Abstract
RNA-templated RNA replication is essential for viral or viroid infection, as well as for regulation of cellular gene expression. Specific RNA motifs likely regulate various aspects of this replication. Viroids of the Pospiviroidae family, as represented by the Potato spindle tuber viroid (PSTVd), replicate in the nucleus by utilizing DNA-dependent RNA polymerase II. We investigated the role of the loop E (sarcin/ricin) motif of the PSTVd genomic RNA in replication. A tertiary-structural model of this motif, inferred by comparative sequence analysis and comparison with nuclear magnetic resonance and X-ray crystal structures of loop E motifs in other RNAs, is presented in which core non-Watson-Crick base pairs are precisely specified. Isostericity matrix analysis of these base pairs showed that the model accounts for the reported natural sequence variations and viable experimental mutations in loop E motifs of PSTVd and other viroids. Furthermore, isostericity matrix analysis allowed us to design disruptive, as well as compensatory, mutations of PSTVd loop E. Functional analyses of such mutants by in vitro and in vivo experiments demonstrated that loop E structural integrity is crucial for replication, specifically during transcription. Our results suggest that the PSTVd loop E motif exists and functions in vivo and provide loss-of-function genetic evidence for the essential role of a viroid RNA three-dimensional motif in rolling-circle replication. The use of isostericity matrix analysis of non-Watson-Crick base pairing to rationalize mutagenesis of tertiary motifs and systematic in vitro and in vivo functional assays of mutants offers a novel, comprehensive approach to elucidate the tertiary-structure-function relationships for RNA motifs of general biological significance.
According to the “RNA world” scenario, the appearance of RNA molecules simultaneously capable of self-replication and information storage signaled a major milestone in the evolution of life (14, 24, 25). In modern biology, RNA replication is central to viral or viroid infection, as well as to the regulation of cellular gene expression. Virus-encoded RNA-dependent RNA polymerases play a major role in the replication of RNA viruses (10, 23). Cellular RNA-dependent RNA polymerases generate double-stranded RNAs as triggers for RNA silencing (1). Intriguingly, the DNA-dependent cellular RNA polymerases can also transcribe at least two types of RNA templates: viroid RNAs (11, 15, 59) and the human hepatitis delta virus (HDV) RNA (28, 60). The replication of viroid and HDV RNAs raises the question of whether the DNA-templated transcription machinery also replicates other cellular RNAs yet to be identified. Elucidating the replication mechanisms of these infectious RNAs should help address this question of profound biological interest.
Viroids are the smallest known nucleic acid-based infectious agents and self-replicating genetic units. Their “genomes” consist of single-stranded, circular RNAs ranging in size from 250 to 400 nucleotides (23). Viroids can replicate and spread throughout an infected plant, although they do not encode proteins, do not have encapsidation mechanisms, and do not require helper viruses. Furthermore, they cause devastating diseases by altering host gene expression and developmental processes (11, 59). Evidently, viroid RNA genomes contain all of the sequence and structural information needed to mediate or trigger the various functions associated with infection.
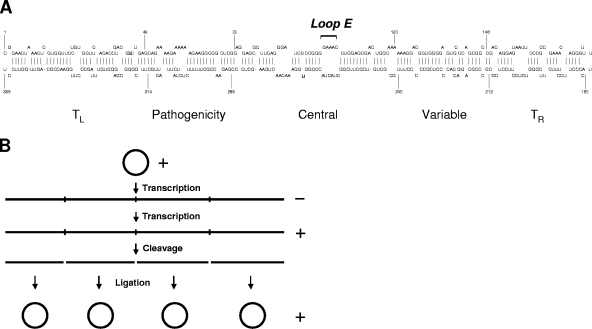
Potato spindle tuber viroid (PSTVd) is the type member of the family Pospiviroidae (8, 12). The PSTVd genome consists of 359 nucleotides and assumes a rod-shaped secondary structure in the native state (48) with five structural domains, as shown in Fig. 1A (26). This secondary structure, which is typical of viroids in the family Pospiviroidae, comprises many loops and bulges flanked by short Watson-Crick helices. Formation of this secondary structure is necessary for infection (62). During asymmetric rolling-circle replication of PSTVd (5), the plus circular strands serve as templates for the synthesis of concatemeric, linear minus strands, which then function as the replication intermediates for the synthesis of concatemeric, linear plus strands. These are subsequently cleaved into monomers and ligated into circular molecules (Fig. 1B). Without encoding proteins, PSTVd replicates in the nucleus of a host cell and therefore presents an ideal model to address the question of how the plant nuclear transcription machinery replicates an RNA template.
FIG. 1.
Secondary structure (A) and replication model (B) of PSTVd. (A) The rod-like secondary structure of PSTVd and the numbering of nucleotides are based on a report by Gross et al. (16). The five structural domains (26) include (i) the left terminal domain (TL, 1 to 46 and 315 to 359), (ii) the pathogenicity domain (47 to 73 and 286 to 314), (iii) the central domain (74 to 120 and 240 to 285), (iv) the variable domain (121 to 148 and 212 to 239), and (v) the right terminal domain (TR, 149 to 211). The loop E motif is in the central conserved region. (B) Asymmetric rolling-circle replication of PSTVd (5). The incoming circular monomeric plus-strand PSTVd serves as the initial template to synthesize linear, concatemeric minus-strand PSTVd. The latter functions as the replication intermediate to direct the synthesis of concatemeric plus-strand PSTVd, which is cleaved into unit length monomers and further ligated into circular forms.
Previous work has attempted to associate particular parts of the viroid RNA with specific steps of the replication cycle. In vitro transcription with potato nuclear extracts and a plus circular PSTVd RNA template suggested that nucleotides U359/C1 in the left terminal loop are the transcription initiation sites on the circular RNA (27). Subsequent site-directed mutagenesis, in combination with tomato infection studies, showed that the C1G mutation is stably maintained whereas U359G reverted to the wild type, suggesting that perhaps U359 is the bona fide initiation site (27). A metastable hairpin II (HPII) encompassing nucleotide sequences 227 to 237 and 318 to 328 was predicted to form during thermal denaturation of the PSTVd secondary structure (20, 46) and has been detected in vitro and in vivo (50). Mutations that are designed to inhibit the formation of HPII were found to always revert to the wild type in mechanically inoculated plants, suggesting that HPII is important for replication, perhaps as a binding site for an unknown cellular factor (6, 36, 41, 45). Many mutations have been mapped throughout the PSTVd genome that inhibit infection in mechanically inoculated plants and often were broadly interpreted to affect replication (19, 21, 39, 41).
The loop E motif, located in the central conserved region (CCR) of the central domain (Fig. 1A) of PSTVd, has been implicated in replication and other functions. This motif is similar to the loop E motifs of archaeal and eucaryal 5S rRNAs, which are also found in the conserved sarcin/ricin loops of 23S rRNA (29, 32). In vitro studies with longer-than-unit length PSTVd transcripts showed that cleavage and ligation occur between G95 and G96, which appear to involve the formation of a metastable tetraloop motif, followed by a conformational change into a stable loop E motif (3). A single nucleotide substitution, C259U, in loop E enhances replication by 5- to10-fold in cultured cells of tobacco (Nicotiana tabacum) (43). The enhanced replication in tobacco cells may account for the adaptation of PSTVd to this host (63, 66). The U257A substitution in the loop E motif also enables PSTVd to infect tobacco (43, 66) and confers lethal symptoms in tomato (42). These in vitro and gain-of-function mutational results suggest that loop E plays one or more crucial roles in viroid-plant interactions.
Several critical issues must be addressed in order to assign a specific function of viroid replication in vivo to a particular viroid structural element. First, whether a motif exists in vivo needs to be demonstrated. Second, reversions of mutated nucleotides to the wild type or gain-of-function mutations do not provide direct evidence for the role of the affected RNA structural element in a biological process. Loss-of-function mutations, on the other hand, will provide definitive proof linking the affected structural element to a function. Third, infection assays on whole plants do not reveal whether failure of infection of a mutant results from defects in a specific step of the replication cycle or in intra- or intercellular trafficking. To address these issues, it is imperative to integrate structural, genetic, cellular, biochemical, and molecular approaches.
Genetic alteration of an RNA structural element for functional studies requires precise understanding of its tertiary structure and the effects of mutations on that structure. However, while the secondary structures of viroids have been well established through thermodynamic, chemical mapping, mutational, and nuclear magnetic resonance (NMR) studies (9, 55, 62), much less is known about their tertiary structures and specific functions. Minimum-free-energy calculations (e.g., mfold; 67), coupled with conventional comparative sequence analysis, reveal RNA secondary structures comprising double-stranded helices formed by Watson-Crick base pairs, punctuated by so-called loops and bulges (hairpin, internal, and junction or multihelix loops) defined by lack of Watson-Crick base pairs. However, most RNA loops in structured, biologically active RNA molecules have been shown by X-ray crystallography and NMR spectroscopy to form specific, well-structured three-dimensional (3D) motifs in which many of the bases form non-Watson-Crick base pairs and/or stacking interactions (31-33). Thus, current thermodynamic calculations generally cannot reveal the nature of non-Watson-Crick base pairing within loops and offer little help in designing mutations that disrupt or restore such base pairing.
