Abstract
Limited data are available on the genotypic and phenotypic resistance profile of the α-(1-2)mannose oligomer-specific prokaryotic lectin cyanovirin (CV-N). Therefore, a more systematic investigation was carried out to obtain a better view of the interaction between CV-N and human immunodeficiency virus type 1 (HIV-1) gp120. When HIV-1-infected CEM cell cultures were exposed to CV-N in a dose-escalating manner, a total of eight different amino acid mutations exclusively located at N-glycosylation sites in the envelope surface gp120 were observed. Six of the eight mutations resulted in the deletion of high-mannose type N-glycans (i.e., at amino acid positions 230, 332, 339, 386, 392, and 448). Two mutations (i.e., at position 136 and 160) deleted a complex type N-glycan in the variable V1/V2 domain of gp120. The level of phenotypic resistance of the mutated virus strains against CV-N generally correlated with the number of glycan deletions in gp120, although deletion of the glycans at N-230, N-392, and N-448 generally afforded a more pronounced CV-N resistance than other N-glycan deletions. However, the extent of the decrease of antiviral activity of CV-N against the mutated virus strains was markedly less pronounced than observed for α(1-3)- and α(1-6)-mannose-specific plant lectins Hippeastrum hybrid agglutinin (HHA) and Galanthus nivalis agglutinin (GNA), which points to the existence of a higher genetic barrier for CV-N. This is in agreement with a more consistent suppression of a wider variety of HIV-1 clades by CV-N than by HHA and GNA. Whereas the antiviral and in vitro antiproliferative activity of CV-N can be efficiently reversed by mannan, the pronounced mitogenic activity of CV-N on peripheral blood mononuclear cells was unaffected by mannan, indicating that some of the observed side effects of CV-N are unrelated to its carbohydrate specificity/activity.
Recently, carbohydrate-binding agents (CBA) received special attention for their potential to act as anti-human immunodeficiency virus (HIV) microbicides. Several plant lectins with specificity for mannose (Man) and/or N-acetylglucosamine (GlcNAc) have been reported to possess favorable properties to qualify as potential microbicidal drugs (1, 5). Also, several CBA of prokaryotic origin or from invertebrates have been isolated and characterized (13, 30). Among them, cyanovirin (CV-N), an 11-kDa protein derived from the cyanobacterium (blue-green alga) Nostoc ellipsosporum, has received by far the most attention in this respect and has already been thoroughly investigated on its structural properties, carbohydrate-binding potential, and associated antiviral activity (8-12, 22, 33). X-ray crystallographic studies of a CV-N-Manα(1,2)Manα disaccharide complex and nuclear magnetic resonance and isothermal titration calorimetry studies revealed that monomeric CV-N recognizes mannose oligomers on two sides of the protein. Both sites are created by structurally equivalent residues belonging to the two domains of CV-N and bind primarily N-linked high-mannose oligosaccharides, such as those found on the viral envelope of HIV type 1 (HIV-1) (11). The first binding site is less selective for the type of mannose oligomers [i.e., Manα(1,2)Manα(1,2)Man, Manα(1,2)Manα(1,3)Man, and Manα(1,2)Manα(1,6)Man are equally recognized with similar KA values]. However, overall, it binds the trimannosides with higher affinities than the second site, which shows a more pronounced discrimination between the different trisaccharides (10). Four sugar-binding sites can be identified in a domain-swapped dimer. It is, however, yet unclear whether CV-N exerts its antiviral activity as a monomer or a dimer.
Very recently, the resistance profiles of the mannose-specific plant lectins derived from Galanthus nivalis (Galanthus nivalis agglutinin [GNA]) and Hippeastrum hybrid (Hippeastrum hybrid agglutinin [HHA]) have been thoroughly investigated (2, 4). Drug-escalating studies in cell culture revealed that these CBA select for mutant HIV-1 strains that predominantly contain deleted N-glycosylation sites in the envelope gp120. Such a drug resistance profile is unprecedented in that none of the known anti-HIV drugs, including entry inhibitors that are in preclinical research, consistently select for such N-glycan deletions in gp120 (3). Consequently, the mutant virus strains do not show measurable cross-resistance against these compounds and vice versa; mutant HIV strains that appeared under drug selection with DS-5000, AMD-3100, and SDF-1 did not lose their sensitivity to be inhibited by the plant lectins irrespective of the (nonglycan) mutations that appeared in the gp120 of the drug-resistant virus strains (2). The appearance of mutations at N-glycosylation sites in HIV gp120 under drug pressure by HHA and GNA was later confirmed to also be the case for the mannose-binding concanavalin A and the prokaryotic lectin CV-N (31). Very recently, a comparable mutational/resistance spectrum was also demonstrated for the N-acetylglucosamine (GlcNAc)-specific plant lectin derived from the stinging nettle Urtica dioica (5).
Although N-glycans on HIV-1 gp120 were consistently deleted under CBA pressure, a wide variety of different N-glycosylation sites on HIV-1 gp120 (20 to 29 or even more, depending on the particular HIV-1 strain) could be affected in CBA-exposed virus isolates. So far only one single virus strain has been isolated under CV-N pressure and genotypically and phenotypically characterized (31). Therefore, we decided to perform a more in-depth study of the mutational pathways that could appear in the gp120 of HIV-1 strains in the presence of increasing concentrations of CV-N to obtain more insights in the interaction of CV-N with HIV-1 gp120. Also the cross-resistance/sensitivity spectrum of CV-N against a wide variety of well-characterized mutant virus strains was investigated.
MATERIALS AND METHODS
Test compounds.
The mannose-specific plant lectins from Galanthus nivalis (GNA), Hippeastrum hybrid (HHA), Listera ovata (Listera ovata agglutinin [LOA]), and Cymbidium hybrid (Cymbidium hybrid agglutinin [CA]) were derived and purified from these plants, as described before (26, 27). AMD3100 was from AnorMed (Langley, BC, Canada), and cyanovirin (CV-N) was from J. B. McMahon (National Institutes of Health, Bethesda, MD) and A. Bolmstedt (Göteborg, Sweden).
Cells.
Human T-lymphocytic CEM cells were obtained from the American Type Culture Collection (Rockville, MD) and cultivated in RPMI 1640 medium supplemented with 10% fetal bovine serum (BioWittaker Europe, Verviers, Belgium), 2 mM l-glutamine, and 0.075 M NaHCO3.
Viruses.
HIV-1(IIIB and BaL) was provided by R. C. Gallo and M. Popovic (at that time at the National Cancer Institute, National Institutes of Health, Bethesda, MD). HIV-1(HE) is a clinical isolate, derived from a Belgian AIDS patient in 1987 and later propagated in MT-4 cells. HIV-2(ROD) was obtained from L. Montagnier (at that time at the Pasteur Institute, Paris, France). Simian immunodeficiency virus strain mac251 (SIVmac251) was provided by C. Bruck (Belgium).
Antiretrovirus assays.
The methodology of the anti-HIV assays has been described previously (1). Briefly, CEM cells (4.5 × 105 cells per ml) were suspended in fresh culture medium and infected with HIV-1 at 100 50% cell culture infective doses per ml of cell suspension. Then, 100 μl of the infected cell suspension was transferred to microplate wells, mixed with 100 μl of the appropriate dilutions of the test compounds, and further incubated at 37°C. After 4 to 5 days, giant cell formation was recorded microscopically in the CEM cell cultures. The 50% cytostatic concentration corresponds to the compound concentrations required to inhibit CEM cell proliferation by 50%. The 50% effective concentration (EC50) corresponds to the compound concentrations required to prevent syncytium formation by 50% in the virus-infected CEM cell cultures. In the experiments where the antiviral activity of CV-N was evaluated in the presence of mannan, 2.5 mg/ml mannan was added to the CV-N dilutions prior to the addition of the virus-infected cell suspension.
