:5-68. [DOI] [PubMed] [Google Scholar]
- 26.Plebanski, M., K. L. Flanagan, E. A. Lee, W. H. Reece, K. Hart, C. Gelder, G. Gillespie, M. Pinder, and A. V. Hill. 1999. Interleukin 10-mediated immunosuppression by a variant CD4 T cell epitope of Plasmodium falciparum. Immunity 10:651-660. [DOI] [PubMed] [Google Scholar]
- 27.Price, M. E., S. P. Fisher-Hoch, R. B. Craven, and J. B. McCormick. 1988. A prospective study of maternal and fetal outcome in acute Lassa fever infection during pregnancy. BMJ 297:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Carreno, M. P., M. S. Nelson, J. Botten, K. Smith-Nixon, M. J. Buchmeier, and J. L. Whitton. 2005. Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology 335:87-98. [DOI] [PubMed] [Google Scholar]
- 29.Ruo, S. L., S. W. Mitchell, M. P. Kiley, L. F. Roumillat, S. P. Fisher-Hoch, and J. B. McCormick. 1991. Antigenic relatedness between arenaviruses defined at the epitope level by monoclonal antibodies. J. Gen. Virol. 72:549-555. [DOI] [PubMed] [Google Scholar]
- 30.Sette, A., and J. Fikes. 2003. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr. Opin. Immunol. 15:461-470. [DOI] [PubMed] [Google Scholar]
- 31.Sette, A., B. Livingston, D. McKinney, E. Appella, J. Fikes, J. Sidney, M. Newman, and R. Chesnut. 2001. The development of multi-epitope vaccines: epitope identification, vaccine design and clinical evaluation. Biologicals 29:271-276. [DOI] [PubMed] [Google Scholar]
- 32.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201-212. [DOI] [PubMed] [Google Scholar]
- 33.Sette, A., A. Vitiello, B. Reherman, P. Fowler, R. Nayersina, W. M. Kast, C. J. M. Melief, C. Oseroff, L. Yuan, J. Ruppert, J. Sidney, M. del Guercio, S. Southwood, R. T. Kubo, R. W. Chesnut, H. M. Grey, and F. V. Chisari. 1994. The relationship between class I binking affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153:5586-5592. [PubMed] [Google Scholar]
- 34.Sidney, J., S. Southwood, C. Oseroff, M. F. Del Guercio, A. Sette, and H. Grey. 1998. Measurement of MHC/peptide interactions by gel filtration. Curr. Protocols Immunol. 18:18.3.1-18.3.19. [DOI] [PubMed] [Google Scholar]
- 35.Snyder, J. T., I. M. Belyakov, A. Dzutsev, F. Lemonnier, and J. A. Berzofsky. 2004. Protection against lethal vaccinia virus challenge in HLA-A2 transgenic mice by immunization with a single CD8+ T-cell peptide epitope of vaccinia and variola viruses. J. Virol. 78:7052-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southern, P. J. 1996. Arenaviridae: the viruses and their replication, p. 1505-1519. In B. N. Fields, D. M. Knipe, and P. M. Howley, et al (ed.), Fields Virology. Lippincott-Raven, Philadelphia, Pa.
- 37.Tangri, S., G. Y. Ishioka, X. Huang, J. Sidney, S. Southwood, J. Fikes, and A. Sette. 2001. Structural features of peptide analogs of human histocompatibility leukocyte antigen class I epitopes that are more potent and immunogenic than wild-type peptide. J. Exp. Med. 194:833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenzer, S., B. Peters, S. Bulik, O. Schoor, C. Lemmel, M. M. Schatz, P. M. Kloetzel, H. G. Rammensee, H. Schild, and H. G. Holzhutter. 2005. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell. Mol. Life Sci. 62:1025-1037. [DOI] [PubMed] [Google Scholar]
- 39.ter Meulen, J., M. Badusche, K. Kuhnt, A. Doetze, J. Satoguina, T. Marti, C. Loeliger, K. Koulemou, L. Koivogui, H. Schmitz, B. Fleischer, and A. Hoerauf. 2000. Characterization of human CD4+ T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J. Virol. 74:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ter Meulen, J., M. Badusche, J. Satoguina, T. Strecker, O. Lenz, C. Loeliger, M. Sakho, K. Koulemou, L. Koivogui, and A. Hoerauf. 2004. Old and New World arenaviruses share a highly conserved epitope in the fusion domain of the glycoprotein 2, which is recognized by Lassa virus-specific human CD4+ T-cell clones. Virology 321:134-143. [DOI] [PubMed] [Google Scholar]
- 41.U.S. Department of Health and Human Services. 1999. Biosafety in microbiological and biomedical laboratories, 4th ed. U.S. Government Printing Office, Washington, D.C.
- 42.van der Most, R. G., K. Murali-Krishna, J. L. Whitton, C. Oseroff, J. Alexander, S. Southwood, J. Sidney, R. W. Chesnut, A. Sette, and R. Ahmed. 1998. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology 240:158-167. [DOI] [PubMed] [Google Scholar]
- 43.Vitiello, A., D. Marchesini, J. Furze, L. A. Sherman, and R. W. Chesnut. 1991. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J. Exp. Med. 173:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, F., M. Yang, and D. D. Eckels. 2005. Interactions between helper T-cell epitopes of hepatitis C virus. Vaccine 23:3572-3580. [DOI] [PubMed] [Google Scholar]