Abstract
Borna disease virus infection of neonatal rats results in a characteristic behavioral syndrome and apoptosis of subsets of neurons in the hippocampus and cerebellum (neonatal Borna disease [NBD]). The cellular mechanisms leading to neurodevelopmental damage in NBD have not been fully elucidated. Insights into this model may have general implications for understanding the pathogenesis of virus-associated neurodevelopmental damage. Here we report the presence of endoplasmic reticulum (ER) stress markers and activation of the unfolded protein response in the NBD hippocampus and cerebellum. Specific findings included enhanced PERK-mediated phosphorylation of eif2α and concomitant regulation of ATF4 translation; IRE1-mediated splicing of XBP1 mRNA; and cleavage of the ATF6 protein in NBD rat brains. We found evidence for regional and cell type-specific divergence in the expression of ER stress-induced proapoptotic and quality control signals. Our results demonstrate that ER stress induction in death-susceptible Purkinje neurons in NBD is associated with the expression of the proapoptotic molecule CHOP in the absence of compensatory expression of the ER quality control molecules Bip and protein disulfide isomerase. In contrast, ER stress in death-resistant astrocytes is associated with complementary expression of CHOP and ER quality control signals. These results implicate an imbalance between ER stress-mediated apoptosis and survival signaling as a critical determinant of neural cell fate in NBD.
Borna disease virus (BDV) is a nonsegmented, negative-sense, single-stranded RNA virus belonging to the virus family Bornaviridae within the order Mononegavirales (5, 11). Experimental infection of adult immunocompetent Lewis rats results in a marked CD8+ immune response (meningoencephalitis) and a progressive movement disorder (20). In contrast, newborn rats infected with BDV (neonatal Borna disease [NBD]) do not mount an overt cellular immune response yet have prominent astrogliosis and microgliosis, altered cytokine, neurotrophic factor, and neurotrophic factor receptor gene expression, abnormal development of brain monoaminergic systems, cerebellar and hippocampal dysgenesis, and disturbances of learning, mood, and behavior (8, 25, 47, 54). Although BDV is noncytolytic, NBD is attended by apoptotic degeneration of neurons in the cerebellum (CBLM) and hippocampus (HC) that undergo substantial postnatal maturation (25, 72). However, the cellular and molecular mechanisms leading to neurodegeneration in NBD remain unclear.
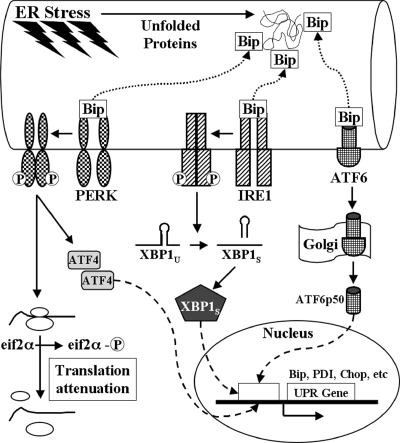
The endoplasmic reticulum (ER) plays a vital role in a variety of cellular functions, including synthesis, folding, and posttranslational modification of secretory and membrane proteins, cytoplasmic and mitochondrial metabolism, homeostasis of intracellular calcium, and apoptosis. The ER is sensitive to nutrient or energy deprivation, disruption of calcium homeostasis, inhibition of N-linked glycosylation, oxidative stress, and accumulation of unfolded, misfolded, or excessive protein (61, 69). Perturbations causing ER stress result in accumulation of unfolded proteins in the ER lumen and initiation of a complex signal transduction cascade, known as the unfolded protein response (UPR) (Fig. 1). The UPR is activated to restore the ER to its normal physiological state by two adaptive mechanisms: (i) increasing the folding capacity of the ER through induction of molecular chaperones and foldases and (ii) decreasing protein burden on the ER through global inhibition of translation and enhancing degradation and clearance of unfolded proteins. Essential to this quality control mechanism is the ATP-dependent ER chaperone, Bip (immunoglobulin heavy chain-binding protein). In the physiological state, Bip binds to and suppresses the activity of three proximal, ER-resident, transmembrane, UPR transducers: PKR-like ER kinase (PERK), inositol-requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6).
FIG. 1.
Schematic illustration of ER stress and UPR activation. ER stresses can result in accumulation of unfolded proteins in the ER. Unfolded proteins are bound by Bip, resulting in release and activation of three ER-resident transmembrane transducers of the UPR: PERK, IRE1, and ATF6. Activated PERK phosphorylates eif2α, attenuating global protein synthesis, and enhances translation of ATF4. Activated IRE1 facilitates splicing of XBP1U, resulting in translation of the transcriptionally active form of XBP1 (XBP1S). Following Bip release, ATF6 translocates to the Golgi, where it is cleaved by proteases, releasing the transcriptionally active form of ATF6 (ATF6p50). ATF4, XBP1S, and ATF6p50 enter the nucleus, where they activate the expression of a distinct set of UPR target genes.
During ER stress, Bip binds misfolded proteins and is thereby sequestered from PERK, IRE1, and ATF6 (3). When released from Bip, the kinase domain of activated PERK phosphorylates Ser51 of the alpha subunit of eukaryotic translation initiation factor 2 (eif2α), thereby attenuating global translation initiation and protein synthesis (17). Paradoxically, PERK-mediated phosphorylation of eif2α preferentially induces translation of activating transcription factor 4 (ATF4) (18). Following Bip release, IRE1 undergoes dimerization and trans-autophosphorylation, activating its endoribonuclease activity; IRE1 then mediates the excision of a 26-nucleotide intron from X-box binding protein 1 (XBP1), resulting in a translational frameshift and formation of a potent transcriptional activator (78). Bip-free ATF6 is transported to the Golgi, where it is cleaved by site-1 and site-2 proteases, generating a 50-kDa bZIP transcription factor (ATF6p50) that migrates to the nucleus (22, 62, 75). The downstream nuclear effectors of the UPR (ATF4, spliced XBP1, and ATF6p50) regulate the expression of genes that enhance the protein folding and degrading capacity of the ER, regulate amino acid metabolism, mediate resistance to oxidative stress, and induce apoptosis (19, 35, 38).
The ER elicits apoptotic signals when protein load exceeds its folding capacity. C/EBP homologous protein (CHOP), also known as GADD153 (growth arrest- and DNA damage-inducible gene 153), is induced by ER stress. Overexpression of CHOP triggers cell cycle arrest and apoptosis, down-regulates the prosurvival molecule Bcl-2, and promotes the production of reactive oxygen species (1, 44-46, 49). In contrast, overexpression of the ER chaperone Bip reduces CHOP induction associated with ER stress and attenuates apoptosis (71). Thus, the cellular fate with ER stress reflects the coordinated interaction of quality control functions and proapoptotic signals. ER stress induction and regulation have been reported following in vitro infections by several viruses, including hepatitis C virus (HCV), Japanese encephalitis virus, bovine viral diarrhea virus, respiratory syncytial virus, human cytomegalovirus (HCMV), and Tula hantavirus (4, 26, 27, 39, 63, 65).
Here we demonstrate that BDV infection of newborn rats results in neural and astrocytic ER stress, induces the UPR, and results in a neural cell-specific imbalance between proapoptotic and survival signaling.
MATERIALS AND METHODS
Animals and virus inoculation.
Lewis rat dams were obtained from Charles River Laboratories (Wilmington, MA). Within 12 h of birth, 50 Lewis rat pups were inoculated into the right cerebral hemisphere with 50 μl of 5 × 103 tissue culture infectious units of BDV strain He/80-1 (NBD) or phosphate-buffered saline (control: sham inoculated). Rats were sacrificed at postnatal days 21, 28, and 84 (PND21, PND28, and PND84) for nucleic acid, protein, and anatomic analyses.
RNA extraction.
At PND28 postinoculation, NBD (n = 7) and control (n = 5) rats were terminally anesthetized with CO2. CBLM and HC were immediately dissected, snap frozen in TRIzol (Invitrogen, Carlsbad, CA), and stored at −80°C. Following extraction using standard protocols, RNA was quantitated by UV spectrophotometry.
DNA microarray analysis.
Equal amounts of total RNA extracted from either CBLM or HC of individual PND28 animals were pooled to create a single control RNA sample (n = 5 control rats) and an NBD RNA pool (n = 7 NBD rats) for use in DNA microarray assays. Indirect labeling and cDNA synthesis were performed using 5 μg of pooled RNA and the 3DNA Array 350 expression array detection kit for microarrays (Genisphere, Hatfield, PA). Cy3 and Cy5 capture sequence-labeled products were hybridized to a rat 8,000-gene oligonucleotide array (Public Health Research Institute, Newark, NJ) in sodium dodecyl sulfate (SDS)-based hybridization buffer containing denatured human Cot-1 (1 μg/40 μl; Invitrogen/Gibco LTI, Gaithersburg, MD) for 18 h at 53°C. Posthybridization washes were followed by an additional hybridization with fluorescent Cy3-3DNA and Cy5-3DNA capture reagents in SDS hybridization buffer for 3 h at 62°C, followed by sequential washes. Fluorescence-labeled slides were dried by centrifugation and scanned with a GenePix 4000B scanner (Axon Instruments, Inc., Union City, CA) at 5-μm resolution. For each spot, background-subtracted signal intensity, acquired using GenePix Pro 4.1 software, was normalized for both Cy3 and Cy5 expression by scaling individual spot intensities in each channel to the total intensity of all spots on the array. Experiments using CBLM or HC RNA sets were performed in triplicate. Data were paired for each array and analyzed using the Cyber T web interface (http://visitor.ics.uci.edu/genex/cybert/). Differences in gene hybridization signal that met the following criteria were considered to be significant: n-fold change greater than 1.5 or less than −1.5 and P value of <0.05 (paired t test).
Quantitative SYBR Green real-time PCR.