Antiviral activity of test compounds against HIV-1 clade isolates in PBMC.
Primary clinical isolates representing different HIV-1 clades and an HIV-2 isolate were all kindly provided by L. Lathey from BBI Biotech Research Laboratories, Inc., Gaithersburg, MD, and their coreceptor use (R5 or X4) was determined by us on the astroglioma U87.CD4 cell line transfected with either CCR5 or CXCR4. The following clinical isolates were included in the study: UG273 (clade A, R5), US2 (clade B, R5), ETH2220 (clade C, R5), UG270 (clade D, X4), ID12 (clade A/E, R5), BZ163 (clade F, R5), BCF-DIOUM (clade G, R5), BCF06 (clade O, X4), and HIV-2 BV-5061W (X4). Antiviral testing of these isolates in peripheral blood mononuclear cells (PBMC) was as follows. PBMC from healthy donors were isolated by density gradient centrifugation and stimulated with phytohemagglutinin (PHA) at 2 μg/ml (Sigma, Bornem, Belgium) for 3 days at 37°C. The PHA-stimulated blasts were washed twice with phosphate-buffered saline and counted by trypan blue dye exclusion. The cells were then seeded at 0.5 × 106 cells per well in a 48-well plate containing various concentrations of compound in cell culture medium (RPMI 1640) containing 10% fetal calf serum and interleukin-2 (25 U/ml; R&D Systems Europe, Abingdon, United Kingdom). The virus stocks were diluted in medium and added at a final dose of 250 pg p24 or p27/ml as determined by a viral core antigen (Ag)-specific enzyme-linked immunosorbent assay. Cell supernatant was collected at day 12, and HIV-1 core Ag in the culture supernatant was analyzed by a p24 Ag enzyme-linked immunosorbent assay kit (Perkin Elmer, Zaventem, Belgium). For HIV-2 p27 Ag detection, the INNOTEST from Innogenetics (Temse, Belgium) was used.
Selection and isolation of CV-N-resistant HIV-1 strains.
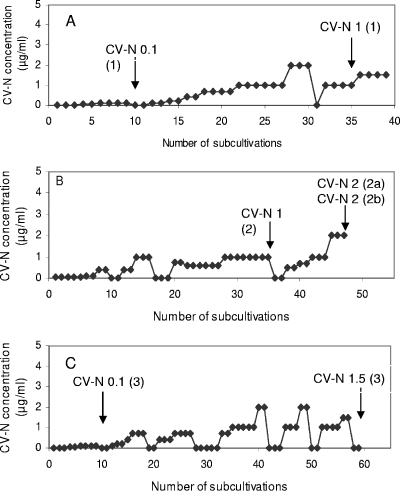
The procedure followed for the selection of drug-resistant virus mutants was essentially as recently described (4, 5). Briefly, HIV-1(IIIB or NL-4.3) was added to CEM cell cultures in 48-well plates in the presence of 0.1 μg/ml CV-N. For the generation of drug-resistant virus mutants, an increased drug concentration was administered when full cytopathicity was obtained in the previous cell culture. The detailed drug resistance selection schedule is depicted in Fig. 1.
FIG. 1.
Resistance selection in HIV-1(IIIB)-infected (A and B) or HIV-1 (NL-4.3)-infected (C) CEM cell cultures in the presence of escalating concentrations of CV-N. Every 4 to 5 days, the cell cultures were subcultivated by adding 100 or 200 μl of the virus-infected cell suspension to 900 or 800 μl fresh culture medium, respectively.
Genotyping of the HIV-1 env region.
Proviral DNA was extracted from cell pellets using the QIAamp blood mini kit (QIAGEN, Hilden, Germany). Both the gp120 and gp41 genes were covered in this assay as recently described (29).
Effect of CV-N on [3H]thymidine incorporation.
PBMC were exposed to different concentrations of CV-N or PHA for 3 days in 96-well microtiter plates. The experiments were performed in triplicate. Then, 0.4 μCi of [3H]thymidine (Amersham Pharmacia, Buckinghamshire, United Kingdom) with a radiospecific activity of 5 Ci/mmol was added to each culture. The plates were harvested 20 h later, and [3H]thymidine incorporation into trichloroacetic acid (TCA)-insoluble cell material, which is a measure of the mitogenic activity of the test compounds, was measured in a scintillation counter (Canberra-Packard, Zellik, Belgium) and expressed as disintegrations per minute. The assays were performed each time in triplicate.
Cytostatic/cytotoxic activity of CV-N against CEM cells and PBMC.
Human lymphocyte CEM cells were seeded at 3 × 105 cells/ml in culture medium in 96-well microtiter plates in the presence of serial dilutions of CV-N. After 3 to 4 days of incubation at 37°C in a CO2-controlled incubator, the cell number was determined in a Coulter counter. The 50% inhibitory concentration was defined as the CV-N concentration required to inhibit CEM cell proliferation by 50%.
Human PBMC were isolated, stimulated with PHA, and seeded in 48-well plates as described above. Different concentrations of CV-N were added to the cell cultures. Three days later, the number of living and dead cells were counted by trypan blue dye exclusion.
Effect of CV-N on cellular activation markers in PBMC.
To study the effect of the CV-N on the expression of cellular activation markers such as CD25, CD69, and HLA-DR antigen, PBMC were incubated with serial fivefold dilutions of CV-N (0.9, 0.18, 0.036, and 0.007 μΜ) or medium in the absence or presence of PHA (2 μg/ml) at 37°C. The monoclonal antibodies (MAbs) against CD4, CD8, CD69, CD25, and HLA-DR were purchased from BD Biosciences (Erembodegem, Belgium).
Cell surface CD antigen expressions were analyzed at day 3 by flow cytometry. Briefly, after washing with phosphate-buffered saline containing 2% fetal calf serum, cells were incubated with fluorescein isothiocyanate-conjugated anti-CD8 MAb and perdinin chlorophyll protein-conjugated anti-CD4 MAb in combination with phycoerythrin-conjugated anti-CD25, -CD69, or -HLA-DR MAbs for 30 min at 4°C. As a negative control for aspecific background staining, cells were stained in parallel with Simultest control γ1/γ2a (BD Biosciences). Then the cells were washed, fixed with 1% formaldehyde solution, and analyzed with a FACSCalibur (BD Biosciences, San Jose, CA). Data were acquired and analyzed with CellQuest software (BD Biosciences).
RESULTS
Antiviral activity of CV-N against HIV strains in cell culture.
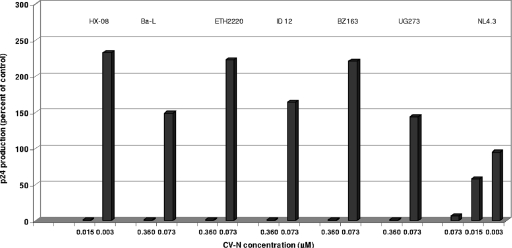
Cyanovirin has been evaluated for its inhibitory activity against a broad variety of retroviral strains, including the laboratory strains HIV-1 IIIB, HE, NL-4.3, and Ba-L; HIV-2 ROD and BV-5061W; SIV Mac251; and representative members of eight different HIV-1 strains belonging to clades A, B, C, D, A/E, F, G, and O. The strains from the D and O clades were X4; the other clade isolates were R5 (Table 1). CV-N showed a consistent suppression of HIV-1, HIV-2, or SIV replication, irrespective of the nature of the virus, the cell type, or the coreceptor tropism of the viral strain. The EC50s ranged between 0.0007 μM and 0.013 μM for HIV-1(IIIB) and HIV-2(ROD) in cell lines and between 0.013 and 0.16 μM for the variety of clinical clade isolates in PBMC. At a CV-N concentration that was fivefold lower than its fully antivirally suppressive concentration, very often a stimulatory effect on HIV-1 p24 production was noted for all (R5-tropic) clade isolates (Fig. 2). However, such an effect was not observed for the X4-tropic NL-4.3 clone. CV-N proved also markedly inhibitory to SIV (Table 1).