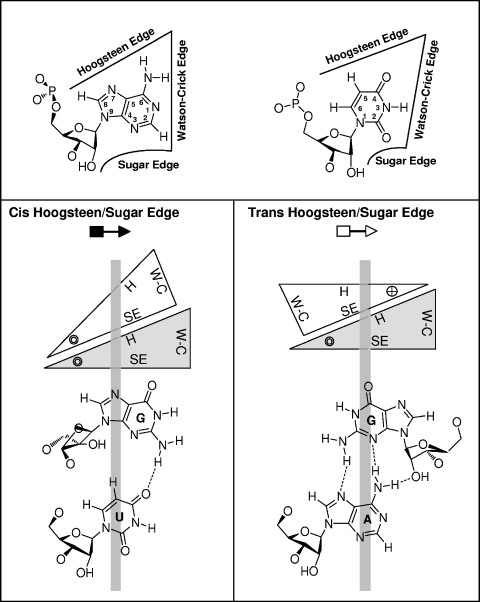
As shown in Fig. 2, RNA bases pair by hydrogen bonding at their Watson-Crick, Hoogsteen, or sugar edge and with their glycosidic bonds oriented cis or trans relative to each other (34). Non-Watson-Crick base pairs result when either or both bases interact at their Hoogsteen or sugar edge. All base pairs, including Watson-Crick and non-Watson-Crick base pairs, can be classified geometrically into 12 families by noting (i) the base edges used to form hydrogen bonds and (ii) the relative orientations (cis or trans) of the glycosidic bonds of the interacting nucleotides (32, 34). Two examples of these families, the cis and trans Hoogsteen/sugar edge base pairs, are shown in the lower part of Fig. 2. In non-Watson-Crick base pairs, the Watson-Crick edges may be available for further interactions—with other RNAs, proteins, or small molecules. In fact, one base can potentially pair with up to three other bases at the same time, allowing the formation of complex motifs involving base triples, quadruples, etc.
FIG. 2.
Geometric classification of RNA base pairing. The upper panel shows that each nucleotide base has three edges (Watson-Crick, Hoogsteen, and sugar) that can potentially hydrogen bond with one of the three edges of another base. Thus, each base can be represented schematically by a triangle and can potentially pair with up to three other bases. The interacting bases can pair with a cis or trans relative orientation of their glycosidic bonds. This is illustrated in the lower panels for the cis and trans orientations of nucleotides pairing at the Hoogsteen edge of one base and the sugar edge of the second base. In these base pairs, the Watson-Crick edges of the interacting bases are available for further interactions—with other RNAs, proteins, or small molecules. The cross and circle in the triangle where the Hoogsteen and sugar edges meet indicate 5′→3′ and 3′→5′ orientations, respectively, of the sugar-phosphate backbones relative to the plane of the page. W-C, Watson-Crick edge; H, Hoogsteen edge; SE, sugar edge. (Adapted from reference 34 with permission from the RNA Society.)
Certain RNA 3D motifs, such as loop E, recur in nonhomologous RNA molecules or in distinct sites of the same RNA molecules and play important roles by interacting with other cellular factors that regulate diverse biological functions (29, 52). Recurrent 3D motifs comprise sets of nucleotides with similar spatial arrangements, including a core of conserved non-Watson-Crick base pairs. The 3D structures of recurrent motifs are more conserved than their sequences (29, 31-33). The set of base substitutions compatible with the 3D structure of a motif is its sequence signature. Like the Watson-Crick base pairs, A/U and G/C, which can substitute for each other in double helices, certain non-Watson-Crick base pairs can substitute for each other in motifs without distorting the 3D structure of the motif; such base pairs are isosteric (29, 31, 37). Isostericity matrices summarize the isosteric relationships for each geometric base-pairing family and provide the basis for analyzing the sequence signatures of RNA motifs (30).
In this paper, we present our studies of the PSTVd loop E tertiary structure and its function in PSTVd replication by combining isostericity matrix analysis with genetic and functional assays. Our results provide the first loss-of-function genetic evidence for the essential role of a viroid 3D motif in replication. They also demonstrate that the integration of rationalized mutagenesis, based on isostericity matrix analysis of non-Watson-Crick base pairing, with in vitro and in vivo functional assays provides a novel, comprehensive approach to investigate the precise tertiary-structure-function relationships for a wide range of RNA motifs in diverse biological processes.
MATERIALS AND METHODS
Isostericity matrix analysis of PSTVd loop E.
In previous work, base pairs belonging to each geometric base-pairing family were grouped into isosteric subgroups and displayed in four-by-four isostericity matrices, one for each geometric family (30). In the isostericity matrices, base pairs belonging to isosteric subgroup n are designated In. For each non-Watson-Crick base pair composing an RNA motif, the isostericity matrices are consulted to determine which mutations result in (i) base pairs belonging to the same isosteric subgroup, (ii) base pairs belonging to nearly isosteric groups, (iii) base juxtapositions that cannot form base pairs of the same geometric type, or (iv) base juxtapositions that can form base pairs but which are nonisosteric. In case iii or iv, the 3D structure of the motif will be altered or even disrupted. In cases i and ii, if one or more bases participate in a second or third base pair, the isostericity matrices corresponding to those base pair types must also be consulted to make sure that the mutation is compatible with all possible interactions. Three criteria were used to group base pairs in isosteric subgroups. The first criterion is that all base pairs share the same relative orientation (cis or trans) of the glycosidic bond vectors. The second criterion is that the distance between the C1′ atoms of the paired nucleotides be the same (nominally, within about 1 Å). The third criterion is that hydrogen bonding takes place between equivalent base positions. Base pairs that meet all three criteria are considered isosteric, whereas base pairs that meet only the first two criteria are called nearly isosteric (35). Nearly isosteric base pairs, such as the wobble base pairs in the cis Watson-Crick/Watson-Crick geometric family, exhibit generally small lateral shifts at the interacting edges and, while not exactly isosteric, can sometimes substitute for each other without major disruption of the structure of a motif.
Additional examples of isosteric base pairs published since the original compilation of Leontis et al. (30) were obtained with the geometrical RNA motif search program FR3D (Find RNA 3D; M. Sarver, J. Stombaugh, A. Mokdad, C. L. Zirbel, and N. Leontis, submitted for publication).
Plasmid construction.
Plasmid pRZ6-2 carries cDNA of PSTVdInt (16) that is flanked by ribozymes at both ends. It was constructed by Hu et al. (22) and kindly provided by Robert Owens. All mutants were generated by site-directed mutagenesis with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The introduced mutations were confirmed by sequencing.
Plasmids pInter(+) and pInter(−), which were used as templates for in vitro synthesis of riboprobes specific for minus- and plus-strand PSTVd RNAs, respectively, were described by Qi and Ding (43).
The full-length cDNA of enhanced green fluorescent protein (GFP) was amplified by PCR with specific primers and with pEGFP-1 as the template (Clontech, Mountain View, CA) and cloned into pGEM4Z (Promega, Madison, WI) at EcoRI and BamHI sites to give rise to pGEM4Z:EGFP to be used for in vitro transcription.
In vitro transcription.
To prepare a PSTVd inoculum, plasmids containing the PSTVd cDNAs were linearized with HindIII. The linearized plasmids were used as templates for in vitro transcription with a T7 MEGAscript kit (Ambion, Austin, TX) according to the manufacturer's instructions. Because of the nature of the restriction site used for DNA cloning, the PSTVd transcripts after ribozyme self-cleavage have G88 at the 5′ end and G87 at the 3′ end. These unit length PSTVd transcripts were used as the inoculum.
To prepare PSTVd RNA substrates for UV cross-linking or ligation analyses, in vitro transcription was performed with the T7 MAXIscript kit (Ambion) in the presence of [α-32P]UTP. To make GFP transcripts that served as control substrates in UV cross-linking experiments, plasmid pGEM4Z:EGFP was linearized by EcoRI to serve as the template for in vitro transcription. The 32P-labeled PSTVd in vitro transcripts were separated on 5% polyacrylamide-8 M urea gel, and unit length monomeric bands were eluted in 0.3 M NaCl and precipitated with ethanol.
For microinjection, PSTVd transcripts were synthesized with a T7 MAXIscript kit (Ambion) in the presence of Alexa Fluor 488-5-UTP (Molecular Probes, Eugene, OR). Fluorescently labeled 434-nucleotide vector RNA transcripts were synthesized from NaeI-digested pGEMT (Promega) template DNA with an SP6 MAXIscript kit (Ambion).
After all transcription reactions, DNA templates were removed by DNase I treatment. RNA transcripts were purified with a MEGAClear kit (Ambion) and further quantified by UV spectrometry or with a scintillation counter (for radioactive riboprobes).
Protoplast inoculation.
Nicotiana benthamiana suspension cells were cultured as described by Sunter and Bisaro (57). Protoplasts were prepared and inoculated with PSTVd transcripts by electroporation as described by Qi and Ding (43). Detailed protocols for the maintenance of N. benthamiana suspension cells, preparation of protoplasts, and electroporation were described by Zhong et al. (65). At 3 days postinoculation, transfected protoplasts were harvested for RNA extraction and Northern blot analysis.
RNA extraction and Northern blotting.