Intron/exon spanning, gene-specific PCR primers specific for rat CHOP and porphobilinogen deaminase (PBGD) as a housekeeping gene control were designed for real-time PCR using Primer Express 1.0 software (Applied Biosystems, Foster City, CA). Published primers were used for XBP1 and Bip assays (Table 1) (36, 51). PCR standards for determining copy numbers of target transcripts were cloned into the vector pGEM-T easy (Promega Corporation, Madison, WI). Linearized plasmids were quantitated by UV spectroscopy, and 10-fold serial dilutions were created in water containing yeast tRNA (1 ng/μl). RNA from CBLM or HC of individual animals was used for real-time PCR assays. cDNA was synthesized using Taqman reverse transcription reagents (Applied Biosystems) from 2 μg unpooled RNA per 100 μl reaction mixture from either the CBLM or HC of each of seven NBD rats and five control rats; each sample was assayed in triplicate. Each 25-μl amplification reaction mixture contained 10 μl template cDNA, 12.5 μl SYBR Green Master Mix (Applied Biosystems), and 300 nm gene-specific primers (Table 1). The thermal cycling profile using a Model 7700 sequence detector system (Applied Biosystems) consisted of the following: stage 1, 1 cycle at 50°C for 2 min; stage 2, 1 cycle at 95°C for 10 min; stage 3, 45 cycles at 95°C for 15 s and 60°C for 1 min (for CHOP amplification); stage 1, 1 cycle at 50°C for 2 min; stage 2, 1 cycle at 95°C for 10 min; stage 3, 45 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s (for Bip amplification); and stage 1, 1 cycle at 50°C for 2 min; stage 2, 1 cycle at 95°C for 10 min; stage 3, 45 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min (for XBP1 amplification). A PBGD fragment was amplified in triplicate reactions by real-time PCR on the same plate as the gene of interest. The mean concentration of PBGD in each sample was used to control for integrity of input RNA and to normalize values of target gene expression to those of the housekeeping gene expression. The final results were expressed as the mean number of copies per 200 ng total RNA for CHOP, Bip, or XBP1, relative to values obtained for PBGD RNA.
TABLE 1.
Primers used in this work
| Gene (accession no.) | Primer pair (5′-3′) (Reaction concn [nm])a | Amplicon size (bp) |
|---|---|---|
| CHOP (U36994) | For: GAAAGCAGAAACCGGTCCAAT (300) | 150 |
| Rev: GGATGAGATATAGGTGCCCCC (300) | ||
| XBP1 (BC079450) | For: AAACAGAGTAGCAGCGCAGACTGC (300) | 601 |
| Rev: GGATCTCTAAAACTAGAGGCTTGGTG (300) | ||
| Bip (M14050) | For: CTGGGTACATTTGATCTGACTGG (300) | 397 |
| Rev: GCATCCTGGTGGCTTTCCAGCCATTC (300) | ||
| PBGD (X06827) | For: ATTCGGGGAAACCTCAACACC (300) | 157 |
| Rev: CTGACCCACAGCATACATGCAT (300) |
For, forward; Rev, reverse.
Western blot analysis.
Individual CBLM and HC were dissected from PND28 NBD (n = 4) and control (n = 4) rats and homogenized in ice-cold hypotonic cell lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA) containing protease inhibitors (Complete Mini, EDTA-free tablets; Roche Molecular Biochemicals, Indianapolis, IN) and the serine-threonine phosphatase inhibitor β-glycerophosphate (17.5 mM, EMD Biosciences, San Diego, CA) and incubated on ice for 15 min. Homogenates were centrifuged at 8,000 × g for 15 min at 4°C. Supernatants containing cytoplasmic proteins were collected, and protein concentrations estimated by Bradford assay (Bio-Rad, Hercules, CA). Cell pellets were washed five times with cell lysis buffer and incubated for 30 min on ice with nuclear lysis buffer (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1 mM dithiothreitol) containing protease and phosphatase inhibitors, followed by centrifugation for 20 min at 15,000 × g. Cleared supernatants containing nuclear extracts were collected and protein concentration estimated by Bradford assay (Bio-Rad). Cytoplasmic or nuclear proteins from regional brain homogenate (20 μg) in sample buffer (10 mM Tris-Hcl, pH 7.5, 10 mM EDTA, 20% [vol/vol] glycerol, 1% [wt/vol] SDS, 0.005% [wt/vol] bromphenol blue, 100 mM dithiothreitol) were boiled for 5 min and size fractionated by 10% SDS-polyacrylamide gel electrophoresis. SYPRO Ruby staining of cytoplasmic proteins was carried out according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Proteins were transferred to nitrocellulose membranes using a semidry blotting apparatus (Owl Separation Systems, Portsmouth, NH). Membranes were blocked in 2% nonfat milk powder (for CHOP, ATF4, ATF6, XBP1, and glyceraldehyde phosphate-3-dehydrogenase [GAPDH]) or 2% bovine serum albumin (for phospho-eif2α) in TTBS (20 mM Tris-Hcl, pH 7.6, 137 mM NaCl, 0.1% Tween 20) overnight at room temperature and incubated with mouse monoclonal anti-CHOP (1:100; Santa Cruz Biotechnology), mouse monoclonal anti-PDI (1:500; Affinity BioReagents, Golden, CO), rabbit anti-XBP1 (1:100; M-186; Santa Cruz Biotechnology), mouse monoclonal anti-ATF6 (1:200; IMGENEX, San Diego, CA), or rabbit anti-ATF4 (1:50; C-20; Santa Cruz Biotechnology) in TTBS with 1% nonfat milk or rabbit monoclonal anti-phospho-eif2α (Ser51) (1:300; Cell Signaling Technology, Danvers, MA) in TTBS with 2% bovine serum albumin for 2 h at room temperature. Membranes were washed three times for 10 min each with TTBS prior to incubation with peroxidase-conjugated goat antimouse immunoglobulin G (IgG) (1:2,000; Bio-Rad) or goat antirabbit IgG (1:2,000; Bio-Rad) in TTBS with 1% nonfat dry milk for 1 h at room temperature. Membranes were developed using the ECL Western blot detection system (Amersham Biosciences, Arlington Heights, IL) and scanned for chemiluminescence using a Storm 840 imager (Molecular Dynamics, Sunnyvale, CA). Blots were stripped and reprobed with mouse anti-GAPDH monoclonal antibody (Ambion, Austin, TX), as a housekeeping gene, for normalization of cytoplasmic proteins. Protein bands were quantified using Image Quant software (v.1.0; Molecular Dynamics).
XBP1 RT-PCR splicing analysis.
ER stress-induced processing of XBP1 mRNA was evaluated by PCR and restriction site analysis. Total RNA (2 μg) extracts from CBLM and HC of individual PND28 NBD (n = 7) and control (n = 5) rats were reverse transcribed using MultiScribe reverse transcriptase (RT) and random hexamer primers (Applied Biosystems, Foster City, CA). The 601-bp PCR product, encompassing the IRE1 cleavage site of XBP1, was amplified using the following XBP1 primers: upper strand primer, 5′-AAACAGAGTAGCAGCGCAGACTGC-3′; and lower strand primer, 5′-GGATCTCTAAAACTAGAGGCTTGGTG-3′. The thermal cycling profile consisted of 30 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. PCR products were either directly resolved on a 2% agarose gel or incubated with PstI at 37°C for 5 h followed by separation on 2% agarose gels. Bands representing spliced (XBP1S) and unspliced (XBP1U) forms of XBP1 from seven NBD rats were extracted from agarose and subcloned into pGEM-T Easy Vector (Promega Corporation) for sequencing.
Histological analysis and immunofluorescence.
Under CO2 anesthesia, PND21, PND28, and PND84 NBD (n = 5/age group) and control (n = 4/age group) rats were perfused via left ventricular puncture with phosphate-buffered saline (1 ml/g body weight), followed by buffered 4% paraformaldehyde (1 ml/g body weight). Brains were postfixed in 4% paraformaldehyde overnight at 4°C and cryoprotected with graded sucrose solutions. Cryostat sections (14 μm) were collected onto glass slides (Super Frost Plus; Fisher Scientific, Pittsburgh, PA). Tissue sections were stained with hematoxylin and eosin (H&E) for histological analysis. Immunofluorescence microscopy was carried out as previously described (74), using the following primary antibodies: rabbit anti-BDVp40 (nucleoprotein; 1:1500) (6), mouse monoclonal anti-CHOP (1:50; Santa Cruz), rabbit anti-P-eif2α (Ser51) monoclonal antibody (1:50; Cell Signaling), rabbit anti-ATF4 (1:50; Santa Cruz), rabbit anti-Bip (1:50; Santa Cruz), mouse monoclonal anti-PDI (1:500; Affinity BioReagents), rabbit anti-glial fibrillary acidic protein (GFAP) antibody (1:100; DAKO Cytomation, Carpinteria, CA), mouse anti-GFAP monoclonal antibody cocktail (1:30; BD Pharmingen, San Diego, CA), rabbit anti-calbindin D-28K (1:100; Chemicon, Temecula, CA). Secondary antibodies were Cy3-conjugated antimouse or antirabbit IgG (1:200; Jackson Immunoresearch Laboratories, Inc., West Grove, PA) and/or Cy2-conjugated antimouse or antirabbit IgG (1:200; Jackson Immunoresearch).
Statistical analysis.
The significance of observed differences among NBD and control groups was assessed by paired t test for microarray experiments or analysis of variance (ANOVA) for real-time PCR and Western immunoblot analysis. Analysis was carried out using StatView software (v.5.0.1; SAS Institute Inc., Cary, NC). Values were considered to be significant when P values were <0.05.
RESULTS
Microarray analysis reveals differential expression of ER stress genes in NBD.
Intracranial inoculation of Lewis rats with BDV within the first 24 h of life results in persistent infection, behavioral deficits, and progressive apoptosis of neurons that undergo postnatal maturation. To identify specific molecular and cellular pathways that mediate these effects, we profiled mRNA expression in brains of PND28 rats, a time point that corresponds with peak histologic evidence of apoptotic neural damage in CBLM and HC (25, 72).