TABLE 1.
Antiviral activity of CV-N against a variety of retroviral strains in cell culture
| Viral strain(s) | Cell culture type | EC50 (μM) |
|---|---|---|
| HIV-1 | ||
| HE | CEM | 0.005 |
| IIIB | CEM | 0.002 |
| NL4.3 | CEM | 0.0007 |
| PBMC | 0.014 | |
| BaL | M/Mb | 0.16 |
| HIV-2 | ||
| ROD | CEM | 0.002 |
| BV-5061W | PBMC | 0.033 |
| SIV Mac251 | MT-4 | 0.017 |
| PBMC | 0.16 | |
| HIV-1 clade isolatesa (A, B, C, D, A/E, F, G, O) | PBMC | 0.013-0.16 |
The representative members of each HIV-1 clade and their X4 and/or R5 tropism have been indicated in Materials and Methods.
M/M, monocyte/macrophage.
FIG. 2.
HIV-1 p24 production in PBMC cultures that are infected by different HIV-1 strains in the presence of various CV-N concentrations. The NL-4.3 strain is X4-tropic; the Ba-L, ETH2220, ID 12, BZ163, and UG273 strains are R5-tropic. The p24 levels in the PBMC supernatants are expressed as percentages of p24 production in the untreated virus-infected control cultures.
Selection of CV-N-resistant HIV-1 strains.
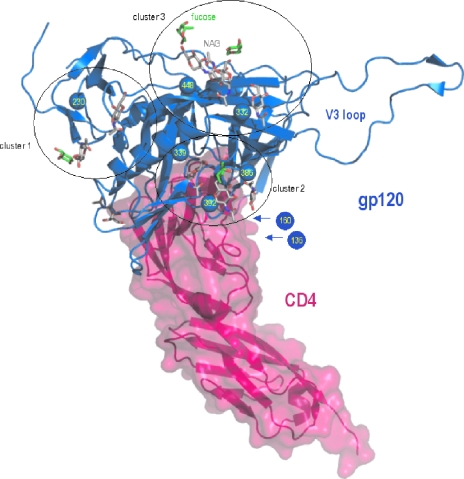
To reveal the drug resistance profile of CV-N, three independent series of CV-N drug resistance selections were performed using an escalating drug selection schedule (Fig. 1). The highest possible CV-N concentration was 2 μg/ml due to toxicity of the drug at higher concentrations in the cell cultures. Selection series A and B were performed with HIV-1(IIIB) and selection series C with HIV-1 (NL-4.3). A total of seven virus isolates were taken during the selection process. A long time period was required to allow the continuous presence of higher drug levels without losing the virus upon passage of the cell cultures. In the first HIV-1(IIIB) selection series (Fig. 1A), the virus isolate taken at 0.1 μg/ml CV-N did not show mutations in gp120, but when taken at 1 μg/ml, four mutations at N-glycosylation sites in gp120 were observed (Table 2). In the second HIV-1(IIIB) selection series (Fig. 1B), an isolate in the presence of 1 μg/ml CV-N contained three mutations. When two additional but independent isolates were taken late in the selection process (2 μg/ml CV-N), three additional N-glycosylation site mutations had appeared. In the third HIV-1(NL-4.3) selection series (Fig. 1C), one mutation was found under 0.1 μg/ml CV-N exposure conditions in one of the two independent isolates. Two mutations at N-glycosylation sites were detected in the gp120 of the virus isolate taken in the presence of 1.5 μg/ml CV-N. Taking all virus isolates together, a total of eight CV-N-related N-glycosylation site mutations in gp120 were found. Six of them deleted a high-mannose type N-glycan. Two mutations deleted a complex/hybrid type N-glycan according to Leonard et al. (21) and Gallaher et al. (16) (i.e., the glycan on positions N-136 and N-160) (Table 2). The deleted N-glycans are plotted on the crystallographic structure of the HIV-1 gp120/CD4 complex according to Kwong et al. (19) and Huang et al. (18) (Fig. 3).
TABLE 2.
N-glycosylation sites in HIV-1 gp120 deleted in mutant HIV-1(IIIB or NL4.3) strains that emerged under escalating CV-N exposurea
| N-glycosylation sites in HIV-1 gp120b | Glycan naturec | Mutation(s) detected at CV-N concn (μg/ml) (selection series, code no.)
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0.1 (1, 107-10) | 1 (1, 113-24) | 1 (2, 108-4) | 2 (2a, 113-14) | 2 (2b, 113-15) | 0.1 (3, 107-9) | 1.5 (3, 113-23) | ||
| 136NDT138 | H/C | 136N/K | ||||||
| 160NIS162 | H/C | 162S/N | 162S/N | 162S/N | ||||
| 230NKT232 | M | 232T/M | 232T/M | |||||
| 332NIS334 | M | 332I | 334N | 334N | 334N | |||
| 339NNT341 | M | 339S | 339S | 339S | 339S | 341A | ||
| 386NST388 | M | 386D | ||||||
| 392NST394 | M | 392N/T | 392N/T | |||||
| 448NIT450 | M | 448N/Y | 448S | 448S | 448S | |||
The wild-type virus used for the drug resistance selections was HIV-1(IIIB) for selection series 1 and 2 and HIV-1(NL4.3) for selection series 3.
Numbering is according to the method of Kwong et al. (19). Boldface type indicates the nature of the amino acids in the glycocylation motifs.
FIG. 3.
Ribbon representation of the HIV-1 gp120 envelope glycoprotein (blue) bound to CD4 receptor (pink), as found in a composite of X-ray crystal structures with Protein Data Bank identification codes 1GC1 (19, 32) and 2B4C (18). Sugar moieties are displayed as sticks, with C atoms colored white for N-acetyl-d-glucosamine and green for fucose. The Cbeta atoms of the glycosylated N residues that are mutated in the presence of CV-N are displayed as spheres and have been labeled. The PyMOL molecular graphics system (http://www.pymol.org; DeLano Scientific LLC, San Carlos, CA) was used for visualization and picture creation. Clustering was done manually on the basis of proximity of the glycosylated sites. Residues 127 to 192 making up loops V1 and V2 are absent in the crystal structures of gp120. Therefore, the approximate positions of the N-136 and N-160 residues are indicated by arrows.
Wild-type HIV-1-infected cell cultures that were subject to subcultivations under similar experimental conditions did not show any random glycan deletions at N-glycosylation sites of gp120. These observations point to the highly specific appearance of glycan deletions under CV-N drug pressure.
Phenotypic sensitivity/resistance determination of virus strains containing N-linked glycan deletions in gp120.
Seven virus isolates were taken during the CV-N escalating selection process, and their gp120 proteins were genotypically characterized. To reveal the phenotypic sensitivity/resistance profile of the mutant virus isolates against CV-N and other mannose-specific lectins, five of the six isolates that contained N-glycan deletions in the gp120 envelope were further investigated and phenotypically characterized (Table 3).
TABLE 3.