Total RNAs were isolated from infected plants and protoplasts with an RNeasy Plant Mini Kit (QIAGEN, Valencia, CA). The procedures were essentially the same as those described by Qi et al. (44). Ten-microgram RNA samples were run on 5% polyacrylamide-8 M urea gel, transferred to a Hybond-XL nylon membrane (Amersham Biosciences, Piscataway, NJ) with a vacuum blotting system (Amersham), and immobilized by UV-cross-linking. Hybridization with [α-32P]UTP-labeled riboprobes was carried out at 65°C with ULTRAhyb reagent (Ambion). After overnight hybridization, the membranes were washed twice at 65°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) and twice in 0.2× SSC-0.1% SDS and exposed to a storage phosphor screen (Kodak, Rochester, NY). Hybridization signals were quantified with a Molecular Imager FX with Quantity One-4.1.1 software (Bio-Rad, Hercules, CA).
Sequencing of RNA progeny.
The protocols for preparing cDNAs of the PSTVd progeny isolated from protoplasts were performed essentially as described by Qi and Ding (43). Briefly, cDNAs of PSTVd RNA were reverse transcription (RT)-PCR amplified and sequenced in both directions with an ABI 377 DNA sequencer (Perkin-Elmer, Boston, MA) at the DNA Sequencing Facility at Ohio State University.
Preparation of N. benthamiana nuclear extract (NbNE).
NbNE was prepared as described by Baumstark and Riesner (2) and Roberts and Okita (47), with slight modifications. All steps were performed on ice. Protoplasts prepared from cultured cells of N. benthamiana (see above) were first resuspended in 15 ml of buffer A (20 mM morpholineethanesulfonic acid-KOH [pH 5.8], 20 mM K-acetate, 15% Ficoll 400, 0.15 mM spermine, 0.5 mM spermidine, 10 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, 0.15 μM pepstatin, 0.6 μM leupeptin) and then lysed in a Dounce homogenizer with 10 to 15 strokes. 4′,6′-Diamidino-2-phenylindole (DAPI) staining was used to monitor the release of nuclei. The nuclei were purified by loading the homogenates onto a two-step gradient consisting of 8 ml of buffer B (87.6% [vol/vol] Percoll, 0.62× buffer C [buffer A minus Ficoll]) and 12 ml of buffer D (buffer A with 18% instead of 15% Ficoll 400). The mixtures were centrifuged at 4,000 × g for 60 min at 4°C. The interphase containing the nuclei was collected with a glass pipette, and 1.5 volumes of buffer C was added. The mixtures were centrifuged at 1,400 × g for 30 min at 4°C, and the pelleted nuclei were resuspended in buffer E (60 mM HEPES-KOH [pH 7.9], 0.12 mM EDTA, 0.84 mM Mg-acetate, 0.72 μM leupeptin, 0.18 μM pepstatin, 12 mM β-mercaptoethanol, 0.6 M KCl). The sample was stirred gently for 1 h at 4°C and centrifuged at 36,000 × g for 60 min at 4°C with an L8-80 M Ultracentrifuge with an SW41 rotor (Beckman, Fullerton, CA). Saturated (NH4)2SO4 was slowly added to the resulting supernatant to a final concentration of 75% and centrifuged at 36,000 × g for 30 min at 4°C to precipitate proteins. The pellet was resuspended in buffer F (20 mM HEPES-KOH [pH 7.9], 10 mM Mg-acetate, 50 mM K-acetate, 5 mM EDTA, 12 mM β-mercaptoethanol, 25% glycerol) and dialyzed overnight at 4°C in buffer F. Aliquots were quickly frozen in liquid nitrogen and stored at −80°C.
RNA circularization assay.
The RNA circularization assay was conducted by incubating 32P-labeled and gel-purified unit length PSTVd in vitro transcripts and control RNAs in NbNE. In a reaction mixture with a final volume of 20 μl, 1 × 105 cpm of linear unit length plus-strand PSTVd transcripts was mixed with 4 μl of 5× ligation buffer (100 mM Tris-HCl [pH 8.0], 30 mM Mg-acetate, 1 mM spermidine, 2 mM EDTA) (40), 1 μl of RNaseOut (Invitrogen), and 10 μl of NbNE and incubated at 37°C for 2 to 3 h. The reaction was terminated by addition of 260 μl of 2× proteinase K buffer (200 mM Tris-HCl [pH 8.0], 25 mM EDTA [pH 8.], 1% SDS) and 10 μl of 20-mg/ml proteinase K and incubation at 65°C for 30 min, followed by phenol extractions (phenol was saturated with 10 mM Tris-HCl buffer [pH 4.0]). The reaction products were ethanol precipitated and analyzed on 5% polyacrylamide-8 M urea gel.
UV-cross-linking to analyze PSTVd loop E.
UV cross-linking was performed as described by Schrader et al. (49) under conditions that favor the formation of a stable PSTVd secondary-structure (equivalent to the ExL conformation of their substrate) containing loop E. Briefly, 32P-labeded in vitro transcripts of gel-purified unit length PSTVd transcripts (∼105 cpm) were preincubated in high-ionic-strength buffer (500 mM NaCl, 4 M urea, 1 mM Na-cacodylate, 0.1 mM EDTA [pH 7.9]) at 40°C for 45 min, followed by slow cooling to room temperature overnight. The reaction mixtures in Eppendorf tubes were placed on ice and irradiated with 258-nm UV light at different time intervals with a Stratalinker (model 1800; Stratagene) in a cold room. The samples were ethanol precipitated and analyzed on 5% polyacrylamide-8 M urea gel.
Microinjection and microscopy.
A small patch of epidermis was removed from the lower surface of an N. benthamiana leaf and covered with distilled water immediately. The leaf was mounted onto a glass plate to fit the stage of a Nikon E600 epifluorescence microscope (Nikon, Tokyo, Japan). Fluorescently labeled RNA transcripts were loaded into glass pipettes made from thin-walled glass tubes (World Precision Instruments, Sarasota, FL) on a pipette puller (model PB-7; Narishige, Tokyo, Japan). Injection was performed with an MMO-203 micromanipulator (Narishige) and a 1 M-5B injector (Narishige). Subcellular localization of the injected RNA transcripts was visualized with a filter set consisting of a 450- to 490-nm excitation filter, a 510-nm dichroic mirror, and a 520- to 560-nm barrier filter. Images were recorded and processed with a SPOT 2 Slider charge-coupled device camera and the associated software (Diagnostics Instruments Inc., Sterling Heights, MI). DAPI was injected into the same cells to visualize the nuclei after imaging of RNA localization. DAPI fluorescence was visualized with a filter set consisting of a 330- to 380-nm excitation filter, a 400-nm dichroic mirror, and a 435- to 485-nm barrier filter and recorded as described above.
Quantitative real-time RT-PCR.
Total RNA isolated from N. benthamiana protoplasts was treated with a TURBO DNA-free kit (Ambion) according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed with the ThermoScript RT-PCR system (Invitrogen) with primer (+)-97-125 (5′-CCTTTTTTGCCAGTTCGCTCCAGGTTTCC-3′) for plus-strand PSTVd, primer (−)-13-39 (5′-CGTGGTTCCTGTGGTTCACACCTGACC-3′) for minus-strand PSTVd, and primer 1093-1114R (5′-CCCGGAACCCAAAAACTTTG-3′) for 18S rRNA. The ThermoScript reverse transcriptase was inactivated by incubating the mixture at 85°C for 5 min. The template RNA was removed by RNase H treatment at 37°C for 1 h. Real-time PCR was performed with a QuantiTect SYBR green PCR kit (QIAGEN) in a PCR mixture containing a 2-μl cDNA sample (the RT products diluted fourfold), 0.5 μM forward and reverse primers, and 1× QuantiTect SYBR green PCR Master Mix. The primer combinations used for real-time PCR were as follows: primer (+)-97-125 and primer (+)-13-39 for amplification of either plus- or minus-strand PSTVd cDNA templates and primer 1093-1114 R and primer 1000-1020 F (5′-GATCAGATACCGTCCTAGTC-3′) for amplification of the 18S rRNA cDNA template. The real-time cycler conditions for the LightCycler system (Roche, Indianapolis, IN) were 10 min at 95°C for activation of HotStar Taq DNA polymerase, followed by 40 cycles of reactions (95°C for 30 s, 55°C for 30 s, and 72°C for 30 s). The melting curve analysis was performed to determine the specificity of the amplified products. Positive (standard-curve) and negative (no-template) controls were measured in each of the PCR runs. The standard curve was generated by serial dilutions ranging from 100 to 1010 copies of pRZ:PSTVdInt. 18S rRNA was used for normalization of PSTVd RNA accumulation levels. PSTVd copy numbers were calculated with the standard curve. The PCR results were analyzed with LightCycler software version 3.5 (Roche, Indianapolis, IN). For wild-type PSTVd and each of the mutant forms, results from six replicate RT-PCR experiments (three from samples of each of the two biological replicates) were used to calculate the mean and standard error. To facilitate comparison, the level of PSTVdInt accumulation has been arbitrarily set to a value of 1 and the levels of the mutants are presented as relative values.
RESULTS
Tertiary-structural model of the PSTVd loop E motif.