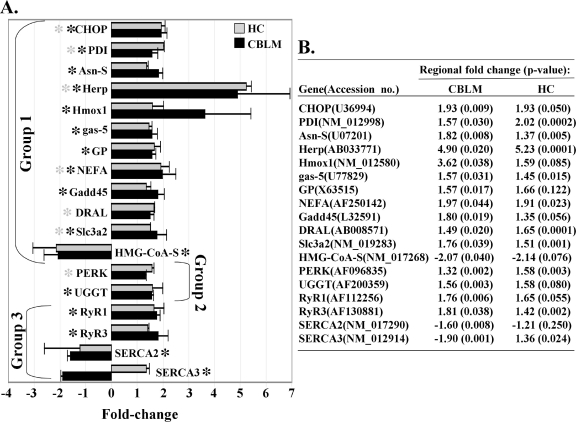
Microarray analysis of mRNA extracted from the CBLM and HC of PND28 NBD and control rats revealed altered mRNA levels for 11 genes previously reported to be induced by ER stress and one ER stress-repressed gene (Fig. 2A) (hereafter referenced as group 1 genes) (19, 35, 48, 56, 60). In some instances, genes met microarray criteria in either CBLM or HC but not both; where degrees of difference were modest, a trend toward altered gene expression was evident in both regions. Of the group 1 genes, five met microarray criteria in both NBD CBLM and HC (CHOP, PDI, Herp [homocysteine-induced ER protein], NEFA [novel DNA-binding/EF-hand/leucine zipper protein], and Slc3a2 [4F2 antigen heavy chain-amino acid transporter]); six specifically met criteria in CBLM (Asn-S [asparagine synthetase], Hmox1 [heme oxygenase], gas-5 [growth arrest-specific 5], GP [glycogen phosphorylase], Gadd45 [growth arrest and DNA-damage-inducible 45], and HMG-CoA-S [3-hydroxy-3-methylglutaryl-coenzyme A synthase]); and only one gene specifically met criteria in the HC (DRAL [down-regulated in rhabdomyasarcoma LIM]). Microarray experiments also identified alterations in genes involved in UPR activation (PERK) or reglucosylation of nonnative glycoproteins in the ER (UGGT [UDP-glucose-glycoprotein glucosyltransferase]) (Fig. 2A) (hereafter referenced as group 2 genes) and ER-resident calcium transporters (RyR1 [ryanodine receptor type 1], RyR3 [ryanodine receptor type 3], SERCA2 [sarcoplasmic reticulum Ca2+-ATPase 2], and SERCA3 [sarcoplasmic reticulum Ca2+-ATPase 3]) (Fig. 2A; hereafter referenced as group 3 genes) (58, 69). Interestingly, all four calcium transporters specifically met microarray criteria in the NBD CBLM, with increased expression of RyR1 and RyR3 and decreased expression of SERCA2 and SERCA3. n-fold change and P values for these genes are given for comparison (Fig. 2B).
FIG. 2.
Identification of ER stress genes in PND28 NBD rats by microarray analysis. Microarray analysis was conducted using pooled total RNA isolated from dissected CBLM and HC of PND28 NBD (n = 7) and PND28 control (n = 5) rats. (A) Results represent mean n-fold change ± standard deviation for genes from three independent experiments comparing RNA from NBD CBLM versus control CBLM (black bars) and mean n-fold change ± standard deviation for genes in NBD HC (gray bars) versus control HC. Genes were considered significant when n-fold induction or repression in NBD rats was >1.5 or <−1.5 and the P value was <0.05 compared to control results. Black asterisks denote genes meeting these criteria in NBD CBLM; gray asterisks denote genes meeting these criteria in NBD HC. Genes are grouped into known ER stress-responsive genes (group 1), genes involved in ER stress response (group 2), and regulators of ER calcium homeostasis (group 3). (B) Table showing n-fold induction or fold repression and P values for ER stress genes (accession no. provided) for NBD rat CBLM and HC. For gene abbreviations, see the text.
Phosphorylation of eif2α in NBD rat brain and increased translation of ATF4.
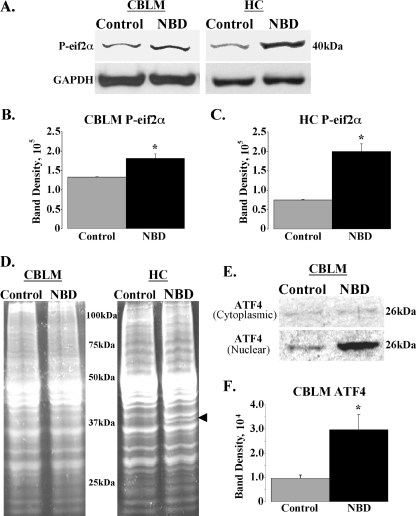
ER stress in mammalian cells leads to the activation of PERK. Activated PERK inhibits general protein biosynthesis through phosphorylation of eif2α, limiting further protein overload in the ER (17). Activation of the PERK pathway in PND28 NBD rats was assessed in Western blots of cytoplasmic extracts of CBLM and HC using antibodies that bind the phosphorylated form of eif2α (P-eif2α) and GAPDH (loading control). P-eif2α antibodies detected a single band of 40 kDa corresponding to the expected size of P-eif2α in both control and NBD CBLM and HC extracts (Fig. 3A). Quantitation of P-eif2α band densities normalized to GAPDH revealed a 1.36-fold increase in P-eif2α in NBD CBLM (ANOVA, P = 0.0136) (Fig. 3B). Levels of P-eif2α were increased 2.67-fold in NBD HC (ANOVA, P = 0.0007) (Fig. 3C).
FIG. 3.
Activation of the PERK/eif2α/ATF4 arm of the UPR in NBD rats. (A) Cytoplasmic lysates (20 μg) from CBLM and HC of control and NBD rats were evaluated by Western analysis using antibodies specific for the phosphorylated form of eif2α (P-eif2α; upper panels) relative to levels of GAPDH (housekeeping gene; lower panels). (B and C) Determination of P-eif2α band density relative to that for GAPDH from control (n = 4) or NBD (n = 4) rat CBLM (B) (1.36-fold increase in NBD; ANOVA, P = 0.0136) or HC (C) (2.67-fold increase in NBD; ANOVA, P = 0.0007). (D) Sypro Ruby stain of protein extract (20 μg) from CBLM and HC of control and NBD rats. Note that relative levels of proteins are similar between control and NBD rats in both brain regions, with the exception of a single band in NBD lysates from HC corresponding to the BDV nucleoprotein (arrowhead). (E) Cytoplasmic (upper panel) and nuclear (lower panel) lysates from CBLM of control and NBD rats were evaluated by Western analysis using antibodies specific for ATF4. Note equal amounts of ATF4 protein in cytoplasmic lysates and higher ATF4 band signal in nuclear extracts from NBD rats compared to results for controls. (F) Determination of ATF4 band density from control (n = 4) or NBD (n = 4) nuclear protein extracts revealed increased ATF4 levels in NBD CBLM (3.06-fold increase in NBD; ANOVA, P = 0.0223). Asterisks indicate P values of <0.05.
Phosphorylation of eif2α prevents the formation of the ternary translation initiation complex (eif2/GTP/Met-tRNAiMet), thereby attenuating global protein synthesis (17). We assessed total protein levels in CBLM and HC from control and NBD rats using SYPRO Ruby staining of SDS-polyacrylamide gel electrophoresis gels loaded with equal amounts of cytoplasmic protein extracts. No differences were detected between NBD and controls in either CBLM or HC except for the presence of a protein band in NBD subsequently identified by Western blot analysis as the BDV nucleoprotein (Fig. 3D, arrowhead).
PERK-mediated phosphorylation of eif2α inhibits general protein translation but selectively promotes translation of ATF4 mRNA during ER stress. In unstressed cells, upstream open reading frames in the 5′ untranslated region of ATF4 mRNA repress the translation of ATF4. During ER stress, eif2α phosphorylation allows ribosomal bypass scanning of the ATF4 mRNA upstream open reading frames, resulting in efficient ATF4 translation (18). ATF4 activates the transcription of genes involved in amino acid metabolism and transport (i.e., genes encoding Asn-S and Slc3a2), oxidative stress responses (i.e., that encoding Hmox-1), and ER stress-induced apoptosis (i.e., that encoding CHOP) (19). Given microarray results indicating increased levels of transcripts corresponding to ATF4-regulated genes in NBD, we assessed levels of the ATF4 protein in cytoplasmic and nuclear extracts of PND28 NBD and control CBLM and HC. ATF4 (26 kDa) levels were 3.06-fold higher in nuclear extracts of NBD CBLM (ANOVA, P = 0.0223) (Fig. 3E and F). No significant differences in ATF4 were observed between NBD and the controls in cytoplasmic or nuclear extracts from HC (data not shown).
XBP1 splicing in NBD rats.
ER stress results in Bip release from IRE1 and activation of IRE1 RNase activity. XBP1 mRNA serves as a specific substrate for IRE1 RNase (78). Thus, splicing of XBP1 is commonly used as a marker of IRE1 activation. We assessed the activation state of IRE1 by investigating XBP1 mRNA splicing and expression, as well as XBP1 protein expression, in PND28 NBD and control rats.