Phenotypic sensitivity of mutant virus strains to CV-N and other entry inhibitors
| Entry inhibitor | EC50 (μM) for:
|
||||||
|---|---|---|---|---|---|---|---|
| HIV-1(IIIB) (WT)a | HIV-1(NL-4.3) (WT) | CV-N concn (μg/ml) (selection series, code no.) (HIV-1 strain)
|
|||||
| 1 (1, 113-24) (IIIB) | 1 (2, 108-4) (IIIB) | 2 (2a, 113-14) (IIIB) | 2 (2b, 113-15) (IIIB) | 1.5 (3, 113-23) (NL-4.3) | |||
| CV-N | 0.002 ± 0.001 | 0.0007 ± 0.0005 | 0.005 ± 0.003 | 0.016 ± 0.003 | 0.040 ± 0.021 | 0.039 ± 0.023 | 0.005 ± 0.0 |
| GNA | 0.005 ± 0.001 | 0.011 ± 0.009 | 0.017 ± 0.01 | 0.140 ± 0.030 | 0.116 ± 0.120 | 0.076 ± 0.064 | 0.018 ± 0.028 |
| HHA | 0.004 ± 0.002 | 0.013 ± 0.004 | 0.020 ± 0.005 | 0.130 ± 0.015 | 0.068 ± 0.018 | 0.042 ± 0.017 | 0.056 ± 0.036 |
| LOA | 0.003 ± 0.001 | 0.014 ± 0.003 | 0.071 ± 0.028 | 0.031 ± 0.0 | 0.030 ± 0.026 | ||
| UDA | 0.138 ± 0.041 | 0.053 ± 0.004 | 0.213 ± 0.056 | 0.287 ± 0.172 | 0.115 ± 0.0 | 0.218 ± 0.016 | 0.138 ± 0.056 |
| CA | 0.021 ± 0.021 | 0.082 ± 0.044 | 0.400 ± 0.0 | 0.220 ± 0.084 | 0.320 ± 0.112 | 0.160 ± 0.112 | |
| 2G12b | 1.1 ± 0.12 | >50 | >50 | >50 | >50 | ||
| T-20b | 0.031 ± 0.017 | 0.015 ± 0.0 | 0.0092 ± 0.004 | 0.045 ± 0.007 | 0.012 ± 0.005 | 0.011 ± 0.00 | 0.0060 ± 0.0028 |
| DS 5000b | 0.33 ± 0.12 | 0.25 ± 0.07 | 0.3 ± 0.0 | 1.4 ± 0.21 | 1.3 ± 0.35 | 1.3 ± 0.35 | 1.35 ± 0.21 |
| AMD3100b | 0.05 ± 0.03 | 0.0072 ± 0.0011 | 0.021 ± 0.016 | 0.070 ± 0.028 | 0.06 ± 0.0 | 0.065 ± 0.007 | 0.014 ± 0.006 |
WT, wild type.
Data are expressed in μg/ml.
The phenotypic resistance of CV-N against the five mutant virus isolates ranged from 2- to 3-fold (low-level resistance) to 10- to 20-fold (high-level resistance). The virus strains with the lowest resistance profile contained combinations of deletions of glycans at N positions 160, 332, 339, 386, and 448. It should be noted that the mutant amino acids at positions 160 and 448 were mixtures with their wild-type amino acids. The most pronounced degree of resistance occurred when additional glycan deletions appeared at N positions 230 (mixture with wild type) and 392 (mixture with wild type). In these virus strains, the glycan deletion at N-448 was invariably present as a pure amino acid mutation, as was also the case for HIV-1 isolate CV-N-1.0 (2), which contained the intact N-glycan at position 160 but lost the glycan at N-448. From these data, N positions 230, 392, and 448 seem to be important to provoke significant CV-N resistance.
The HIV-1 isolates with low-level (2- to 3-fold) CV-N resistance showed a 2- to 10-fold degree of resistance to the mannose-specific plant lectins, depending on the nature of the lectin (GNA, HHA, LOA, CA), but kept full sensitivity to the GlcNAc-specific Urtica dioica agglutinin (UDA) plant lectin (Table 3). Interestingly, the MAb 2G12 completely lost its inhibitory potential at 50 μg/ml (>50-fold resistance) against these mutant virus strains. The other entry inhibitors generally kept their antiviral potential or even became more inhibitory, a trend that was particularly striking for T-20 and AMD3100 (Table 3). The virus isolates that showed the highest levels of CV-N resistance (10- to 20-fold) were also 10- to 30-fold cross-resistant to the mannose-specific plant lectins and showed again the previously noted >50-fold resistance for 2G12. In 2 of 3 cases, T-20 still showed a threefold-more increased inhibitory potential than against the wild type. DS-5000 was four- to fivefold less inhibitory than against the wild type (Table 3). Again, the GlcNAc-specific UDA fully kept its inhibitory activity against all high-level CV-N-resistant virus strains.
Genotypic and phenotypic properties of a broad variety of 18 HIV-1 strains with deletions of N-glycans in their gp120 envelopes.
It is interesting not only to reveal the resistance/sensitivity profile of mutant HIV-1 strains that appeared under CV-N drug pressure but also to determine the resistance/sensitivity profile of CV-N against mutant virus strains that were selected in the presence of other CBAs. Therefore, a variety of 18 HIV-1(IIIB) strains that have been previously isolated in drug-escalating selection experiments that were carried out in the presence of the mannose-specific plant lectins HHA and GNA, the GlcNAc-specific plant lectin UDA, and the low-molecular weight mannose-specific artificial lectin pradimycin A (PRMA) (2, 4-6) were included in the study. All of these virus strains contained deletions in 4 to 9 N-glycans of their gp120 envelopes. The genotypic profile and relative resistance to CV-N and four mannose-specific plant lectins were shown in Tables 4 and 5. Generally, those virus strains that showed the highest resistance to CV-N (29- to 83-fold) also proved to be the most resistant to the plant lectins (32- to 926-fold). Interestingly, the virus strains that were isolated in the presence of GNA and HHA (isolates 14 to 18) generally showed a much more pronounced phenotypic resistance spectrum against the plant lectin than against CV-N. It may not be a coincidence that the N-448 glycan was invariably deleted in those virus isolates that showed the most pronounced CV-N resistance (i.e., isolates 18, 9, 11, 12, and 13) in addition to the N-230 and N-392 glycan deletions (Tables 4 and 5). When we plotted the relative resistance values obtained for CV-N against those obtained for GNA, HHA, LOA, and CA, we observed no significant correlation (r range between 0.085 and 0.330) (Table 6). However, when the relative resistance values obtained for the different mannose-specific plant lectins were plotted against each other, strong correlation coefficients were observed (r range between 0.785 and 0.972).
TABLE 4.
Genotypic properties of HIV-1 strains mutated at glycosylation sites in gp120
| N-glycan position | Result for HIV-1(IIIB) isolatea:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| 88 | +/− | +/− | + | + | + | + | + | + | +/− | + | +/− | |||||||
| 136 | ||||||||||||||||||
| 141 | ||||||||||||||||||
| 156 | ||||||||||||||||||
| 160 | + | + | ||||||||||||||||
| 186 | + | |||||||||||||||||
| 197 | ||||||||||||||||||
| 230 | +/− | +/− | +/− | + | +/− | +/− | +/− | + | + | + | + | +/− | + | + | ||||
| 234 | +/− | +/− | +/− | +/− | + | + | + | + | + | + | + | + | + | + | + | |||
| 241 | ||||||||||||||||||
| 262 | ||||||||||||||||||
| 276 | +/− | +/− | ||||||||||||||||
| 289 | + | + | + | + | +/− | + | + | + | +/− | + | +/− | |||||||
| 295 | +/− | + | + | + | +/− | + | +/− | + | + | |||||||||
| 301 | +/− | + | + | + | + | + | + | + | + | + | + | |||||||
| 332 | +/− | + | + | + | + | + | + | + | +/− | + | + | |||||||
| 339 | + | + | + | + | + | + | + | +/− | + | + | + | |||||||
| 356 | ||||||||||||||||||
| 386 | + | |||||||||||||||||
| 392 | + | + | +/− | + | + | + | +/− | + | + | |||||||||
| 397 | ||||||||||||||||||
| 406 | + | + | + | |||||||||||||||
| 448 | + | +/− | +/− | + | + | + | ||||||||||||
| 463 | ||||||||||||||||||
1, PRMA-8-1; 2, PRMA-30-1; 3, PRMA-8-2; 4, PRMA-30-2; 5, PRMA-30-3; 6, PRMA-30-4; 7, UDA-20; 8, UDA-25-1; 9, UDA-50-1; 10, UDA-15-2; 11, UDA-50-2; 12, UDA-100-2; 13, UDA-200-2; 14, GNA-2.1; 15, HHA-2.2; 16, HHA-500CS; 17, HHA-500SN; 18, GNA-500CS; +/−, mixture of wild-type and mutant virus at the indicated amino acid position; +, deletion of an N-glycan at the indicated amino acid position.