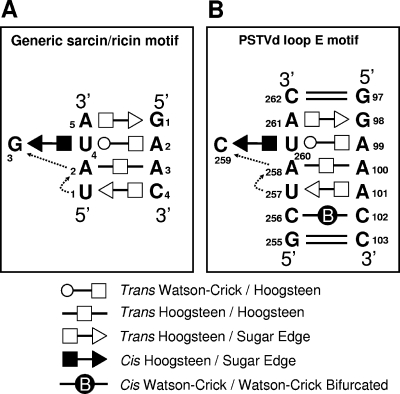
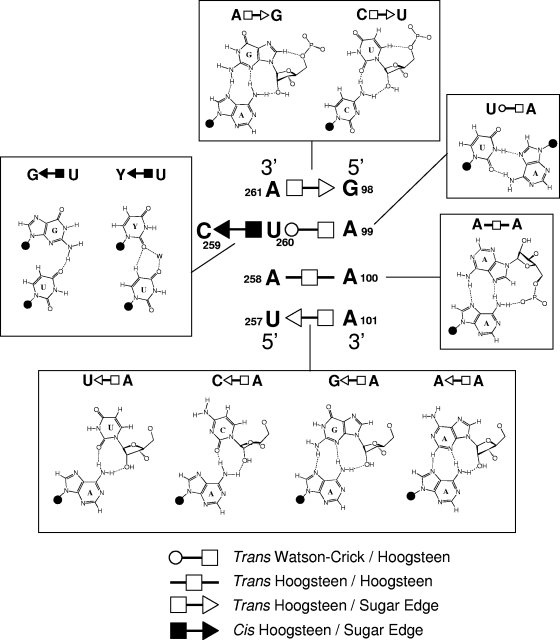
To facilitate genetic studies of PSTVd loop E function, we first analyzed the tertiary structure of this motif by comparative sequence analysis with isostericity matrices (30). Sequence analysis has identified recurrent loop E or sarcin/ricin motifs in different RNA molecules (29, 32). This motif comprises five core non-Watson-Crick base pairs, as shown in Fig. 3A. On the basis of UV cross-linking (4), nuclease and chemical mapping (13), and sequence comparison, it was concluded that a loop E (sarcin/ricin) motif also exists in PSTVd at the position indicated in Fig. 3B. Figure 4 shows the inferred base pair structures with hydrogen bonds for the five core non-Watson-Crick base pairs (A261/G98, U260/A99, U260/C259, A258/A100, and A101/U257) in PSTVd loop E. The structures of some common isosteric sequence variants are also shown. Each non-Watson-Crick base pair and its sequence variations in the PSTVd loop E motif are discussed below. Following the convention (34), the edges that participate in the interactions for each base pair are presented in the following order: Watson-Crick edge, Hoogsteen edge, and sugar edge.
FIG. 3.
Tertiary-structural model of PSTVd loop E. (A) Paradigmatic sarcin/ricin motif based on X-ray crystal structures (adapted from reference 29 with permission from Elsevier). (B) Inferred PSTVd loop E structural model. The dashed arrows indicate local changes in the strand orientation. All symbols that denote non-Watson-Crick base pairs and strand orientations are based on a report by Leontis and Westhof (34). Circles, squares, and triangles indicate the participation of Watson-Crick, Hoogsteen, and sugar edges, respectively. Open symbols indicate base pairs with a trans orientation of the glycosidic bonds, and closed symbols indicate base pairs with a cis orientation.
FIG. 4.
The five non-Watson-Crick base pairs that form the core of the PSTVd loop E motif. Examples of natural variants or experimentally designed mutations that can form isosteric base pairs are also illustrated. Y, pyrimidine (U or C); W, water molecule. Ribose is represented by a closed circle, except when it is involved in hydrogen bonding.
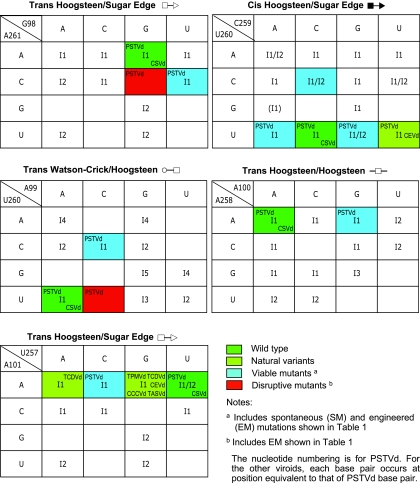
A261/G98 trans Hoogsteen/sugar edge pair.
The Hoogsteen edge of A261 forms hydrogen bonds with the sugar edge of G98 in the trans orientation. The trans Hoogsteen/sugar edge geometric family comprises two isosteric subfamilies (I1 and I2) (30; see Materials and Methods for the specific criteria used for base pair classifications). The A261/G98 pair belongs to I1 (see Fig. 5). The isosteric C/U double mutant, discussed below, is shown in Fig. 4.
FIG. 5.
The functional status of viroid mutants and natural variants mapped onto the respective isostericity matrices for each base pair, according to the geometric family to which it belongs (30). I1 to I5 denote subfamilies within each of which the base pairs are isosteric (see Materials and Methods for the definition of isostericity and reference 30 for details of the classification of subfamilies). Each blank and red box indicates that non-Watson-Crick base pairing between the two nucleotides in the family has not been reported in X-ray crystal structures or cannot be formed on the basis of isosteric predictions (30). Chrysanthemum stunt viroid (CSVd) has sequences identical to those of PSTVd in loop E. For sources of data and abbreviations of all other viroids, see Table 1.
U260/C259 cis Hoogsteen/sugar edge pair.
In the PSTVd sequence, U260 and C259 may form a cis Hoogsteen/sugar edge base pair. In most loop E (sarcin/ricin) motifs, the corresponding bases are U and G, which form a cis Hoogsteen/sugar edge base pair stabilized by a strong hydrogen bond between U (O4; oxygen at position 4 of uracil) and G (N2; nitrogen at position 2 of guanine). cis Hoogsteen/sugar edge base pairs between successive U/U, U/C, and U/A pairs are observed infrequently in crystal structures. While they have C1′-C1′ distances comparable to that of the U/G pair, they lack the strong hydrogen bond. Consequently, these pairs are usually stabilized by interactions with other bases, forming triples, or with proteins. The isosteric U/U cis Hoogsteen/sugar edge pair has been observed in several crystal structures, including the 23S rRNA (PDB file 1S72, U831/U832 from the Protein Data Bank), where it is part of a base triple. On the basis of observed U/U pairs, the isosteric U/C pair was proposed (30) and has since been observed (for example, U223/C222, chain A, PDB file 1ET4). The U/U and U/C pairs are stabilized in part by weak hydrogen bonds between U (C5; carbon atom at position 5 of uracil) and Y (O2; oxygen at position 2 of pyrimidine).
The U/A pair can form by weak hydrogen bonding between A (C2; carbon at position 2 of adenine) and U (O4) or between A (O2′; the 2′ OH of adenosine) and U (C5). One example of this type of base pair occurs in association with a protein and indicates that U/A has a C1′-C1′ distance of 6.8 Å (PDB file 1EC6, U12/A11 in C chain) and thus can also substitute isosterically for U/U, U/C, and U/G. The RNA structural search programs used at the time of the original compilation of base pairs (30) did not detect the U/A pair.
U260/A99 trans Watson-Crick/Hoogsteen edge pair.
The Watson-Crick edge of U260 hydrogen bonds with the Hoogsteen edge of A99 in the trans orientation. The U at this position is very conserved in most loop E motifs because it also uses its Hoogsteen edge to form a second base pair with G (i.e., a cis Hoogsteen/sugar edge pair). In other contexts, however, C can substitute for U in trans Watson-Crick/Hoogsteen base pairs to form isosteric C/C, or nearly isosteric C/A or C/G, base pairs of this type (30).
A258/A100 trans Hoogsteen/Hoogsteen edge pair.
The Hoogsteen edges of A258 and A100 interact to form a symmetric trans Hoogsteen/Hoogsteen base pair. Mutation of A100 to G still allows for hydrogen bonding with A258 by the formation of a hydrogen bond between A (N6; nitrogen at position 6 of adenine) and G (O6; oxygen at position 6 of guanine). This substitution has been observed in a composite loop E motif in the 23S rRNA of Haloarcula marismortui (PDB file 1S72, A913/G1071) with a C1′-C1′ distance of 12.2 Å. Thus, the A/G base pair is nearly isosteric with A/A.
A101/U257 trans Hoogsteen/sugar edge pair.
The Hoogsteen edge of A101 hydrogen bonds with the sugar edge of U257. Note that this base pair is oriented oppositely to trans Hoogsteen/sugar edge pair A261/G98, as shown by the symbols annotating these base pairs in Fig. 4. The structures of three isosteric variants, A/G, A/C, and A/A, are shown in Fig. 4. Very often, the isosteric C/U base pair variant occurs at this position, as shown for the generic sarcin/ricin loop in Fig. 3A. The structure of the C/U base pair is shown in the upper inset of Fig. 4.
C256/C102 cis Watson-Crick/Watson-Crick bifurcated pair.