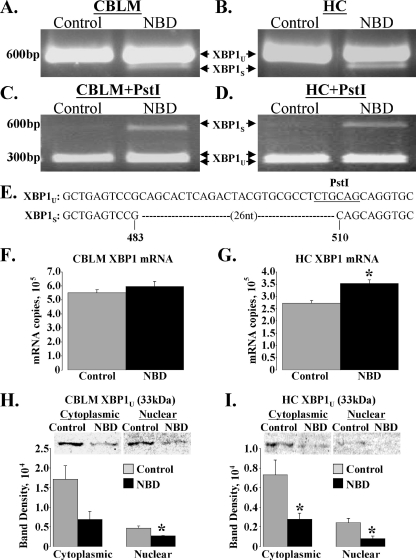
XBP1 splicing was evaluated by RT-PCR using primers that amplify products spanning the 26-nucleotide excision site. Agarose gel analysis of XBP1 cDNA amplified from CBLM (Fig. 4A) and HC (Fig. 4B) mRNA of individual controls (n = 5) revealed a single, abundant product of approximately 601 bp, the expected amplicon size of unspliced XBP1 (XBP1U). In NBD, an additional lower-molecular-weight band of approximately 575 bp (the expected size of XBP1S) was observed in CBLM and HC (Fig. 4A and B, respectively). Splicing of XBP1 disrupts a PstI restriction site localized within the 26-nucleotide excision site. Thus, PstI digestion is expected to completely cut XBP1U into two fragments of 312 bp and 289 bp, while XBP1S should be resistant to digestion. As expected, XBP1 PCR products were completely digested by PstI in CBLM and HC of all controls (Fig. 4C and D, respectively) while XBP1S was resistant to PstI digestion in all NBD rats (Fig. 4C and D). Sequencing of gel-extracted PCR fragments from NBD and control rats confirmed that the PCR products represented the expected sequences for the XBP1U and XBP1S forms of XBP1 (Fig. 4E).
FIG. 4.
XBP1 mRNA is selectively spliced in NBD rats. RNA extracted from CBLM and HC of PND28 control (n = 5) and NBD (n = 7) rats was analyzed by RT-PCR using primers that amplify both unspliced XBP1 (XBP1U) and spliced (XBP1S) XBP1. (A and B) Representative gel images of XBP1 RT-PCR. Note detection of XBP1U in control rats and both XBP1U and XBP1S in NBD rat CBLM (A) or HC (B). (C and D) PstI digestion of XBP1 RT-PCR products. Note PstI-insensitive XBP1S band in NBD but not control rat CBLM (C) and HC (D). (E) Sequencing of XBP1U and XBP1S from CBLM and HC confirms excision of the 26-nucleotide intron from XBP1S in NBD rats. (F and G) SYBR Green real-time PCR analysis of total XBP1 mRNA in CBLM and HC of control (n = 5) and NBD (n = 7) rats. Note that XBP1 mRNA expression levels are unchanged in NBD CBLM (F) but are significantly increased in NBD HC (G: 1.3-fold increase in NBD; ANOVA, P = 0.005). (H and I) Western blot analysis of XBP1U (33 kDa) protein levels in CBLM and HC cytoplasmic and nuclear extracts from control (n = 4) and NBD (n = 4) rats. Representative blots are shown. (H) Relative band densities for CBLM extracts revealed a trend toward decreased XBP1U protein in cytoplasmic extracts (2.49-fold decrease in NBD; ANOVA, P = 0.065) and a significant decrease in nuclear extracts (1.72-fold decrease in NBD; ANOVA, P = 0.017) from NBD rats compared to controls. (I) Relative band density for HC extracts revealed significant decreases in XBP1U protein in both cytoplasmic (2.63-fold decrease in NBD; ANOVA, P = 0.033) and nuclear (3.02-fold decrease in NBD; ANOVA, P = 0.018) extracts from NBD rats compared to results for controls. Asterisks indicate P values of <0.05.
XBP1 mRNA levels are also induced during ER stress through an ER stress response element (ERSE) in the XBP1 promoter (77). To detect total XBP1 mRNA, we performed real-time PCR using primers that amplify both unspliced and spliced transcripts of XBP1 mRNA. We did not find differences in total XBP1 mRNA in NBD CBLM (Fig. 4F), but total XBP1 mRNA was increased in NBD HC (1.3-fold increase; ANOVA, P = 0.005) (Fig. 4G).
We assessed XBP1 protein levels in cytoplasmic and nuclear extracts from control and NBD CBLM and HC in Western blots using antibodies reactive with both constitutively expressed XBP1U (33 kDa) and XBP1S (54 kDa). In NBD CBLM there was a trend toward decreased XBP1U protein in the cytoplasm (2.49-fold decrease; ANOVA, P = 0.065) and a significant decrease in the nucleus (1.72-fold decrease; ANOVA, P = 0.017) (Fig. 4H). In NBD HC, XBP1U protein was significantly decreased in both the cytoplasm (2.63-fold decrease; ANOVA, P = 0.033) and the nucleus (3.02-fold decrease; ANOVA, P = 0.018) (Fig. 4I). XBP1S was not detected in any extract.
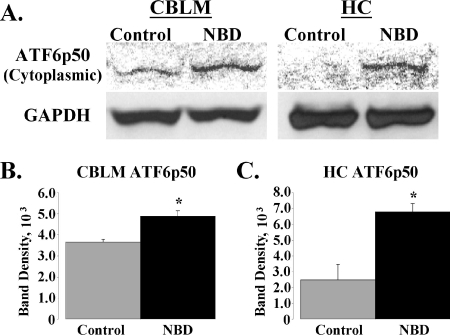
ATF6 cleavage in NBD rats.
Accumulation of unfolded or misfolded proteins in the ER results in release of ATF6 from Bip and permits ATF6 transport to the Golgi compartment. Cleavage of ATF6 by site-1 and site-2 proteases generates the activated cytosolic form of ATF6 (ATF6p50), which translocates to the nucleus to orchestrate transcriptional activation of genes encoding ER chaperones and folding enzymes (22, 48, 62, 75). We assessed the cleavage status of ATF6 in protein extracts from NBD CBLM and HC by Western blotting (Fig. 5A). ATF6p50 protein levels were significantly increased in NBD CBLM (Fig. 5B) (1.33-fold increase; ANOVA, P = 0.0073) and HC (Fig. 5C) (2.74-fold increase; ANOVA, P = 0.0084). ATF6p50 was not detectable in NBD or control CBLM or HC nuclear extracts (data not shown).
FIG. 5.
Enhanced cleavage of ATF6 in NBD rats. (A) Western immunoblots of ATF6 cleavage (ATF6p50) in cytoplasmic extracts from PND28 control and NBD CBLM (upper left panel) or HC (upper right panel). Corresponding signals for the GAPDH housekeeping gene are shown (lower panels). (B) ATF6p50 band density, relative to that for GAPDH, from control (n = 4) or NBD (n = 4) rat CBLM was significantly higher for NBD rats than for controls (1.33-fold increase for NBD; ANOVA, P = 0.0073). (C) ATF6p50 band density, relative to that for GAPDH, from control (n = 4) or NBD (n = 4) rat HC was significantly higher for NBD rats than for controls (2.74-fold increase in NBD; ANOVA, P = 0.0084). Asterisks indicate P values of <0.05.
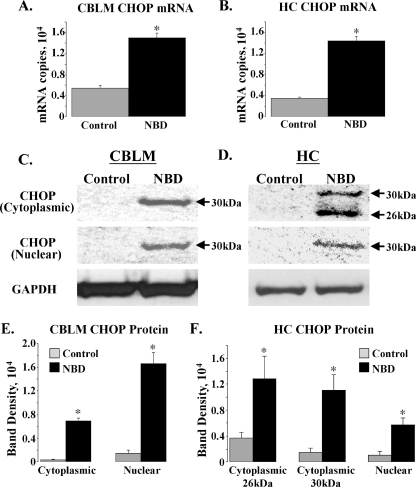
CHOP mRNA and protein expression in NBD rat brain.
The CHOP promoter contains cis-acting elements (ERSEs and amino acid regulatory elements) that regulate transcriptional activation. The downstream UPR effectors, ATF4, XBP1S, and ATF6p50, converge on elements in the CHOP promoter to induce CHOP transcription (49). SYBR Green real-time PCR confirmed increases in CHOP mRNA in PND28 NBD CBLM (Fig. 6A) (2.77-fold increase; ANOVA, P < 0.0001) and HC (Fig. 6B) (4.14-fold increase; ANOVA, P < 0.0001).
FIG. 6.
Increased CHOP mRNA and protein in NBD rats. (A and B) Real-time PCR analysis of CHOP mRNA in CBLM (A) and HC (B) of PND28 control (n = 5) or NBD (n = 7) rats. CHOP mRNA was significantly increased for NBD rats relative to results for control CBLM (A: 2.77-fold increase in NBD; ANOVA, P < 0.0001) or HC (B: 4.14-fold increase in NBD; ANOVA, P < 0.0001). (C and D) Representative Western immunoblots for CHOP protein in cytoplasmic (top panel) or nuclear (middle panel) extracts from control or NBD rat CBLM (C) or HC (D). Corresponding cytoplasmic immunoblot signals for GAPDH are shown (bottom panel). (E and F) Determination of CHOP protein band density in cytoplasmic and nuclear extracts from control (n = 4) or NBD (n = 4) rat CBLM (E) or HC (F). CBLM CHOP protein levels were significantly increased in both cytoplasmic (E: 21.4-fold increase in NBD; ANOVA, P < 0.0001) and nuclear (E: 11.34-fold increase in NBD; ANOVA, P = 0.0003) extracts from NBD rats, relative to results for controls. HC CHOP protein levels were significantly increased in both cytoplasmic (F: 26-kDa band, 3.48-fold increase in NBD; ANOVA, P = 0.043; and 30-kDa band, 7.49-fold increase in NBD; ANOVA, P = 0.009) and nuclear (F: 5.47-fold increase in NBD; ANOVA, P = 0.02) extracts from NBD rats, relative to controls. Asterisks indicate P values of <0.05.