TABLE 5.
Antiviral activity of HIV-1 strains mutated at glycosylation sites in gp120
| Compound | EC50 (μM) for wild type | Antiviral activity (fold resistance) for virus isolatea:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | ||
| HHA | 0.006 | 6.3 | 19 | 1.9 | 17 | 55 | 28 | 0.8 | 83 | 83 | 15 | 133 | 110 | 39 | 75 | 67 | 262 | 919 | 490 |
| GNA | 0.010 | 5.2 | 8.3 | 1.9 | 10 | 40 | 27 | 0.3 | 31 | 33 | 6.1 | 39 | 44 | 117 | 72 | 78 | 99 | 926 | 581 |
| CA | 0.024 | 10 | 11 | 8.1 | 6.7 | 27 | 13 | 1.9 | 42 | 45 | 32 | 39 | 32 | 45 | 85 | 58 | 140 | 233 | 133 |
| LOA | 5.8 | 3.8 | 5.0 | 5.5 | 1.3 | 32 | 33 | 10 | 102 | 158 | 80 | 43 | 16 | 109 | 228 | 246 | |||
| CV-N | 0.003 | 1.5 | 7.4 | 0.68 | 4.2 | 7.9 | 6.8 | 0.3 | 43 | 37 | 1.3 | 57 | 50 | 83 | 2.9 | 6.3 | 5.1 | 29 | 11 |
1, PRMA-8-1; 2, PRMA-30-1; 3, PRMA-8-2; 4, PRMA-30-2; 5, PRMA-30-3; 6, PRMA-30-4; 7, UDA-20; 8, UDA-25-1; 9, UDA-50-1; 10, UDA-15-2; 11, UDA-50-2; 12, UDA-100-2; 13, UDA-200-2; 14, GNA-2.1; 15, HHA-2.2; 16, HHA-500CS; 17, HHA-500SN; 18, GNA-500CS.
TABLE 6.
Paired correlations of the relative phenotypic resistance between CV-N and mannose-specific plant lectins against a variety of 18 HIV-1 strains containing glycan deletions in gp120
| Lectin comparison | r |
|---|---|
| CV-N vs GNA | 0.100 |
| CV-N vs HHA | 0.111 |
| CV-N vs LOA | 0.330 |
| CV-N vs CA | 0.085 |
| GNA vs HHA | 0.972 |
| GNA vs LOA | 0.822 |
| GNA vs CA | 0.882 |
| HHA vs CA | 0.932 |
| LOA vs CA | 0.785 |
Effect of mannan on the antiviral and cytostatic/cytotoxic activity of cyanovirin.
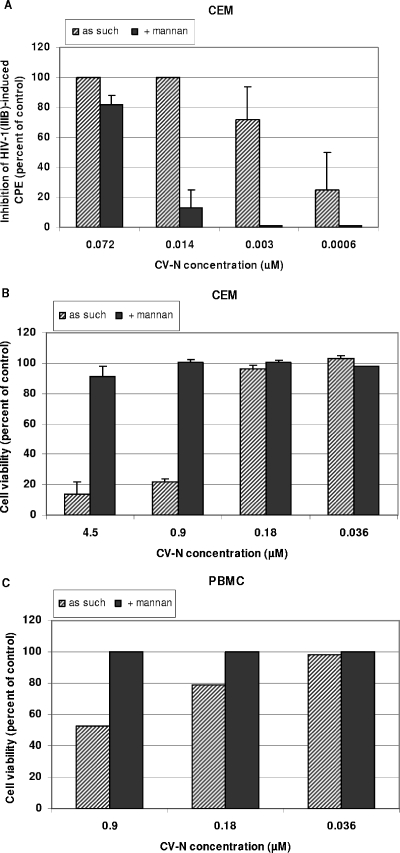
Since CV-N has mannose-recognizing properties, its biological effects are expected to be influenced in the presence of mannan. Therefore, the effect of mannan on the CV-N properties was investigated. The antiviral activity of CV-N against HIV-1 in CEM cell cultures was determined in the absence or presence of 2.5 mg/ml mannan. The addition of mannan decreased the antiviral activity of CV-N by ∼20-fold in CEM cell cultures (Fig. 4A). When the number of cells were determined at 3 to 4 days after seeding of CEM cells or interleukin-2-exposed PBMC in culture medium in the presence of a variety of CV-N concentrations, it was found that CV-N markedly inhibited CEM cell proliferation at relatively low drug concentrations (50% inhibitory concentration, 0.5 to 1 μM) (Fig. 4B). CV-N was also relatively toxic in PBMC cultures, as determined by trypan blue staining at day 3 post CV-N exposure (Fig. 4C). The cytostatic/cytotoxic activity of CV-N in CEM and PBMC cultures could also be markedly reversed in the presence of mannan (Fig. 4B,C).
FIG. 4.
Effect of 2.5 mg/ml mannan on the anti-HIV-1 activity of CV-N in CEM cell cultures (A) and on cell viability in CEM (B) and PBMC (C) cell cultures. CPE, cytopathic effect.
Mitogenic activity of cyanovirin in PBMC cultures.
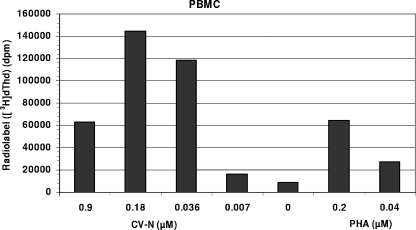
Often, lectins are endowed with mitogenic activity (i.e., PHA). Therefore, we wanted to investigate whether CV-N is also endowed with mitogenic activity. PBMC cultures were exposed to [3H]thymidine for 20 h. After this time period, the TCA-insoluble radiolabel was quantified in the cell cultures (Fig. 5). Unstimulated PBMC incorporated 9,124 disintegrations per minute, whereas the presence of 0.9 μM CV-N inhibited the incorporation but 0.18, 0.036, and 0.007 μM CV-N markedly and dose-dependently enhanced [3H]thymidine incorporation. The dose of 0.9 μM showed toxicity (Fig. 4) and thus provides an explanation for the inhibition of the mitogenic activity by CV-N at this concentration, whereas at 0.18 μM, no toxicity was observed. Thus, at 0.18, 0.036, and 0.007 μM, CV-N had stimulation indices (SI) of 16, 13, and 18, respectively. These data were derived from triplicate experiments for two different CV-N batches. In comparison, when PBMC was stimulated with PHA at 0.2 and 0.04 μM, SI of 12 and 3.1 were obtained (Fig. 5). Such a pronouncedly increased SI was not observed for the mannose-specific plant lectins GNA and HHA at concentrations up to 100 μM (data not shown).
FIG. 5.
Mitogenic activity of CV-N in PBMC. [3H]thymidine ([3H]dThd) incorporation in TCA-insoluble cell material was quantified after a 3-day incubation period of PBMC with CV-N or PHA, followed by 20 h of exposure to [3H]thymidine. dpm, disintegrations per minute.
Effect of cyanovirin on the expression of cellular activation markers in PBMC cultures.