The C256/C102 cis Watson-Crick/Watson-Crick bifurcated pair is outside the conserved core structure of loop E and sarcin/ricin motifs. For C/C to form an edge-to-edge Watson-Crick base pair, one cytosine must be protonated at the N3 position. While this has been observed in some structures, more commonly the cytosines adopt the Watson-Crick bifurcated geometry in which the N4 amino group of one cytosine hydrogen bonds with both N3 and O2 of the other cytosine. This bifurcated geometry results in a longer C1′-C1′ distance of about 10.4 Å, which is nearly isosteric to cis Watson-Crick A/U and G/C base pairs or with wobble Watson-Crick base pair U/G or C/A (30).
The tertiary-structural model of PSTVd loop E accounts for natural sequence variations and viable mutations.
To test the validity of the proposed tertiary-structural model of PSTVd loop E, we first examined whether this model could account for the functional state of the reported natural sequence variations or viable mutations in loop E motifs of PSTVd and other viroid species in the family Pospiviroidae. The natural sequence variants and viable mutants are summarized in Table 1. Figure 4 illustrates the molecular structures of selected sequence variants, mapped onto the annotated non-Watson-Crick base pairing structure that forms the core of the PSTVd loop E motif. Figure 5 shows the functional status of mutants and natural variants mapped onto the respective isostericity matrices for each base pair, according to the geometric family to which it belongs (30).
TABLE 1.
Effects of nucleotide sequence variations on the predicted structure of loop E motifs and the viability of viroidsa
| Loop E base pair (PSTVd wild type) | Mutation(s) (equivalent position in PSTVd) | Origin | Viroidb | Predicted loop E structurec | Viroid viabilityd |
|---|---|---|---|---|---|
| trans H/SE (A261/G98) | A261Ce | EM | PSTVd | − | − |
| A261C/G98Ue | EM | PSTVd | + | + | |
| trans WC/H (U260/A99) | A99Ce | EM | PSTVd | − | − |
| U260C/A99Ce | EM | PSTVd | + | + | |
| cis H/SE (U260/C259) | C259Af | SM | PSTVd | + | + |
| C259Ge,g | EM | PSTVd | + | + | |
| C259Uf,h,i | NV/SM | PSTVd | + | + | |
| C264Uj (259) | NV | CEVd | + | + | |
| trans H/H (A258/A100) | A100Gk | EM | PSTVd | + | +m |
| trans H/SE (A101/U257) | U257Af | SM | PSTVd | + | + |
| U257Cg | EM | PSTVd | + | + | |
| U257Gg | EM | PSTVd | + | + | |
| U258Al (257) | NV | TCDVd | + | + | |
| U262Gj (257) | NV | CEVd | + | + | |
| U255Gl (257) | NV | TPMVd | + | + | |
| U174Gl (257) | NV | CCCVd | + | + | |
| U255Gl (257) | NV | TASVd | + | + |
WC, Watson-Crick edge; EM, engineered mutation; SE, sugar edge; SM, spontaneous mutation; H, Hoogsteen edge; NV, natural variation.
MPVd, Mexican papita viroid; TCDVd, Tomato chlorotic dwarf viroid; CCCVd, Coconut cadang cadang viroid; TPMVd, Tomato planta macho viroid; TASVd, Tomato apical stunt viroid.
+, nondisruptive; −, disruptive.
+, nonlethal; −, lethal.
This study.
Reference 66.
Reference 43.
Reference 38.
Reference 63.
References 51 and 61.
Reference 49.
Reference 53.
In vitro processing.
The C259U substitution is found in a natural PSTVd variant, together with the C256A substitution and several other nucleotide changes outside loop E (38). The C→U substitution at this position is also detected in the loop E-like motif of a natural variant of Citrus exocortis viroid (CEVd), a member of the family Pospiviroidae (51). These are operationally classified as natural variations in Table 1. Furthermore, the C259U substitution was detected in the PSTVd progeny in transgenic tobacco plants expressing the wild-type PSTVd sequences (63, 66). These are classified as spontaneous mutations in Table 1. The experimentally constructed C259G and C259A substitutions, designated engineered mutations in Table 1, retained PSTVd viability as well as the C259U spontaneous mutation in tobacco and N. benthamiana protoplasts (43). These natural variations and viable mutations can all be accounted for by the fact that U260 will form isosteric cis Hoogsteen/sugar edge pairs with A, C, G, or U at position 259 of PSTVd or at the equivalent position within loop E of CEVd. cis Hoogsteen/sugar edge base pairs can occur between immediately successive nucleotides, as in the PSTVd loop E motif, or between nucleotides separated by one or more other nucleotides. The first type comprises isosteric subgroup I1, and the second type comprises subgroup I2. At the time of the 2002 compilation of base pairs (30), the only example of a cis Hoogsteen/sugar edge U/A pair detected by the computation search programs was of type I2. The recently developed FR3D search program (Sarver et al., submitted), however, identified a type I1 U/A cis Hoogsteen/sugar edge base pair (in this case, in complex with a protein) (PDB file 1EC6, U12/A11 in C chain). The C1′-C1′ distance of 6.8 Å makes this base pair isosteric with U/C (7 Å), U/G (6.7 Å), and U/U (7 Å) pairs (30). Therefore, all of these mutations could maintain the correct tertiary structure of loop E, if the corresponding bases are paired (in some cases, the pairing may occur only in the presence of another factor, such as a protein, that stabilizes this interaction). Because loop E motifs in cellular RNAs are well known to serve as important binding sites for RNAs and proteins (29), it will be of great interest to determine in future studies whether binding of a cellular protein to PSTVd loop E contributes to the stabilization of U/C and its viable variant U/U, U/G, and U/A base pairs.
It should be noted that the C259G mutation in loop E was shown to inhibit in vitro 3′ cleavage and ligation of a minicircle RNA derived from the CCR of PSTVd (49). Given that (i) the U/G cis Hoogsteen/sugar edge base pair is common at the equivalent position in other loop E motifs (Fig. 3 shows the generic sarcin/ricin motif) (30), (ii) the C259G mutation is not expected to alter the tertiary structure of PSTVd, and (iii) this mutation does not inhibit PSTVd replication in tobacco and N. benthamiana protoplasts (43) and in potato plants (data not shown), the reason for its effect on the in vitro processing of the minicircle RNA remains to be understood.
Zhu et al. (66) detected a U257A spontaneous mutation in transgenic tobacco expressing the wild-type PSTVd sequence. Experimentally constructed U257G and U257C substitutions retained PSTVd viability in tobacco and N. benthamiana protoplasts (43). Interestingly, a U→G substitution occurs at the equivalent position in loop E motifs of several CEVd natural variants (51, 54, 61), as well as Mexican papita viroid, Tomato planta macho viroid, and Tomato apical stunt viroid (53). Furthermore, a U→C substitution occurs in CEVd-C at this position (17). All of these observations can be accounted for by the fact that A101 can form isosteric trans Hoogsteen/sugar edge pairs with A, C, G, or U at position 257 of PSTVd and the equivalent position in the loop E motifs of all of the other viroids (Table 1; Fig. 4 and 5). All of these base pairs belong to the same isosteric subgroup, I1, of the trans Hoogsteen/sugar edge geometric family.
The A100G substitution does not affect in vitro cleavage-ligation of a minicircle RNA containing repeated sequences of the CCR of PSTVd (49). This observation was at first difficult to explain because the A/G trans Hoogsteen/Hoogsteen base pair has a C1′-C1′ distance of 12.9 Å according to Leontis et al. (30), which places it in a different isosteric subfamily than an A/A trans Hoogsteen/Hoogsteen base pair having a C1′-C1′ distance of 11 Å (30). However, since the publication of the original compilation of base pairs (30), we have identified other examples in higher-resolution structures for some of the base pairs reported. Thus, we have a higher-resolution example (2.4 Å versus 3.1 Å) of an A/G trans Hoogsteen/Hoogsteen base pair—A913/G1071 in the 50S subunit of H. marismortui (PDB file 1S72). In this base pair, the C1′-C1′ distance is 12.2 Å, which is closer to the 11 Å seen for an A/A base pair of this type. More significantly, this base pair is actually part of a loop E motif (30) and thus shows directly that A/G can substitute for A/A at positions 258/100. Whether this substitution affects the biological activities of PSTVd in vivo remains to be determined.
Outside the non-Watson-Crick base pair core of loop E, the C256/C102 cis Watson-Crick/Watson-Crick bifurcated pair also shows viable sequence variations. Owens et al. (38) reported a C256A substitution in a natural variant of PSTVd (together with a C259U substitution). Nucleotide A is also found at the equivalent position in Mexican papita viroid and Tomato apical stunt viroid (53), and A or C is found at the equivalent position in CEVd (51, 61). These variants can be accounted for by the fact that an A at this position in the motif can form a wobble base pair with C102 of PSTVd and with the C at the equivalent position in the other viroids. The C1′-C1′ distance in the C/A wobble Watson-Crick pair is ∼10.4 Å, almost identical to the C1′-C1′ distance in the bifurcated Watson-Crick C/C pair, so the A/C and C/C base pairs are nearly isosteric (30).
In conclusion, we find that all reported biologically viable sequence variations or mutations in viroid loop E motifs can be accounted for by the isosteric base pairing rules as summarized in the isostericity matrices, within the scope of the literature of which we are aware. This supports the notion that an intact loop E motif is required for PSTVd viability. Furthermore, this argues for the functional significance of intact loop E motif in other viroids in which this motif occurs. In particular, the loop E motifs of Chrysanthemum stunt viroid and PSTVd have identical sequence signatures (26).