To test whether increased CHOP mRNA correlated with increased CHOP protein levels and to assess distribution in cellular compartments, cytoplasmic and nuclear extracts from NBD and control CBLM and HC were analyzed by Western blotting. CHOP-specific antibodies detected a single, 30-kDa band in cytoplasmic and nuclear extracts of NBD CBLM and in nuclear extracts of NBD HC; no distinct bands were detected in extracts from controls (Fig. 6C and D). In cytoplasmic fractions of NBD HC, an additional 26-kDa band was present (Fig. 6D) that may represent unphosphorylated CHOP protein (45, 70). Quantitation revealed prominent increases in the 30-kDa protein in both NBD CBLM cytoplasmic (21.4-fold increase; ANOVA, P < 0.0001) and nuclear (11.34-fold increase; ANOVA, P = 0.0003) extracts (Fig. 6E). Significant increases were also found in NBD HC cytoplasmic extracts for the 26-kDa (3.48-fold increase; ANOVA, P = 0.043) and 30-kDa (7.49-fold increase; ANOVA, P = 0.009) CHOP species and in NBD HC nuclear extracts for the 30-kDa nuclear CHOP species (5.47-fold increase; ANOVA, P = 0.02) (Fig. 6F).
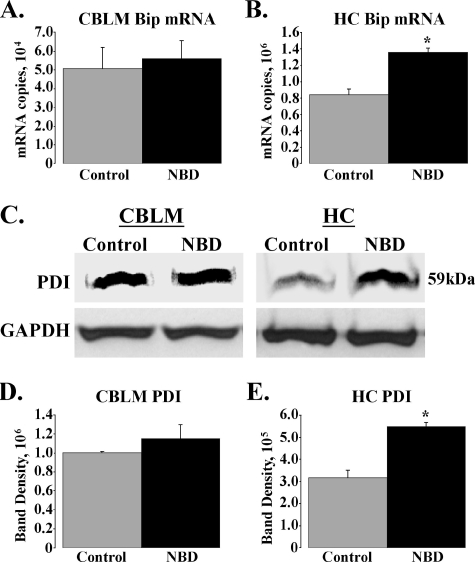
Hippocampus-specific induction of Bip and PDI in PND28 NBD rats.
The ER chaperone, Bip, serves as a key sentinel for ER disturbances, monitoring the folding status of proteins and controlling the activation state of the UPR transducers. Bip is induced at the transcriptional level during ER stress to cope with the accumulation of misfolded proteins in the ER (33). XBP1S and ATF6p50 induce Bip mRNA expression through ERSE elements in the Bip promoter, while ATF4 activates Bip through an ERSE-independent, ATF-CRE-like binding site (42, 76, 78). Thus, Bip induction serves as an indicator of ER stress and UPR induction. Overexpression of Bip reduces ER stress-mediated apoptosis and attenuates CHOP induction (71). Real-time PCR analysis of Bip expression in NBD revealed no alteration in CBLM (Fig. 7A) but a significant elevation in NBD HC relative to results with control rats (Fig. 7B) (1.61-fold increase; ANOVA, P = 0.0001).
FIG. 7.
Hippocampus-specific increases in Bip mRNA and PDI protein levels. (A and B) SYBR green real-time PCR analysis of Bip transcript levels in CBLM (A) and HC (B) of PND28 control (n = 5) or NBD (n = 7) rats. Bip mRNA was not significantly altered in NBD CBLM (A). Note significant increase in HC Bip mRNA in NBD rats relative to controls (B: 1.61-fold increase in NBD; ANOVA, P = 0.0001). (C) Representative Western immunoblot analysis for PDI protein in CBLM and HC of control or NBD rats (upper panels). Corresponding immunoblot signals for the GAPDH housekeeping gene are shown (lower panels). (D and E) Quantitation of PDI immunoblot band density, relative to GAPDH signal, from control (n = 4) or NBD (n = 4) rat CBLM (D: not significant) and HC (E: 1.73-fold increase in NBD; ANOVA, P = 0.0009). Asterisks indicate P values of <0.05.
PDI is induced by ER stress and plays a crucial role in regulating the proper folding of proteins in the ER by catalyzing disulfide bond formation (43). Neuronal overexpression of PDI has also been shown to promote survival in response to stresses that induce ER dysfunction (64). We evaluated protein levels of PDI in extracts from NBD and control rats. PDI-specific antibodies detected the expected 59-kDa species for PDI in both control and NBD CBLM and HC (Fig. 7C). Although PDI protein levels were similar in CBLM extracts from NBD and controls (Fig. 7D), PDI levels were increased in NBD HC (Fig. 7E) (1.73-fold increase; ANOVA, P = 0.0009).
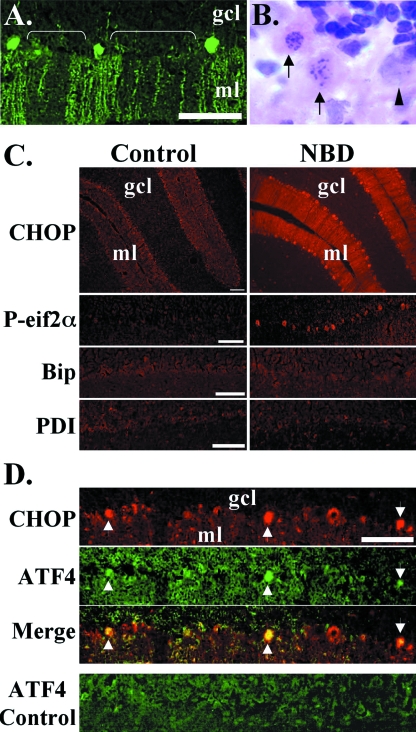
ER stress in PCs.
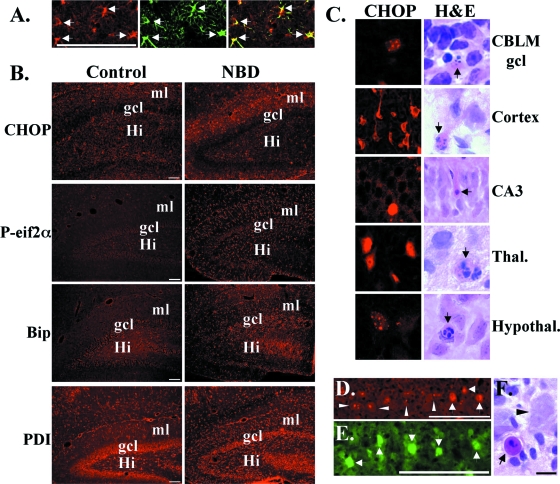
Previous reports have identified apoptotic neurodegeneration of specific neural populations in NBD, including Purkinje cells (PCs) (25, 72). Calbindin staining of PND28 NBD rat CBLM confirmed PC loss (Fig. 8A, brackets). H&E staining identified degenerating PCs in NBD, with pyknotic nuclei or signs of nuclear fragmentation (Fig. 8B). Regional expression of CHOP was assessed in brain sections of PND21, PND28, and PND84 NBD and controls by immunofluorescence microscopy. Specific CHOP staining was not present in control rat CBLM (Fig. 8C). At PND21, a time point preceding loss of PCs, we did not detect differences in the CHOP signal between NBD and the controls (data not shown). However, at PND28, the CHOP signal was dramatically increased in many PC bodies and dendrites in rats with NBD (Fig. 8C). To test whether UPR activation localized to PCs in rats with NBD, we assessed eif2α phosphorylation using antibodies specific to P-eif2α. P-eif2α was observed in PCs of rats with NBD but not in controls (Fig. 8C). We also evaluated Bip and PDI immunofluorescence. There was no detectable difference in PC signal for Bip and PDI between control and NBD rats (Fig. 8C). Faint PDI fluorescence was detected in a few glial cells in the CBLM of NBD rats (data not shown). Double-label immunofluorescence experiments using antibodies to CHOP and ATF4 indicated colocalization in NBD rat PCs and enhanced ATF4 signal in PND28 NBD PCs relative to results with control rats (Fig. 8D).
FIG. 8.
ER stress in NBD rat PCs. (A) Calbindin staining in the CBLM of PND28 NBD rats; note gaps in the PC and molecular layers (brackets). (B) H&E-stained PCs from PND28 NBD rat; note degenerating PCs with pyknotic and fragmented nuclei (arrows), with normal PC shown for comparison (arrowhead). (C) Representative CHOP, P-eif2α, Bip, and PDI immunofluorescence of PND28 control (left panels) or NBD (right panels) rat CBLM. Note intense CHOP and P-eif2α immunoreactivity in PCs of NBD rats compared to results for control rats. Bip and PDI signals are equivalent in control and NBD rat PCs. (D) Double-label immunofluorescence analysis of CHOP (red) and ATF4 (green) in PND28 NBD rat CBLM (merged image shows colocalized signal in PCs, arrowheads). ATF4 immunofluorescence in PND28 control rat CBLM shown for comparison (bottom panel). Abbreviations: gcl, granule cell layer; ml, molecular layer. Micron bars, 100 μm.
ER stress in hippocampal astrocytes.
CHOP induction in NBD HC occurs in conjunction with induction of Bip and PDI. Thus, we examined the distribution of each of these proteins by immunofluorescence microscopy. CHOP was confined to astrocytes (Fig. 9A). CHOP staining was weak in control HC; however, at PND21, PND28, and PND84, NBD HC had marked CHOP signal in astrocytes in the molecular layer of the dentate gyrus, stratum lacunosum moleculare, stratum radiatum, and stratum oriens (Fig. 9B, representative PND28 rats). Surprisingly, dentate gyrus granule cell neurons, which rapidly degenerate between 3 and 5 weeks postinfection in NBD rats, did not overexpress CHOP at PND21 or PND28. Whereas P-eif2α signal was weak in controls, intense P-eif2α immunoreactivity was observed in astrocytes in NBD HC (Fig. 9B). Bip fluorescence was weak in controls and localized primarily to neurons. In NBD rats, however, enhanced Bip staining was readily detected in astrocytes in HC. PDI levels were high in neurons in the HC of controls (i.e., dentate gyrus granule cell neurons and pyramidal neurons of CA3-CA1), but astrocytes were only weakly stained (Fig. 9B). In contrast, PDI signal was increased in numerous astrocytes in the HC of NBD rats (Fig. 9B).
FIG. 9.