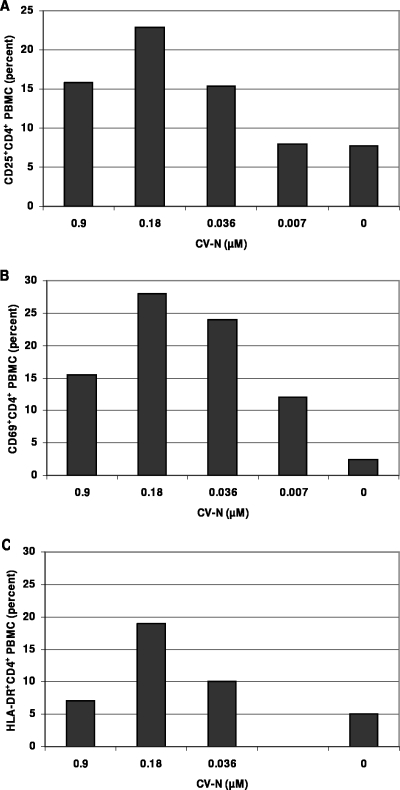
Since mitogenic activity of drugs can result in the induction and expression of activation markers, PBMC were stained for the activation markers CD69, CD25, and HLA-DR. It became apparent that CV-N stimulated the expression of these markers in a dose-dependent manner. Indeed, unstimulated PBMC at 3 days in cell culture contained only 2.3% CD69+ CD4+ T cells, whereas in the presence of 0.9, 0.18, 0.036, and 0.007 μM CV-N, the percentages of activated cells were markedly increased (Fig. 6A). Comparable stimulatory effects were also noticed for CD25 and HLA-DR expression. Indeed, unstimulated PBMC at 3 days in culture contained 7.7% CD25+ CD4+ cells, whereas in the presence of 0.9, 0.18, 0.036, and 0.007 μM CV-N, this number was substantially increased (Fig. 6B). Unstimulated PBMC contained 5% HLA-DR+ CD4+ cells, whereas in the presence of 0.9, 0.018, 0.036 μM CV-N, this number increased again (Fig. 6C). In all cases, stimulation of the activation markers was less pronounced at the highest CV-N concentration (0.9 μM) than at lower concentrations, obviously due to the masking of the stimulatory effect by the cell toxic effect at 0.9 μM. A similar activation phenomenon has also been observed on CD8+ T cells. In comparison, PHA-stimulated PBMC (at 0.04 μM) contained 13% CD69+ CD4+, 22% CD25+ CD4+, and 8% HLA-DR+ CD4+ cells. Moreover, when PHA and CV-N were added to the cultures, a further enhancement of the activation markers was observed. For example, exposure to PHA solely resulted in 22% CD25+ CD4+ cells, but in combination with CV-N at 0.9, 0.018, and 0.036 μM this percentage changed to 14%, 40%, and 35% CD25+ CD4+ cells, respectively. Thus, CV-N as such has not only mitogenic properties (see above), but in combination with PHA, it can even further enhance cellular activation. These effects were not observed with the mannose-specific plant lectins HHA and GNA up to 10 μM (data not shown).
FIG. 6.
Effect of CV-N on the expression of three activation markers in PBMC. (A) CD25+ CD4+ cell population; (B) CD69+ CD4+ cell population; (C) HLA-DR+ CD4+ cell population.
DISCUSSION
Cyanovirin is the very first mannose-specific lectin that has been isolated from a prokaryotic (Cyanobacterium) organism and found to be endowed with a pronounced anti-HIV activity in cell culture (13). CV-N efficiently and consistently inhibits a wide variety of virus strains irrespective of the coreceptor usage (X4 or R5), the nature of the virus-infected target cell (cell lines, PBMC, macrophages), and the nature of the clinical virus isolate (covering a broad variety of HIV-1 clades and HIV-2 and SIV strains). In this respect, CV-N is superior in antiviral potency to other mannose-specific lectins, such as the Amaryllidaceae lectins GNA and HHA. There exists also a smaller range of variation of CV-N potency versus HHA and GNA potency when the drug sensitivities of the different HIV clades are considered (5). The plant lectins, however, are tetrameric proteins (4 × 12.5 kDa) with specificity for α(1-3)-mannose (GNA) or α(1-3)/α(1-6)-mannose (HHA) oligomers (24, 28). It is unclear whether the superior activity of CV-N both with regard to its antiviral potency and its antiviral variability spectrum is due to its differential mannose oligomer specificity [man α(1-2) oligomers], its relatively small size (a dimer of 2 × 11 kDa), and/or a tighter binding to the gp120 glycans than HHA and GNA. However, the LOA and CA lectins are also dimeric proteins (2 × 12.5 kDa) that are only slightly bigger than CV-N but endowed with a mannose oligomer specificity comparable to that of GNA and HHA. These dimeric plant lectins also showed, like CV-N, a more consistent suppression of the different HIV-1 clade isolates (5). The α(1-2) mannose oligomers are located at the outside area of the high-mannose N-glycans and thus are easily reachable by high-molecular-size drugs. Instead, the α(1-3) and α-(1-6) mannose configurations are rather internally located in the N-glycans and may be less easily reached by large-size CBAs. Therefore, the relatively small size of CV-N together with its α(1-2) mannose oligomer specificity may be a prerequisite for its potent and consistent antiviral suppression of a broad spectrum of HIV variants.
The consistent suppression of a wide variety of virus clade isolates by CV-N is also suggestive of a relatively high genetic barrier of CV-N. This is confirmed by our drug resistance selection experiments. Indeed, it took a long time period before CV-N concentrations could be substantially increased in the dose escalation selection experiments without losing the virus in the cell cultures. Moreover, phenotypic drug resistance determinations with the CV-N-resistant HIV-1 strains revealed that, even in the concomitant presence of 4 N-glycan deletions in gp120, CV-N still showed a pronounced inhibitory potency. The high genetic barrier of CV-N against HIV is also corroborated by the findings that most of the mutant HIV-1 isolates that were obtained earlier during GNA, HHA, UDA, or PRMA resistance selections (2, 4-6) kept pronounced sensitivity for CV-N. Only when more than 5 N-glycans were deleted in the gp120 envelope of HIV-1 did CV-N lose significant antiviral potency (Tables 4 and 5).
Interestingly, HIV-1(IIIB) gp120 consists of ∼24 glycosylation sites, 11 of them carrying high-mannose N-glycans and 13 bearing hybrid or complex type N-glycans according to Leonard et al. (21) and Gallaher et al. (16). The high-mannose type glycans have a core of a α(1-3)/α(1-6) mannose oligomer but an α(1,2) mannose oligomer periphery. The complex/hybrid type glycans mainly lack α(1,2) mannose oligomers. In this perspective, it is striking that from all eight N-glycan deletions observed in a total of five independent CV-N-exposed HIV-1 strains, six were high-mannose type N-glycans and only two were hybrid/complex type N-glycans. In fact, they are part of a cluster of six hybrid/complex mannose type N-glycans in the V1/V2 loop of gp120. Such predominant selectivity to delete high-mannose type glycans under CV-N drug pressure has not been observed for the α(1-3)/α(1-6) mannose-specific plant lectins (HHA and GNA) in previous drug selection studies. Also here, high-mannose type glycans were statistically significantly more affected than complex/hybrid type glycans but not as striking as observed for CV-N (2, 4). Mapping of the N-glycan deletions on the three-dimensional structure of HIV-1 gp120 (18, 19, 32) revealed four clustered groups of glycan deletions under CV-N drug pressure. They are marked as three circles on the gp120 structure in Fig. 3 and two arrows that locate the N-136 and N-160 glycan deletions among a cluster of 6 complex/hybrid mannan type glycans in V1/V2 because they are not clearly resolved in the crystal structure and, thus, their exact location in gp120 is uncertain. Interestingly, each of the three N-glycan deletions that are associated with the highest CV-N drug resistance in our studies (i.e., N-448, N-230, and N-339) belongs to a different clustered area of N-glycans. The role of the N-136 and N-160 glycans, being the only complex mannose type glycans in CV-N resistance, is currently unclear. Interestingly, the single virus strain that was selected in the presence of CV-N by Witvrouw et al. (31) showed glycan deletions at N positions 332, 392, 397, 406, and 448. The deletions at 392, 397, and 406 were due to a deletion of a 13-amino-acid stretch 394-TWFNSTWSTEGSN-406 affecting 3 glycosylation sites at the same time. This virus strain demonstrated a pronounced resistance toward the MAb 2G12 and a slight loss of susceptibility to the HIV adsorption inhibitor DS-5000. No loss of susceptibility was observed for the entry inhibitors AMD3100 and enfuvirtide (T20). Three of the N-glycan deletions observed in the mutant virus strain isolated by Witvrouw et al. (31) were also found in several of the virus isolates reported in our study (N-332, N-392, and N-448), which resulted in a 140-fold phenotypic CV-N resistance (31). Thus, our studies made it clear that HIV-1 has a variety of mutational options to escape CV-N drug pressure. However, there is a predominant preference for high-mannose type glycans, and some of the high-mannose type N-glycans are mutated much more frequently than other N-glycans. It is also clear that deletions of N-glycans in gp120 rather than mutations at other (nonglycosylated) amino acid areas in gp120 are obligatory for the virus to become phenotypically resistant to CV-N. This also explains the cross-resistance of CV-N-exposed mutated virus strains to mannose-specific plant lectins but not to other entry inhibitors, such as the adsorption inhibitor DS-5000, the CXCR4 agonist SDF-1, or the CXCR4 antagonist AMD3100. Also, the gp41 fusion inhibitor T20 is not negatively affected by the glycan deletions in gp120.