Application of isostericity matrix analysis to design disruptive mutations in PSTVd loop E motif.
To investigate the function of the PSTVd loop E motif in replication by loss-of-function genetics, we applied isostericity matrix analysis to design mutations that disrupt this motif, based on the tertiary-structural model in Fig. 3B. Changing A99 to C (mutation A99C) prevents the trans Watson-Crick/Hoogsteen interaction between positions 260 and 99 because the Watson-Crick edge of U260 cannot hydrogen bond with the Hoogsteen edge of C99 (30). This is indicated in the isostericity matrices for trans Watson-Crick/Hoogsteen base pairs by the red box for the matrix element corresponding to the U row and the C column (Fig. 5) (30). Likewise, changing A261 to C prevents the trans Hoogsteen/sugar edge base pair at positions 261/98 from forming because the Hoogsteen edge of C261 cannot hydrogen bond with the sugar edge of G98. This is indicated by the red box at the corresponding positions (row C, column G) of the isostericity matrices for trans Hoogsteen/sugar edge base pairs (Fig. 5) (30). Computer programs that calculate minimum free energy secondary structures, such as mfold, do not predict the formation of non-Watson-Crick base pairs. Therefore, mutations affecting the structures of motifs consisting of ordered arrays of non-Watson-Crick base pairs, such as loop E, are also not correctly accounted for by mfold and similar programs.
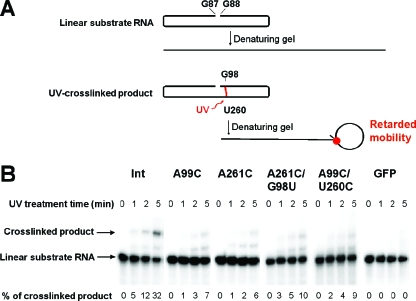
To test physically whether the A99C and A261C mutations each indeed disrupt the tertiary structure of loop E, we subjected the corresponding mutants to UV cross-linking experiments. These mutants are referred to as PSTVdIntA99C and PSTVdIntA261C, respectively. The names indicate that they are derived from the wild-type PSTVd intermediate strain (PSTVdInt) (16). Previous studies have demonstrated that UV treatment specifically cross-links bases G98 and U260 in loop E of PSTVd (3, 4) and of a minicircle RNA derived from the PSTVd CCR (49). It also cross-links the corresponding G and U bases in loop E of the human 5S rRNA from HeLa cells (4). This cross-linking occurs because of a unique partial cross-strand stack of G on U in loop E motifs that gives rise to photoreactivity. This cross-linking causes RNA mobility retardation on denaturing gels (Fig. 6). We used unit length linear in vitro transcripts as substrates. These transcripts are predicted to fold into the same rod-shaped secondary structure as the circular RNA, except for the nick between the two ends (G88 and G87; see Materials and Methods for details). As shown in Fig. 6, the A99C and A261C mutations greatly reduced the accumulation of cross-linked products. For instance, after 1 min of UV irradiation, the cross-linked product was already evident for PSTVdInt RNA but barely visible for the two mutants. After 5 min of UV irradiation, 32% of the wild-type RNA was cross-linked whereas only 6 to 7% of the mutant RNAs were cross-linked. The similar sizes of the cross-linked products in the wild-type and mutant PSTVd RNAs suggest that UV cross-linking occurred in the same region of these RNAs. Furthermore, the specificity of the G98-U260 cross-linking was demonstrated with two mutants that lack specific G98-U260 base stacking in loop E (Fig. 6). The absence of cross-linked products from GFP RNAs further demonstrates the specificity of UV cross-linking (Fig. 6). The quantitative difference indicates that a substantial portion of the PSTVdIntA99C and PSTVdIntA261C mutant RNA molecules failed to form the correct loop E structure, so as to place G98 and U260 into the necessary proximity to allow UV cross-linking, in contrast to the wild-type transcripts under the same incubation conditions. The band observed between the cross-linked product and the substrate RNA was also observed in previous experiments, and its nature remains to be determined (49).
FIG. 6.
Analysis of loop E formation by UV-cross-linking. Gel-purified substrate RNAs were preincubated under conditions that favor loop E formation and subjected to UV-cross-linking as described in Materials and Methods. (A) Diagram showing structural conformations of the substrate and cross-linked RNAs in their native state and in denaturing gels. G88 and G87 are the 5′ and 3′ ends, respectively, of the folded linear substrate RNA. The established cross-linking site (U260/G98) is marked in red. (B) Autoradiography of gel blots showing the presence or absence of UV-cross-linked products from the substrate RNAs as indicated. Efficiency of cross-linking is presented as the percentage of substrate RNAs that is converted into the cross-linked products. Note that UV treatment of the two compensatory mutants PSTVdIntA261C/G98U and PSTVdIntA99C/U260C, which are predicted by isostericity matrix analysis to restore loop E tertiary structure and showed restored replication, resulted in little cross-linked product formation. This indicates that G98/U260 base stacking, in addition to proper loop E tertiary structure, is required for specific UV cross-linking. The data from these two mutants thus provide evidence that UV cross-linking occurs specifically in loop E.
Maintaining the tertiary structure of loop E is necessary for PSTVd accumulation in inoculated protoplasts.
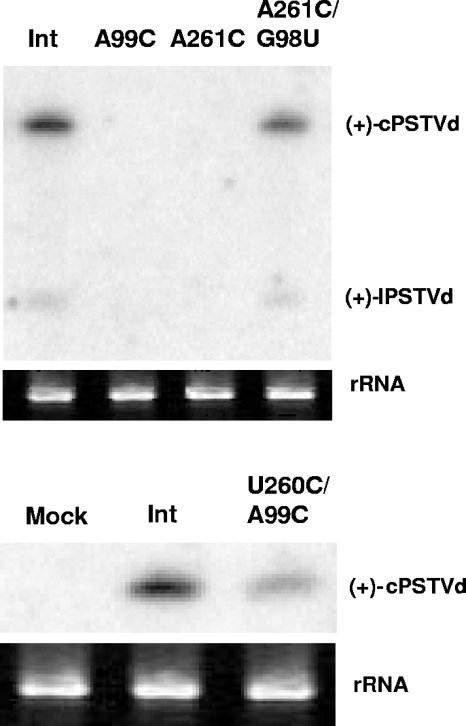
The availability of the loop E-defective mutants allowed us to test whether the intact tertiary structure of loop E is crucial for PSTVd infection. To this end, we inoculated N. benthamiana protoplasts with in vitro transcripts derived from the wild-type PSTVdInt and loop E-defective single mutants PSTVdIntA99C and PSTVdIntA261C, respectively. Northern blot assays, run to detect circular and linear plus-strand PSTVd, showed no visible accumulation of either PSTVdIntA99C or PSTVdIntA261C in protoplasts (Fig. 7), in contrast to PSTVdInt.
FIG. 7.
Northern blot assays showing accumulation of PSTVd RNAs in protoplasts of N. benthamiana. The rRNA served as a loading control. c-PSTVd and l-PSTVd indicate monomeric circular and monomeric linear PSTVd RNAs, respectively.
These data suggest that maintaining the intact tertiary structure of loop E is critical for PSTVd accumulation in inoculated protoplasts. To further test this, we applied isostericity matrix analysis to design a compensatory mutant in which G98 is replaced with U in the PSTVdIntA261C background. In the double mutant PSTVdIntA261C/G98U, the trans Hoogsteen/sugar edge C261/U98 base pair can form. This C261/U98 base pair is isosteric with the original A261/G98 base pair, and similar substitutions have been observed in the crystal structures of related motifs (29, 33) (Fig. 4 and 5). We found that, indeed, this double mutant accumulated as well as PSTVdInt in inoculated protoplasts (Fig. 7). Sequencing of PSTVd progeny confirmed that no other mutations occurred during replication of the double mutant. To extend this finding, we generated a second compensatory mutant in which U260 is replaced by C in the PSTVdIntA99C background. In this double mutant, PSTVdIntU260C/A99C, the trans Watson-Crick/Hoogsteen C260/C99 base pair that is isosteric with the original U260/A99 base pair can form (30). Importantly, C260 and C259 may also form a cis Hoogsteen/sugar edge base pair isosteric with the original U260/C259 base pair (Fig. 5) (30). A protoplast infection assay showed that this double mutant also restored replication function (Fig. 7). Taken together, all of these data provide compelling evidence that the tertiary structure of loop E is essential for PSTVd accumulation in inoculated protoplasts. Furthermore, these experimental results demonstrate the value of isostericity matrix analysis as a tool to design mutations that disrupt or restore non-Watson-Crick base pairs in functional studies of RNA motifs.
A99C and A261C mutations do not impair nuclear import function.