Induction of ER stress in hippocampal astrocytes (A and B) and Chop expression in diverse, degenerating neural populations (C and D) in NBD rats. (A) Double-label immunofluorescence for CHOP (left panel, red) and GFAP (middle panel, green) in HC of NBD rats (right panel, merged image). (B) CHOP, P-eif2α, Bip, and PDI immunofluorescence in dentate gyrus of PND28 control (left panels) or NBD (right panels) rats. Note intense immunoreactivity for CHOP, P-eif2α, Bip, and PDI in NBD rat astrocytes in dentate gyrus molecular layer and hilus compared to controls. (C) Chop immunofluorescence (left panels) and degenerative morphology (arrows in right panels) in cerebellar granule cell neurons (CBLM gcl), cortical neurons (Cortex), hippocampal CA3 pyramidal neurons (CA3), thalamic neurons (Thal.), and hypothalamic neurons (Hypothal.). (D) Chop immunofluorescence in mitral cells in the olfactory bulb of PND28 NBD rat. Note intense nuclear staining (wide arrowheads) and localization to nuclear bodies (narrow arrowheads). (E) Anti-BDV nucleoprotein staining of mitral cells in NBD rats. (F) H&E staining shows degenerating mitral cell (arrow) and normal mitral cell (arrowhead) in NBD rat. Abbreviations: gcl, granule cell layer; ml, molecular layer; Hi, hilus. Micron bars, 100 μm.
CHOP expression in other brain regions.
The CBLM and HC are not the only brain regions reported to be affected in NBD (72); thus, we investigated CHOP expression throughout the brains of NBD rats. Neuronal CHOP expression was not restricted to PCs. Subcellular localization of CHOP varied in individual neurons, including PCs, localizing to the cytoplasm only, the nucleus only, both the cytoplasm and the nucleus, or intensely stained nuclear bodies. Scattered cerebellar granule cell neurons, numerous cortical neurons, rare CA3 pyramidal neurons, and numerous thalamic and hypothalamic neurons with a strong CHOP signal were found in NBD rat brains (Fig. 9C). We also detected clear signs of neurodegeneration in H&E-stained sections from these same regions (Fig. 9C; note nuclear pyknosis and fragmentation in neurons [arrows]). CHOP staining was detected in numerous mitral cells in the olfactory bulb of NBD rats (Fig. 9D). Mitral cells stained intensely with antibodies for BDV nucleoprotein (Fig. 9E), and some showed classic histological signs of neurodegeneration (Fig. 9F; note mitral cell with condensed chromatin; normal mitral cell is shown for comparison). While the density of astrocytes expressing CHOP was highest in the HC of NBD rats, CHOP-positive astrocytes were observed in the cortex, striatum, and thalamic and hypothalamic regions (data not shown). Neither neuronal nor astrocytic CHOP expression was detected in corresponding regions of control rats (data not shown) or in regions of NBD rats where histological signs of neurodegeneration were not evident.
DISCUSSION
Viral infections have been shown to induce apoptosis through ER stress. However, the pathological consequences of ER stress associated with viral disease have not been well established, since most reports have focused on in vitro infections. In this study we demonstrate that UPR activation and ER stress are correlated temporally and anatomically with neural damage in NBD. Cell death occurs when ER quality control mechanisms are overwhelmed by excessive or prolonged stress; the balance between apoptotic and quality control UPR signaling likely determines cell fate. Our investigation of the ER stress-induced proapoptotic gene CHOP and the ER quality control chaperones Bip and PDI show cell type and regional discordance. In CBLM, increased expression of CHOP in degenerating PCs was not associated with compensatory increases in Bip and PDI. In contrast, astrocytes, a cell type that survives in NBD, expressed Bip and PDI as well as CHOP. These results suggest that distinct cell types may harbor unique, deficient, or developmentally regulated UPR signals that modulate their response to ER stress in central nervous system infection.
UPR activation in NBD.
Microarray experiments in NBD revealed differential expression of ER stress genes and prompted investigations into the activation state of the three transducers of the UPR: PERK, IRE1, and ATF6. PERK-mediated phosphorylation of eif2α during ER stress inhibits global translation, reducing the burden of newly synthesized proteins in the ER (17). Several viruses have developed mechanisms to circumvent translational shutdown. Infection of fibroblasts with HCMV activates ER stress signals, including PERK, but has only minimal impact on eif2α phosphorylation (26). The hepatitis C virus E2 protein binds activated PERK, preventing eif2α phosphorylation, and the herpes simplex virus type 1 γ34.5 protein stimulates eif2α dephosphorylation, resulting in blockade of P-eif2α-mediated translational attenuation (23, 52). In NBD, infection was associated with eif2α phosphorylation in CBLM and HC but did not result in a detectable reduction in overall protein synthesis. Although it is tempting to speculate that BDV has evolved strategies to circumvent translation attenuation despite eif2α phosphorylation, ER stress is limited to discrete cellular populations in NBD and the sensitivity of our assay may be insufficient to detect protein attenuation in heterogeneous cellular homogenates.
CBLM-specific increases in nuclear ATF4 were detected in NBD, and the ATF4 protein localized to PCs. Consistent with this finding, expression of the Asn-S, Hmox1, and Slc3a2 genes, genes regulated by ATF4 (19), were increased in CBLM microarray experiments. Double-labeling experiments colocalized ATF4 and CHOP in PCs. ER stress-regulated ATF4 translation and CHOP induction are markedly attenuated in PERK null and eif2αS51A cells, suggesting that the PERK signaling arm of the UPR may be dominant over IRE1 and ATF6 (18, 60). ATF4 was not detected in NBD HC astrocytes despite increased P-eif2α immunoreactivity. These results suggest divergent, cell-type- and region-specific regulation of ATF4 translation in NBD.
XBP1S encodes a potent transcriptional activator that induces the expression of ER chaperones and genes involved in the degradation of misfolded proteins (35). XBP1S mRNA was detected in NBD CBLM and HC but not in controls. We were unable to detect XBP1S protein in NBD extracts. This result may suggest that XBP1S protein levels are below the level of detection or that XBP1S translation is actively inhibited by viral infection. HCMV-infected cells and HCV replicon-expressing cells exhibit enhanced XBP1 splicing and increased XBP1 mRNA levels; however, XBP1S trans-activating activity is suppressed (26, 66). XBP1U protein levels were significantly decreased in NBD CBLM and HC. While it is an indirect measurement, we suspect that decreased XBP1U protein is a result of increased splicing of XBP1 and the resultant shift and extension of the open reading frame of XBP1 mRNA.
Proteolysis of ATF6 in the Golgi results in release of its amino-terminal domain (ATF6p50) into the cytoplasm, which translocates to the nucleus to induce ER chaperone and folding enzyme gene expression. We detected significant increases in cytoplasmic ATF6p50 in both the CBLM and HC of NBD rats, although the degree of increase in HC was greater than that in CBLM. Surprisingly, we did not detect ATF6p50 in nuclear extracts. Evidence suggests that ATF6-mediated transcription is linked to proteasomal degradation of ATF6 (67). Hence, rapid degradation through association with nuclear proteasomal machinery may account for low or undetectable levels of nuclear ATF6p50 in NBD. Alternatively, undefined protein-protein interactions could account for cytoplasmic accumulation and nuclear exclusion of ATF6p50 in this model. ATF6p50 can bind to the ERSE in the XBP1 promoter to induce transcription of XBP1 mRNA (77). This is thought to increase XBP1 mRNA substrate availability for IRE1-mediated splicing. Consistent with the large increase in ATF6p50 in NBD HC, we detected significant increases in HC XBP1 mRNA.
In summary, all three arms of the UPR are activated in NBD with regional discordance in the levels of individual downstream effectors. Although significant in both NBD CBLM and HC, levels of eif2α phosphorylation and ATF6 cleavage were higher in HC. In contrast, enhanced ATF4 translation was specific to CBLM, while XBP1 splicing was prominent in both regions. We speculate that regional differences in UPR-regulated gene expression (i.e., Bip and PDI) may reflect region-specific contributions of these effectors.
CHOP, ER stress, and neurodegeneration.
The importance of CHOP in facilitating ER stress-induced apoptosis is well documented, and ER stress mediates apoptosis following in vitro infection by several viruses (27, 39, 49, 63). In Japanese encephalitis virus infection, CHOP levels correlate with the extent of apoptosis, and overexpression of Bcl-2 or inhibition of p38 mitogen-activated protein kinase attenuates apoptosis (63). In the NBD CBLM, virus is found almost exclusively in PCs (2). We found increased levels of CHOP mRNA and protein in NBD CBLM where CHOP protein is localized to PCs. Although the increase in CHOP levels was associated with increased levels of the UPR effectors P-eif2α and ATF4, the expression levels of the quality control genes Bip and PDI were not increased in PCs. Overexpression of Bip and PDI inhibits ER stress-mediated apoptosis (64, 71). Thus, our results suggest that ER stress in PCs is associated with enhancement of apoptotic ER signals but not ER survival molecules, shifting the balance toward neural death.
PCs are particularly vulnerable to toxic insults, including ischemic and excitotoxic stress (73). The ER stress protein, oxygen-regulated protein 150, plays a crucial role in PC vulnerability during neural development (30). PC death in two spontaneous mouse mutants (Purkinje cell degeneration and woozy mutants) is associated with ER stress (34, 79). Thus, PCs may be particularly sensitive to ER disturbances. ER stress has also been implicated in pathological processes resulting in neural death in various degenerative diseases and acute disorders, including Alzheimer's disease, Parkinson disease, Huntington disease, amyotrophic lateral sclerosis, prion disease, ischemic insult, and brain trauma (21, 41). Hence, ER stress may represent a common pathway culminating in neural cell death.
Survival of astrocytes and ER stress.