The MAb 2G12 has also been shown to exert an α(1-2) mannose oligomer specificity (14, 23). However, whereas a large number of N-glycan deletions are required to afford a moderate decrease of CV-N sensitivity against the mutated virus strains, one or two glycan deletions were already sufficient for 2G12 to completely lose its antiviral potential. These observations demonstrate that neutralizing antibodies such as 2G12 have a low genetic barrier. This is obviously due to their high specificity and selectivity of interaction with HIV-1 gp120. In fact, the 2G12 epitope comprises N-glycans located at N-332 and N-339. These two sites are precisely those two glycans that were present on the silent face of gp120 and that were deleted in the CV-N-resistant virus strains as well (23). However, it is clear that CV-N binds to many more areas on gp120 than does 2G12 (Fig. 2), likely including additional epitopes on gp120 for CV-N interaction. This is in agreement with the observation that CV-N can fully block binding of 2G12 to gp120 but not vice versa (15, 23). Therefore, the antiviral activity of CBAs such as CV-N should be endowed with a broad universally neutralizing capacity against a variety of wild-type and mutant viruses. Instead, the specific interaction of any monoclonal antibody with its target glycoprotein can be easily compromised and annihilated by only one or two well-defined amino acid mutations in gp120.
Prokaryotic, fungal, invertebrate, and plant lectins have all been suggested as potential microbicidal drugs (7). In fact, CV-N is one of the very few entry inhibitors that have been evaluated for its microbicidal action in simian human immunodeficiency virus-infected female Macaca fascicularis monkeys through vaginal virus transmission (25). CV-N was found to be highly protective in this in vivo study. CV-N is also considered to be produced by commensal bacteria (i.e., Lactobacillus sp.) that should colonize the female vagina, thereby creating a microbicidal environment (17, 20). Lectins such as CV-N and HHA have the advantage of binding tightly to the HIV gp120 envelope, which results in a virtually irreversible inactivation of the virus. However, due to their protein nature, it may be expensive to produce proteins such as CV-N in large quantities. Besides, it would be important to make a careful choice on the nature of the lectin that should be selected as a microbicidal drug candidate. Indeed, some lectins have been shown to be endowed with unfavorable properties such as human red blood cell agglutination, mitogenic stimulation of PBMC, inflammatory activity, and cellular toxicity, etc. (for an overview, see references 24 and 28). In our study, CV-N was found to inhibit lymphocyte cell proliferation at relatively low concentrations (i.e., 2 to 10 μg/ml), that is, at a concentration which is at least 1 to 2 orders of magnitude lower than that of several plant lectins (such as HHA, GNA, and UDA) (1, 5). Intriguingly, at concentrations that were ∼5- to 10-fold lower than those fully suppressing virus infection in PBMC cultures, viral p24 production was rather stimulated. Such a phenomenon was consistently seen for clinical isolates but not for NL-4.3 (Fig. 2). Moreover, CV-N also proved mitogenic at (subtoxic) compound concentrations that are almost comparable with those of PHA. This is another lectin with complex sugar specificity that is commonly used for stimulation (activation) of human PBMC (Fig. 5). Whereas the antiproliferative activity of CV-N in CEM (and MT-4) cell cultures and PBMC can be reversed by the addition of mannan, we found that the mitogenic activity of CV-N could not be reversed by mannan (data not shown), thus raising the possibility that the latter is independent of the carbohydrate specificity of the lectin. Thus, our observations indicate that CV-N exerts carbohydrate-independent mitogenic activity at relatively low concentrations (i.e., 0.18 μM). Higher drug concentrations (i.e., starting from ∼1 μM and higher) may mask the mitogenic activity due to a carbohydrate-related antiproliferative activity of the drug. The latter effect was observed in both CEM/MT-4 and PBMC cultures and confirmed for two different CV-N batches. The plant lectins HHA, GNA, and UDA do not show significant mitogenic nor antiproliferative activity in cell culture (28; and data not shown). These observations indicate that the antiviral activity of lectins can be dissected from some other biological activities depending on the nature of the lectin. The highly potent antiviral activity of CV-N in cell culture occurs at concentrations that are ∼50-fold lower than those necessary for its antiproliferative and mitogenic activities. However, the application of CV-N as a microbicidal drug will probably require careful monitoring of the drug from a safety perspective, especially in the light of its pronounced mitogenic activity and stimulatory effect on cellular differentiation markers.
In conclusion, CV-N was found to be a potent inhibitor of infection of lymphocytes and macrophages by a broad variety of virus isolates. Its resistance profile is rather unique and differs from other nonlectin entry inhibitors in that it selects for predominantly high-mannose type glycan deletions in the HIV gp120 envelope. HIV can follow different mutational patterns to escape from CV-N, although deletion of certain high-mannose type N-glycosylation sites is preferred. Although CV-N has a potent antiviral activity and a high genetic barrier, it is not devoid of potential side effects that are unrelated to its carbohydrate binding properties. Careful monitoring of potential side effects should be required if CV-N is to be applied for treatment of HIV-exposed/infected individuals.
Acknowledgments
We are grateful to Ann Absillis, Lizette van Berckelaer, Yoeri Schrooten, Sandra Claes, Rebecca Provinciael, and Eric Fonteyn for excellent technical assistance and Christiane Callebaut for dedicated editorial assistance.
The research was supported by the Geconcerteerde Onderzoeksacties (GOA no. 05/19), the Flemish Foundation for Scientific Research (no. G-0267-04), the European Commission (René Descartes Prize-2001 HPAW-2002-90001 and EMPRO 503558 of the 6th Frame Work Programme), and the Centers of Excellence from the K.U. Leuven (EF/05/15).