Following delivery into the cytoplasm of protoplasts and leaf cells, the viroid transcripts must first enter the nucleus to be replicated. To determine whether the diminished accumulation of mutants PSTVdIntA99C and PSTVdIntA261C in the inoculated protoplasts is due to defects in nuclear import, we injected fluorescently labeled plus-strand in vitro transcripts into the cytoplasm of leaf cells. Both single mutants, like the wild type, appeared in the nucleus to visible levels under a fluorescence microscope 20 to 30 min after injection (Fig. 8; Table 2). Fluorescently labeled in vitro transcripts derived from vector sequences did not show nuclear accumulation (Fig. 8; Table 2), indicating specificity of nuclear import of the PSTVd RNAs. Therefore, the diminished accumulation of the mutants in the inoculated protoplasts can be attributed to their impaired capacity to replicate within the nucleus, rather than to defects in nuclear import. These results demonstrate that the intact tertiary structure of loop E is required for PSTVd replication in the nucleus. Furthermore, the fact that the fluorescently labeled transcripts of PSTVdIntA99C, PSTVdIntA261C, and PSTVdInt are imported into the nuclei and accumulate there, unlike the cytoplasmic accumulation of vector transcripts, indicates that A99C and A261C mutations do not make PSTVd RNA more vulnerable to degradation.
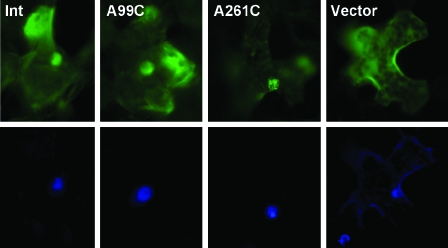
FIG. 8.
Nuclear import of fluorescently labeled in vitro transcripts of PSTVd. The transcripts of the wild type (Int) and the two mutants (A99C and A261C) appear in the nucleus within 30 min of injection into the cytoplasm (upper row). The vector transcripts do not show nuclear accumulation. DAPI staining (lower row) shows the position of the nucleus in each injected cell.
TABLE 2.
Nuclear import of PSTVd RNAs
| Injected RNA | No. of injected cells showing nuclear import/total no. of injected cells (% of cells showing import) |
|---|---|
| PSTVdInt | 14/16 (89) |
| PSTVdIntA99C | 9/9 (100) |
| PSTVdIntA261C | 10/12 (83) |
| Vector | 0/15 (0) |
The A99C and A261C mutations do not affect circularization of unit length inoculum transcripts.
According to the rolling-circle replication model, incoming circular PSTVd molecules serve as templates for the synthesis of minus strands. Therefore, upon entering the nucleus, the linear, unit length in vitro transcripts of PSTVd may need to be ligated into circular molecules prior to serving as the initial transcription templates. Alternatively, it may be possible for the PSTVd-transcription enzyme complex to transcribe the linear RNA transcripts by jumping across the nick with a template-switching mechanism, as does the HDV-transcription enzyme complex during rolling-circle replication in mammalian cells (7). Assuming, however, that circularization of the input PSTVd transcripts is necessary for transcription, one possible reason for the failure of single mutants PSTVdIntA99C and PSTVdIntA261C to replicate is that their linear in vitro transcripts cannot be ligated at G88 and G87 (the 5′ and 3′ ends, respectively, of the transcripts; see Materials and Methods for details) to form circular molecules. To test the circularization of PSTVdIntA99C and PSTVdIntA261C, we incubated gel-purified unit length linear plus-strand PSTVd RNA transcripts of the wild type and mutants in NbNE. As shown in Fig. 9, all of the RNA substrates were properly circularized. As a control, GFP RNAs showed no circularization. Therefore, the two mutations do not affect the circularization of PSTVd RNA inocula.
FIG. 9.
Circularization of loop E mutants in NbNE. Gel-purified and radiolabeled unit length linear plus-strand PSTVd transcripts were incubated in NbNE for 2 h and analyzed by urea-polyacrylamide gel electrophoresis. c-PSTVd and l-PSTVd indicate monomeric circular and monomeric linear PSTVd RNAs, respectively.
The A99C and A261C mutations inhibit the accumulation of minus-strand PSTVd.
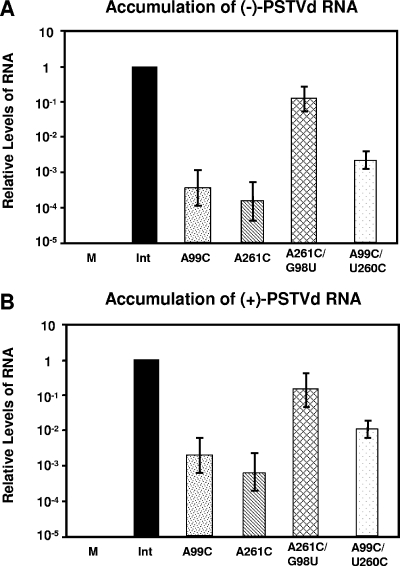
Given that PSTVdIntA99C and PSTVdIntA261C proved to be competent for nuclear import and ligation, we asked whether they were defective in transcription. One method to address this question was to examine the accumulation of minus-strand PSTVd RNA, the replication intermediate. Northern blot assays did not detect the presence of the minus-strand PSTVd RNA for either mutant (data not shown). Because the minus-strand PSTVd RNA is present at low levels during replication of even wild-type PSTVdInt (43), we used the more sensitive quantitative real-time RT-PCR method to quantify the minus strands. The 18S rRNA served as an internal control template in each reaction mixture. As shown in Fig. 10, the accumulation levels of the minus-strand RNAs from PSTVdIntA99C and PSTVdIntA261C were approximately 1,000-fold lower, respectively, than that of the wild type. In contrast, the accumulation level of minus-strand PSTVdIntA261C/G98U was approximately fivefold lower than that of wild-type PSTVdInt. The accumulation level of PSTVdIntU260C/A99C was approximately 10-fold higher than that of PSTVdIntA99C, showing notable restoration of accumulation. Similar accumulation patterns were detected for the plus-strand PSTVd RNAs, which is consistent with Northern blot analysis. These data suggested that the plus-strand PSTVdIntA99C and PSTVdIntA261C transcripts inoculated into the protoplasts were inefficiently transcribed into the minus strands, which further led to reduced synthesis and accumulation of the plus-strand RNAs.
FIG. 10.
Determination of minus-strand PSTVd (A) and plus-strand PSTVd (B) RNA levels in infected N. benthamiana protoplasts by quantitative real-time RT-PCR. For details, see Materials and Methods. M, mock inoculation. To facilitate comparison, the accumulation level of the wild type (Int) was arbitrarily set to a value of 1 and the levels of the mutants are presented as relative values on a log scale.
Whether transcription of plus-strand RNA from concatemeric minus-strand template RNA is affected by the mutations remains unknown. This question can be addressed once methods are developed to construct a concatemeric RNA template that contains identical mutations in each of the unit length monomers for transcription analyses.
DISCUSSION
Viroid infection represents a remarkable case in which a small infectious RNA molecule, without encoding any proteins, contains all of the genetic information and forms the appropriate 3D structure to interact with cellular factors so as to ensure its replication and systemic trafficking throughout a host plant. It is unlikely that plants have evolved mechanisms for RNA-templated RNA replication and systemic RNA trafficking simply to serve viroid RNA infection. Rather, the viroid RNA must have evolved to adapt to and exploit existing cellular mechanisms used for endogenous functions. Therefore, the biology of viroid infection likely mimics the biological roles of endogenous RNAs and mechanisms for their synthesis, processing, and intra- and intercellular transport. Elucidating how PSTVd replicates should therefore shed new light on the structure and function of the eucaryal nuclear transcription machinery, on the evolution of RNA structure-function relationships, and on the evolution of RNA-based infectious agents.
We have developed a novel approach that integrates comparative sequence analysis of non-Watson-Crick base pairs with isostericity matrices, mutagenesis, and in vitro, as well as in vivo, functional assays to dissect the role of one conserved RNA motif in the rolling-circle replication of PSTVd. This systematic approach is necessary to pinpoint the process(es) affected by mutations in a noncoding and multifunctional RNA so as to determine the specific biological role of an RNA structural motif.
A tertiary-structural model for viroid loop E that can guide functional studies.
Branch et al. (4) first recognized that PSTVd contains an RNA motif very similar or identical to loop E of eucaryal (or archaeal) 5S rRNA on the basis of specific UV cross-linking between the conserved U260 and G98 bases. On the basis of nuclease and chemical mapping data and comparisons with the NMR structures of Xenopus laevis (eucaryal) 5S rRNA loop E (64) and of the sarcin/ricin loop in 28S rRNA (58), Gast et al. (13) suggested a scheme of non-Watson-Crick base-pairing interactions in the PSTVd loop that is similar to these rRNA motifs. The Gast model depicted the A261/G98, A99/U260, and A100/A258 base pairs essentially as the same as in our model. However, it depicted A101/U257 as a canonical Watson-Crick base pair and did not specify the nature of the C102/C256 base pair or the possible existence of the cis Hoogsteen/sugar edge U260/C259 interactions. Our isostericity matrix analysis is based on high-resolution X-ray crystal structures of other loop E motifs and the principles of isosteric non-Watson-Crick base pairing (30). The tertiary-structural model we present here specifies the precise nature of the edge-to-edge interactions for all of the non-Watson-Crick base pairs. The model is currently well supported by several lines of biological and physical data. First, this model can explain mechanistically why certain mutations in loop E do not affect PSTVd viability (43, 49, 63, 66). Second, it can account for the naturally occurring nucleotide sequence variations in the loop E motifs of PSTVd and other viroids in the family Pospiviroidae (17, 38, 51, 53, 54, 61). Third, UV cross-linking data provide further evidence that the A99C and A261C substitutions disrupt the loop E structure by preventing the formation of isosteric non-Watson-Crick base pairs. Fourth, as discussed below, the functional analyses pinpointed the deleterious effects of these mutations on PSTVd transcription. Finally, the two compensatory mutations that are predicted to restore the loop E tertiary structure on the basis of isosteric base-pairing principles and on the basis of the X-ray crystal structures of similar base pairs also restored PSTVd replication. Further studies on the effects of other nucleotide mutations within loop E should provide additional tests of the validity of this structural model. All of these data also suggest strongly that the PSTVd loop E motif exists and functions in vivo.