Analysis of ER stress markers in the NBD HC revealed different results than in CBLM. In this study, we found that astrocytes in NBD HC express classic markers of ER stress. Although astrocytes are infected by BDV, NBD results in neuronal but not astrocytic apoptosis (7). In fact, astrocytes proliferate in the brains of NBD rats. Astrocyte resistance in NBD may reflect an intrinsic capacity to adapt to ER stress. In ischemia and hypoxic ER stress paradigms, neurons are vulnerable but astrocytes are resistant (24, 37, 64). Astrocytes possess a unique ER stress transducer, OASIS (old astrocyte specifically induced substance) that induces Bip and PDI expression and attenuates ER stress-induced apoptosis (31). In NBD, both PCs and astrocytes show increased expression of CHOP and P-eif2α; however, astrocytes also have increased levels of the quality control molecules, Bip and PDI. Thus, we suggest that in astrocytes the balance between apoptosis and survival may favor survival. We did not detect increased levels of ER stress markers (i.e., CHOP, P-eif2α, Bip, and PDI) in HC dentate gyrus granule cell neurons, which are severely affected in NBD. However, we have recently shown that disturbances in HC zinc physiology appear to mediate HC granule cell demise (74). Thus, regionally discordant mechanisms may account for neural damage in NBD. Alternatively, a different complement of ER stress mediators may regulate apoptosis in HC granule cell neurons. In fact, caspase-12 and not CHOP is reported to mediate ER stress-induced apoptosis of cultured hippocampal neurons (32). We also observed CHOP induction and histological signs of neurodegeneration in other neural populations previously reported to undergo apoptosis in NBD rats, such as cerebellar granule cells and cortical neurons (72). In addition we report CHOP induction and signs of neurodegeneration in neural populations not previously reported to be affected in NBD (i.e., CA3 pyramidal neurons, thalamic and hypothalamic neurons, and mitral cells in the olfactory bulb). These results not only show that ER stress-regulated CHOP expression is present in other NBD brain regions but suggest that neurodegeneration may not be as restricted as previously described.
ER stress initiation in NBD.
The initiator signals leading to neuronal and astrocytic ER stress in NBD may be associated with viral protein-induced disturbances or extracellular signals (i.e., cytokines) from infected or activated glia (8, 25). Viral proteins known to directly induce ER stress include the E2 protein of hepatitis C virus and the Us11 glycoprotein of HCMV (40, 68). The viral glycoprotein of BDV (gp94) seems a likely candidate for induction of ER stress in NBD. It is translocated across the ER, inserted in the ER membrane, cotranslationally N glycosylated in the ER lumen, and inefficiently transported to the cell surface and accumulates in the ER/cis-Golgi region (14, 57). These properties may play a role in viral persistence by allowing BDV to evade host immune surveillance. Further studies will be needed to elucidate whether BDV gp94 can be implicated in the ER stress response.
Infections with a chimeric neuropathogenic murine retrovirus (FrCasE) and a temperature-sensitive mutant of Moloney murine leukemia virus (ts1) cause ER stress and spongiform neurodegeneration in mice. These viruses infect glia but not neurons, suggesting an indirect link to neuropathology that may be associated with impaired neuronal support by glia or secretion of proinflammatory cytokines and neurotoxins (12, 13, 29, 55). Altered cytokine levels in NBD rat brains have been reported by several groups (25, 53, 59). Tumor necrosis factor alpha mRNA levels are increased in NBD, and tumor necrosis factor alpha has been shown to increase ER Ca2+ release and induce hepatoma cell apoptosis (28); interleukin 1 beta, in combination with gamma interferon, induces ER stress in pancreatic β-cells through downregulation of the ER Ca2+ pump SERCA2B and resultant depletion of ER Ca2+ (9). Of relevance to PC degeneration in NBD, CBLM microarray analysis detected alterations in ER calcium transporters (RyR1, RyR3, SERCA2, and SERCA3). The ER plays a vital role in maintaining intracellular Ca2+ homeostasis. Ca2+ storage status of the ER and free Ca2+ levels in the cytoplasm are major determinants of cell death (10, 15, 16, 50).
Conclusions.
Pre- and perinatal viral infections are important causes of neurodevelopmental damage; however, the molecular mechanisms by which damage can be mediated are poorly understood. Clues to pathogenesis may be found in NBD, where differences in vulnerability of specific cell populations may reflect differences in response to dysregulation of ER dynamics and ER stress pathways.
Acknowledgments
We thank Mady Hornig and Thomas Briese for helpful comments and Kavitha Yaddanapudi for technical assistance.
This work was supported by NIH awards NS29425 and HD37546.
REFERENCES
- 1.Barone, M. V., A. Crozat, A. Tabaee, L. Philipson, and D. Ron. 1994. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 8:453-464. [DOI] [PubMed] [Google Scholar]
- 2.Bautista, J. R., S. A. Rubin, T. H. Moran, G. J. Schwartz, and K. M. Carbone. 1995. Developmental injury to the cerebellum following perinatal Borna disease virus infection. Brain Res. Dev. Brain Res. 90:45-53. [DOI] [PubMed] [Google Scholar]
- 3.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 4.Bitko, V., and S. Barik. 2001. An endoplasmic reticulum-specific stress-activated caspase (caspase-12) is implicated in the apoptosis of A549 epithelial cells by respiratory syncytial virus. J. Cell. Biochem. 80:441-454. [DOI] [PubMed] [Google Scholar]
- 5.Briese, T., A. Schneemann, A. J. Lewis, Y. S. Park, S. Kim, H. Ludwig, and W. I. Lipkin. 1994. Genomic organization of Borna disease virus. Proc. Natl. Acad. Sci. USA 91:4362-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briese, T., C. G. Hatalski, S. Kliche, Y. S. Park, and W. I. Lipkin. 1995. Enzyme-linked immunosorbent assay for detecting antibodies to Borna disease virus-specific proteins. J. Clin. Microbiol. 33:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carbone, K. M., B. D. Trapp, J. W. Griffin, C. S. Duchala, and O. Narayan. 1989. Astrocytes and Schwann cells are virus-host cells in the nervous system of rats with Borna disease. J. Neuropathol. Exp. Neurol. 48:631-644. [DOI] [PubMed] [Google Scholar]
- 8.Carbone, K. M., S. A. Rubin, Y. Nishino, and M. V. Pletnikov. 2001. Borna disease: virus-induced neurobehavioral disease pathogenesis. Curr. Opin. Microbiol. 4:467-475. [DOI] [PubMed] [Google Scholar]
- 9.Cardozo, A. K., F. Ortis, J. Storling, Y. M. Feng, J. Rasschaert, M. Tonnesen, F. Van Eylen, T. Mandrup-Poulsen, A. Herchuelz, and D. L. Eizirik. 2005. Cytokines downregulate the sarcoendoplasmic reticulum pump Ca2+ ATPase 2b and deplete endoplasmic reticulum Ca2+, leading to induction of endoplasmic reticulum stress in pancreatic beta-cells. Diabetes 54:452-461. [DOI] [PubMed] [Google Scholar]
- 10.Corbett, E. F., K. Oikawa, P. Francois, D. C. Tessier, C. Kay, J. J. Bergeron, D. Y. Thomas, K. H. Krause, and M. Michalak. 1999. Ca2+ regulation of interactions between endoplasmic reticulum chaperones. J. Biol. Chem. 274:6203-6211. [DOI] [PubMed] [Google Scholar]
- 11.de la Torre, J. C. 1994. Molecular biology of borna disease virus: prototype of a new group of animal viruses. J. Virol. 68:7669-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimcheff, D. E., S. Askovic, A. H. Baker, C. Johnson-Fowler, and J. L. Portis. 2003. Endoplasmic reticulum stress is a determinant of retrovirus-induced spongiform neurodegeneration. J. Virol. 77:12617-12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimcheff, D. E., M. A. Faasse, F. J. McAtee, and J. L. Portis. 2004. Endoplasmic reticulum (ER) stress induced by a neurovirulent mouse retrovirus is associated with prolonged BiP binding and retention of a viral protein in the ER. J. Biol. Chem. 279:33782-33790. [DOI] [PubMed] [Google Scholar]
- 14.Eickmann, M., S. Kiermayer, I. Kraus, M. Gossl, J. A. Richt, and W. Garten. 2005. Maturation of Borna disease virus glycoprotein. FEBS Lett. 579:4751-4756. [DOI] [PubMed] [Google Scholar]
- 15.George, C. H., G. V. Higgs, J. J. Mackrill, and F. A. Lai. 2003. Dysregulated ryanodine receptors mediate cellular toxicity: restoration of normal phenotype by FKBP12.6. J. Biol. Chem. 278:28856-28864. [DOI] [PubMed] [Google Scholar]
- 16.Groenendyk, J., and M. Michalak. 2005. Endoplasmic reticulum quality control and apoptosis. Acta Biochim. Pol. 52:381-395. [PubMed] [Google Scholar]
- 17.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 18.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 19.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 20.Hatalski, C. G., W. F. Hickey, and W. I. Lipkin. 1998. Evolution of the immune response in the central nervous system following infection with Borna disease virus. J. Neuroimmunol. 90:137-142. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi, T., A. Saito, S. Okuno, M. Ferrand-Drake, R. L. Dodd, and P. H. Chan. 2005. Damage to the endoplasmic reticulum and activation of apoptotic machinery by oxidative stress in ischemic neurons. J. Cereb. Blood Flow Metab. 25:41-53. [DOI] [PubMed] [Google Scholar]
- 22.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He, B., M. Gross, and B. Roizman. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hori, O., M. Matsumoto, K. Kuwabara, Y. Maeda, H. Ueda, T. Ohtsuki, T. Kinoshita, S. Ogawa, D. M. Stern, and T. Kamada. 1996. Exposure of astrocytes to hypoxia/reoxygenation enhances expression of glucose-regulated protein 78 facilitating astrocyte release of the neuroprotective cytokine interleukin 6. J. Neurochem. 66:973-979. [DOI] [PubMed] [Google Scholar]