REFERENCES
- 1.Balzarini, J., S. Hatse, K. Vermeire, K. Princen, S. Aquaro, C.-F Perno, E. De Clercq, H. Egberink, G. Vanden Mooter, W. Peumans, E. Van Damme, and D. Schols. 2004. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrob. Agents Chemother. 48:3858-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzarini, J., K. Van Laethem, S. Hatse, K. Vermeire, E. De Clercq, W. Peumans, E. Van Damme, A.-M. Vandamme, A. Bolmstedt, and D. Schols. 2004. Profile of resistance of human immunodeficiency virus to mannose-specific plant lectins. J. Virol. 78:10617-10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzarini, J. 2005. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infect. Dis. 5:726-731. [DOI] [PubMed] [Google Scholar]
- 4.Balzarini, J., K. Van Laethem, S. Hatse, M. Froeyen, E. Van Damme, W. Peumans, E. De Clercq, and D. Schols. 2005. Marked depletion of glycosylation sites in HIV-1 gp120 under selection pressure by the mannose-specific plant lectins of Hippeastrum hybrid and Galanthus nivalis. Mol. Pharmacol. 67:1556-1565. [DOI] [PubMed] [Google Scholar]
- 5.Balzarini, J., K. Van Laethem, S. Hatse, M. Froeyen, W. Peumans, E. Van Damme, and D. Schols. 2005. Carbohydrate-binding agents cause deletions of highly conserved glycosylation sites in HIV gp120. A new therapeutic concept to hit the Achilles heel of HIV. J. Biol. Chem. 280:41005-41014. [DOI] [PubMed] [Google Scholar]
- 6.Balzarini, J., K. Van Laethem, S. Hatse, A. Bugatti, M. Rusnati, S. Aquaro, C.-F. Perno, Y. Igarashi, T. Oki, and D. Schols. 2006. Chemotherapy of human immunodeficiency virus by pradimicin A: a novel therapeutic concept for treatment of glycosylated enveloped viruses. Abstr. 19th Int. Conf. Antivir. Res., abstr. 6.
- 7.Balzarini, J. Large-molecular-weight carbohydrate-binding agents as HIV entry inhibitors targeting glycoprotein gp120. Curr. Opin. HIV AIDS, in press. [DOI] [PubMed]
- 8.Bewley, C. A., K. R. Gustafson, M. R. Boyd, D. G. Covell, A. Bax, G. M. Clore, and A. M. Gronenborn. 1998. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol. 5:571-578. [DOI] [PubMed] [Google Scholar]
- 9.Bewley, C. A. 2001. Solution structure of a cyanovirin-N:Man alpha 1-2Man alpha complex: structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure 9:931-940. [DOI] [PubMed] [Google Scholar]
- 10.Bewley, C. A., S. Kiyonaka, and I. Hamachi. 2002. Site-specific discrimination by cyanovirin-N for α-linked trisaccharides comprising the three arms of Man(8) and Man(9). J. Mol. Biol. 322:881-889. [DOI] [PubMed] [Google Scholar]
- 11.Bolmstedt, A. J., B. R. O'Keefe, S. R. Shenoy, J. B. McMahon, and M. R. Boyd. 2001. Cyanovirin-N defines a new class of antiviral agent targeting N-linked, high-mannose glycans in an oligosaccharide-specific manner. Mol. Pharmacol. 59:949-954. [DOI] [PubMed] [Google Scholar]
- 12.Botos, I., B. R. O'Keefe, S. R. Shenoy, L. K. Cartner, D. M. Ratner, P. H. Seeberger, M. R. Boyd, and A. Wlodawer. 2002. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J. Biol. Chem. 277:34336-34342. [DOI] [PubMed] [Google Scholar]
- 13.Boyd, M. R., K. R. Gustafson, J. B. McMahon, R. H. Shoemaker, B. R. O'Keefe, T. Mori, R. J. Gulakowski, L. Wu, M. I. Rivera, C. M. Laurencot, M. J. Currens, J. H. Cardellina II, R. W. Buckheit, Jr., P. L. Nara, L. K. Pannell, R. C. Sowder II, and L. E. Henderson. 1997. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob. Agents Chemother. 41:1521-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Buron, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 15.Dwek, R. A., T. D. Butters, F. M. Platt, and N. Zitzmann. 2002. Targetting glycosylation as a therapeutic approach. Nat. Rev. Drug Discov. 1:65-75. [DOI] [PubMed] [Google Scholar]
- 16.Gallaher, W. R., J. M. Ball, R. F. Garry, A. M. Martin-Amedee, and R. C. Montelaro. 1995. A general model for the surface glycoproteins of HIV and other retroviruses. AIDS Res. Hum. Retrovir. 11:191-202. [DOI] [PubMed] [Google Scholar]
- 17.Giomarelli, B., R. Provvedi, F. Meacci, T. Maggi, D. Medaglini, G. Pozzi, T. Mori, J. B. McMahon, R. Gardella, and M. R. Boyd. 2002. The microbicide cyanovirin-N expressed on the surface of commensal bacterium Streptococcus gordonii captures HIV-1. AIDS 16:1351-1356. [DOI] [PubMed] [Google Scholar]
- 18.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagenaur, L. A., X. Liu, D. A. Simpson, K. Essenmacher, C. Parker, C.-H. Chang, D. Tsai, S. Rao, D. Hamer, T. P. Parks, P. P. Lee, and Q. Xu. 2005. Development of vaginal lactobacilli for mucosal delivery of a topical microbicide, cyanovirin-N (CV-N). Abstr. Int. Meet. Inst. Hum. Virol., abstr. S93.
- 21.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 22.Mori, T., R. H. Shoemaker, R. J. Gulakowski, B. L. Krepps, J. B. McMahon, K. R. Gustafson, L. K. Pannell, and M. R. Boyd. 1997. Biochem. Biophys. Res. Commun. 238:218-222. [DOI] [PubMed] [Google Scholar]
- 23.Scanlan, C. N., R. Pantophlet, M. R. Wormald, S. E. Ollmann, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha 1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon, N., and H. Lis (ed.). 2003. Lectins, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 25.Tsai, C. C., P. Emau, Y. Jiang, M. B. Agy, R. J. Shattock, A. Schmidt, W. R. Morton, K. R. Gustafson, and M. R. Boyd. 2004. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. AIDS Res. Hum. Retrovir. 20:11-18. [DOI] [PubMed] [Google Scholar]
- 26.Van Damme, E. J. M., A. K. Allen, and W. J. Peumans. 1987. Leaves of the orchid twayblade (Listera ovata) contain a mannose-specific lectin. Plant Physiol. 85:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Damme, E. J. M., A. K. Allen, and W. J. Peumans. 1988. Related mannose-specific lectins from different species of the family Amaryllidaceae. Physiol. Plant 73:52-57. [Google Scholar]
- 28.Van Damme, E. J. M., W. J. Peumans, A. Pusztai, and S. Bardocz (ed.). 1998. Handbook of plant lectins: properties and biomedical applications. John Wiley & Sons, Chichester, United Kingdom.
- 29.Van Laethem, K., Y. Schrooten, P. Lemey, E. Van Wijngaerden, S. De Wit, M. Van Ranst, and A.-M. Vandamme. 2005. A genotypic resistance assay for the detection of drug resistance in the human immunodeficiency virus type 1 envelope gene. J. Virol. Methods 123:25-34. [DOI] [PubMed] [Google Scholar]
- 30.Williams, D. C., Jr., J. Y. Lee, M. Cai, C. A. Bewley, and G. M. Clore. 2005. Crystal structures of the HIV-1 inhibitory cyanobacterial protein MVL free and bound to Man3GlcNAc2: structural basis for specificity and high-affinity binding to the core pentasaccharide from N-linked oligomannoside. J. Biol. Chem. 280:29269-29276. [DOI] [PubMed] [Google Scholar]
- 31.Witvrouw, M., V. Fikkert, A. Hantson, C. Pannecouque, B. R. O'Keefe, J. McMahon, L. Stamatatos, E. De Clercq, and A. Bolmstedt. 2005. Resistance of human immunodeficiency virus type 1 to the high-mannose binding agents cyanovirin N and concanavalin A. J. Virol. 79:7777-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 33.Yang, F., C. A. Bewley, J. M. Louis, K. R. Gustafson, M. R. Boyd, A. M. Gronenborn, M. Clore, and A. Wlodawer. 1999. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J. Mol. Biol. 288:403-412. [DOI] [PubMed] [Google Scholar]