- 25.Hornig, M., H. Weissenbock, N. Horscroft, and W. I. Lipkin. 1999. An infection-based model of neurodevelopmental damage. Proc. Natl. Acad. Sci. USA 96:12102-12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan, R., L. Wang, T. M. Graczyk, T. M. Block, and P. R. Romano. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 76:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, B. C., H. T. Kim, M. Mamura, I. S. Ambudkar, K. S. Choi, and S. J. Kim. 2002. Tumor necrosis factor induces apoptosis in hepatoma cells by increasing Ca(2+) release from the endoplasmic reticulum and suppressing Bcl-2 expression. J. Biol. Chem. 277:31381-31389. [DOI] [PubMed] [Google Scholar]
- 29.Kim, H. T., K. Waters, G. Stoica, W. Qiang, N. Liu, V. L. Scofield, and P. K. Wong. 2004. Activation of endoplasmic reticulum stress signaling pathway is associated with neuronal degeneration in MoMuLV-ts1-induced spongiform encephalomyelopathy. Lab. Investig. 84:816-827. [DOI] [PubMed] [Google Scholar]
- 30.Kitao, Y., K. Hashimoto, T. Matsuyama, H. Iso, T. Tamatani, O. Hori, D. M. Stern, M. Kano, K. Ozawa, and S. Ogawa. 2004. ORP150/HSP12A regulates Purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. J. Neurosci. 24:1486-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo, S., T. Murakami, K. Tatsumi, M. Ogata, S. Kanemoto, K. Otori, K. Iseki, A. Wanaka, and K. Imaizumi. 2005. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7:186-194. [DOI] [PubMed] [Google Scholar]
- 32.Kosuge, Y., T. Sakikubo, K. Ishige, and Y. Ito. 2006. Comparative study of endoplasmic reticulum stress-induced neuronal death in rat cultured hippocampal and cerebellar granule neurons. Neurochem. Int. [Epub ahead of print.] [DOI] [PubMed]
- 33.Kozutsumi, Y., M. Segal, K. Normington, M. J. Gething, and J. Sambrook. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462-464. [DOI] [PubMed] [Google Scholar]
- 34.Kyuhou, S. I., N. Kato, and H. Gemba. 2006. Emergence of endoplasmic reticulum stress and activated microglia in Purkinje cell degeneration mice. Neurosci. Lett. 396:91-96. [DOI] [PubMed] [Google Scholar]
- 35.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, A. S. 2005. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35:373-381. [DOI] [PubMed] [Google Scholar]
- 37.Li, F., T. Hayashi, G. Jin, K. Deguchi, S. Nagotani, I. Nagano, M. Shoji, P. H. Chan, and K. Abe. 2005. The protective effect of dantrolene on ischemic neuronal cell death is associated with reduced expression of endoplasmic reticulum stress markers. Brain Res. 1048:59-68. [DOI] [PubMed] [Google Scholar]
- 38.Li, M., P. Baumeister, B. Roy, T. Phan, D. Foti, S. Luo, and A. S. Lee. 2000. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, X. D., H. Lankinen, N. Putkuri, O. Vapalahti, and A. Vaheri. 2005. Tula hantavirus triggers pro-apoptotic signals of ER stress in Vero E6 cells. Virology 333:180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liberman, E., Y. L. Fong, M. J. Selby, Q. L. Choo, L. Cousens, M. Houghton, and T. S. Yen. 1999. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J. Virol. 73:3718-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindholm, D., H. Wootz, and L. Korhonen. 2006. ER stress and neurodegenerative diseases. Cell Death Differ. 13:385-392. [DOI] [PubMed] [Google Scholar]
- 42.Luo, S., P. Baumeister, S. Yang, S. F. Abcouwer, and A. S. Lee. 2003. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through an upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J. Biol. Chem. 278:37375-37385. [DOI] [PubMed] [Google Scholar]
- 43.Lyles, M. M., and H. F. Gilbert. 1991. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry 30:613-619. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto, M., M. Minami, K. Takeda, Y. Sakao, and S. Akira. 1996. Ectopic expression of CHOP (GADD153) induces apoptosis in M1 myeloblastic leukemia cells. FEBS Lett. 395:143-147. [DOI] [PubMed] [Google Scholar]
- 45.Maytin, E. V., M. Ubeda, J. C. Lin, and J. F. Habener. 2001. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp. Cell. Res. 267:193-204. [DOI] [PubMed] [Google Scholar]
- 46.McCullough, K. D., J. L. Martindale, L. O. Klotz, T. Y. Aw, and N. J. Holbrook. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21:1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narayan, O., S. Herzog, K. Frese, H. Scheefers, and R. Rott. 1983. Behavioral disease in rats caused by immunopathological responses to persistent borna virus in the brain. Science 220:1401-1403. [DOI] [PubMed] [Google Scholar]
- 48.Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oyadomari, S., and M. Mori. 2004. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11:381-389. [DOI] [PubMed] [Google Scholar]
- 50.Pan, Z., D. Damron, A. L. Nieminen, M. B. Bhat, and J. Ma. 2000. Depletion of intracellular Ca2+ by caffeine and ryanodine induces apoptosis of Chinese hamster ovary cells transfected with ryanodine receptor. J. Biol. Chem. 275:19978-19984. [DOI] [PubMed] [Google Scholar]
- 51.Paschen, W., S. Hotop, and C. Aufenberg. 2003. Loading neurons with BAPTA-AM activates xbp1 processing indicative of induction of endoplasmic reticulum stress. Cell Calcium 33:83-89. [DOI] [PubMed] [Google Scholar]
- 52.Pavio, N., P. R. Romano, T. M. Graczyk, S. M. Feinstone, and D. R. Taylor. 2003. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2α kinase PERK. J. Virol. 77:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plata-Salaman, C. R., S. E. Ilyin, D. Gayle, A. Romanovitch, and K. M. Carbone. 1999. Persistent Borna disease virus infection of neonatal rats causes brain regional changes of mRNAs for cytokines, cytokine receptor components and neuropeptides. Brain Res. Bull. 49:441-451. [DOI] [PubMed] [Google Scholar]
- 54.Pletnikov, M. V., S. A. Rubin, G. J. Schwartz, K. M. Carbone, and T. H. Moran. 2000. Effects of neonatal rat Borna disease virus (BDV) infection on the postnatal development of the brain monoaminergic systems. Brain Res. Dev. Brain Res. 119:179-185. [DOI] [PubMed] [Google Scholar]
- 55.Qiang, W., X. Kuang, J. Liu, N. Liu, V. L. Scofield, A. J. Reid, Y. Jiang, G. Stoica, W. S. Lynn, and P. K. Wong. 2006. Astrocytes survive chronic infection and cytopathic effects of the ts1 mutant of the retrovirus Moloney murine leukemia virus by upregulation of antioxidant defenses. J. Virol. 80:3273-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reimertz, C., D. Kogel, A. Rami, T. Chittenden, and J. H. Prehn. 2003. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J. Cell Biol. 162:587-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richt, J. A., T. Furbringer, A. Koch, I. Pfeuffer, C. Herden, I. Bause-Niedrig, and W. Garten. 1998. Processing of the Borna disease virus glycoprotein gp94 by the subtilisin-like endoprotease furin. J. Virol. 72:4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritter, C., K. Quirin, M. Kowarik, and A. Helenius. 2005. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 24:1730-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sauder, C., and J. C. de la Torre. 1999. Cytokine expression in the rat central nervous system following perinatal Borna disease virus infection. J. Neuroimmunol. 96:29-45. [DOI] [PubMed] [Google Scholar]
- 60.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 61.Schroder, M., and R. J. Kaufman. 2005. ER stress and the unfolded protein response. Mutat. Res. 569:29-63. [DOI] [PubMed] [Google Scholar]
- 62.Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- 63.Su, H. L., C. L. Liao, and Y. L. Lin. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 76:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka, S., T. Uehara, and Y. Nomura. 2000. Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J. Biol. Chem. 275:10388-10393. [DOI] [PubMed] [Google Scholar]
- 65.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76:7453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tardif, K. D., K. Mori, R. J. Kaufman, and A. Siddiqui. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 279:17158-17164. [DOI] [PubMed] [Google Scholar]
- 67.Thuerauf, D. J., L. E. Morrison, H. Hoover, and C. C. Glembotski. 2002. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J. Biol. Chem. 277:20734-20739. [DOI] [PubMed] [Google Scholar]
- 68.Tirosh, B., N. N. Iwakoshi, B. N. Lilley, A. H. Lee, L. H. Glimcher, and H. L. Ploegh. 2005. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 79:2768-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verkhratsky, A. 2004. Endoplasmic reticulum calcium signaling in nerve cells. Biol. Res. 37:693-699. [DOI] [PubMed] [Google Scholar]
- 70.Wang, X. Z., and D. Ron. 1996. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science 272:1347-1349. [DOI] [PubMed] [Google Scholar]
- 71.Wang, X. Z., B. Lawson, J. W. Brewer, H. Zinszner, A. Sanjay, L. J. Mi, R. Boorstein, G. Kreibich, L. M. Hendershot, and D. Ron. 1996. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 16:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weissenbock, H., M. Hornig, W. F. Hickey, and W. I. Lipkin. 2000. Microglial activation and neuronal apoptosis in Bornavirus infected neonatal Lewis rats. Brain Pathol. 10:260-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welsh, J. P., G. Yuen, D. G. Placantonakis, Y. Q. Vu, F. Haiss, E. O'Hearn, M. E. Molliver, and S. A. Aicher. 2002. Why do Purkinje cells die so easily after global brain ischemia? Aldolase C, EAAT4, and the cerebellar contribution to posthypoxic myoclonus. Adv. Neurol. 89:331-359. [PubMed] [Google Scholar]
- 74.Williams, B. L., K. Yaddanapudi, C. M. Kirk, A. Soman, M. Hornig, and W. I. Lipkin. 2006. Metallothioneins and zinc dysregulation contribute to neurodevelopmental damage in a model of perinatal viral infection. Brain Pathol. 16:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 76.Yoshida, H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741-33749. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 79.Zhao, L., C. Longo-Guess, B. S. Harris, J. W. Lee, and S. L. Ackerman. 2005. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 37:974-979. [DOI] [PubMed] [Google Scholar]