PSTVd loop E plays a crucial role in RNA-templated RNA transcription.
Our functional analyses of the PSTVd loop E mutations provide new insights into the role of this motif in PSTVd replication at the cellular level. Our experiments showed that the A99C and A261C mutations, which disrupt the loop E structure, inhibit accumulation of PSTVd progeny in inoculated protoplasts. Further analyses indicated that these mutations do not impair nuclear import function or circularization of the linear inoculum transcripts. Therefore, the failure of the PSTVd mutants to replicate and accumulate in inoculated protoplasts likely results from a defect(s) in one or more of the steps of rolling-circle replication. The requirement for intact loop E in replication is further supported by the observation that compensatory mutations predicted to restore non-Watson-Crick base pairs within the motif were observed to restore the replication function.
The A99C and A261C loss-of-function mutations could inhibit the transcription, cleavage, or ligation step of rolling-circle replication. Although our present data cannot rule out completely the possibility that these mutations affect cleavage-ligation, several observations suggest strongly that they mainly inhibit transcription. First, the A99C and A261C mutations severely inhibit the synthesis of minus-strand PSTVd RNA. Second, if these mutations in fact inhibited cleavage rather than transcription, one would expect to see accumulation of high levels of the multimeric plus-strand PSTVd RNAs. This was not observed in Northern blot assays. Third, if these mutations inhibited ligation, rather than transcription or cleavage, one would expect to see accumulation of high levels of the linear monomeric plus-strand PSTVd RNAs. This was not observed either in Northern blot assays. On the basis of these considerations, we propose that the loop E motif plays a crucial role in transcription, in addition to its well-documented role in cleavage-ligation (3), during PSTVd replication. The multifunctionality of loop E is discussed below.
How might loop E be involved in transcription? Loop E motifs in cellular RNAs are well known to serve as important binding sites for RNAs and proteins (29). The loop E motif of PSTVd may serve as a binding site for the RNA polymerase or for a cellular factor(s) that recruits the RNA polymerase for transcription. Alternatively, loop E may interact with a cellular factor to localize the PSTVd RNA to a particular subnuclear site to facilitate access to the nuclear transcription machinery. It is also possible that this motif is required for interacting with a cellular factor that leads to a conformational change in the viroid RNA, allowing it to be recognized by the transcription machinery. These possibilities can be tested by further experimental studies.
It should be noted that a HPI is predicted to form as a thermodynamically metastable structure through pairing of nucleotides 79CGCUUCAGG87 and nucleotides 110GCGAGGUCC102 with nucleotides 88 to 101, forming a 14-nucleotide loop as a result of HPI formation (46). Whether the mutations we generated would affect the structure and function of HPI requires further analyses. In fact, it remains to be determined whether HPI exists in vivo and has any biological functions. A putative tetraloop containing A99 was postulated by Baumstark et al. (3) to be important for in vitro processing. Whether such a tetraloop structure exists and functions in vivo and whether A99C would affect its structure and function remain to be investigated.
PSTVd loop E is a useful model to study RNA structure-function relationships of broad significance.
Loop E is one of the most extensively studied RNA structural motifs (29, 31). Besides serving as a model to dissect PSTVd replication and the general mechanisms of RNA-templated replication by the nuclear transcription machinery, the PSTVd loop E motif offers an attractive system to study RNA structure-function relationships of general significance. First, the multifunctionality of this motif is biologically significant. This motif is involved in transcription (this study), processing (3), pathogenicity (42), and host adaptation (43, 63, 66). To accomplish such diverse functions, submotifs of loop E may interact with distinct cellular factors. Elucidating the underlying mechanisms will greatly expand our understanding of the capacity of a single RNA motif to regulate multiple biological processes. Such multifunctionality of a single RNA motif may be important for a wide range of RNA-based pathogens to expand their genome functions during evolution without increases in genome size.
Second, loop E is a recurrent motif found in many RNAs, including 5S, 16S, and 23S rRNAs; group I and group II introns; bacterial RNase P; ribozyme of Tobacco ringspot virus satellite RNA (reviewed in reference 32); and lysine riboswitches (18, 56), where it plays critical roles in RNA-RNA and RNA-protein interactions. While mutational analyses of cellular RNA motifs may be limited by the potential lethality of motif-disruptive mutations to an organism or by the high structural complexity of large cellular RNAs, such limitations do not apply to PSTVd, a host parasite. Therefore, further mutagenesis can be performed on PSTVd loop E and other motifs to determine their sequence signatures and gain detailed insights into RNA structure-function relationships that may not be feasible with cellular RNA. Furthermore, the recurrent nature of this motif allows broad application of the knowledge obtained.
An integrative approach to address structure-function relationships of RNA motifs.
Our findings from PSTVd loop E structure and function studies demonstrate that integration of isostericity matrix analysis, rationalized mutagenesis, and systematic functional studies can provide important insights into the structure-function relationship of an RNA motif and further into the elaborate cellular control over this relationship. Isostericity matrix analysis can be used to predict base substitutions that will prevent the formation of isosteric (or nearly isosteric) base pairs, thereby disrupting the 3D structure of an RNA motif. If the structural integrity of the motif is critical for function, these mutations are likely to result in loss of function. This is well demonstrated by the replication defects of PSTVd carrying either the A99C or the A261C mutation that disrupts loop E. Isostericity matrix analysis can also predict which base substitutions can retain or restore the tertiary structure of a motif, providing a useful means to infer the tertiary structure of RNA motifs. This analysis alone, however, may not always predict a priori which isosteric or nearly isosteric base substitutions in a motif will retain or restore the function of a given RNA in the cellular environment, because the functioning of the motif requires interactions with other factors that can only be elucidated through biological assays. This is illustrated well by the results from the two compensatory mutants, PSTVdIntA261C/G98U and PSTVdIntU260C/A99C. Both mutants are predicted by isostericity matrix analysis to have restored the loop E tertiary structure. However, while the replication level of PSTVdIntA261C/G98U is close to that of the wild type, the replication level of PSTVdIntU260C/A99C is less than that of the wild type. Presumably, the loop E motif with the C259/C260/C99 base triple in the compensatory mutant PSTVdIntU260C/A99C is sufficient, but not as optimal as the motif with the C259/U260/A99 base triple as in the wild type, to be recognized by the cellular machinery for replication. These results therefore demonstrate that a combination of isostericity matrix analysis and genetic and biological experiments can provide a foundation to elucidate the elaborate RNA motif-cellular factor interactions that regulate diverse biological processes and to address the fundamental question of how sophisticated features of RNA structures have evolved to achieve optimal functions.
Acknowledgments
We thank Ying Wang, Ryuta Takeda, and Xiaorong Tao for technical assistance. We thank Venkat Gopalan, John M. Burke, and Joyce Heckman for constructive discussions of UV cross-linking and Jesse Stombaugh for helpful discussions of base pair analyses. We are indebted to Yuhua Lu for technical assistance with the preparation of Fig. 2.
This work was supported by grants from the National Science Foundation (IBN-0238412 and IOB-0515745) and from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (2004-35304-15005) to B.D., by National Institutes of Health grant 3R15-GM55898 to N.L., and by National Institutes of Health grant CA97024 to K.B.L.
REFERENCES
- 1.Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 2.Baumstark, T., and D. Riesner. 1995. Only one of four possible secondary structures of the central conserved region of potato spindle tuber viroid is a substrate for processing in a potato nuclear extract. Nucleic Acids Res. 23:4246-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumstark, T., A. R. Schroder, and D. Riesner. 1997. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branch, A. D., B. J. Benenfeld, and H. D. Robertson. 1985. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. USA 82:6590-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNA's. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 6.Candresse, T., A. Gora-Sochacka, and W. Zagorski. 2001. Restoration of secondary hairpin II is associated with restoration of infectivity of a non-viable recombinant viroid. Virus Res. 75:29-34. [DOI] [PubMed] [Google Scholar]
- 7.Chang, J., and J. Taylor. 2002. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J. 21:157-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diener, T. O. 1971. Potato spindle tuber “virus”. IV. Replicating, low molecular weight RNA. Virology 45:411-428. [DOI] [PubMed] [Google Scholar]
- 9.Dingley, A. J., G. Steger, B. Esters, D. Riesner, and S. Grzesiek. 2003. Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J. Mol. Biol. 334:751-767. [DOI] [PubMed] [Google Scholar]
- 10.Flint, S. J., L. W. Enquist, V. R. Racaniello, and A. M. Skalka. 2004. Principles of virology, 2nd ed. ASM Press, Washington, D.C.
- 11.Flores, R., C. Hernandez, A. E. Martinez de Alba, J. A. Daros, and F. Di Serio. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 12.Flores, R., J. W. Randles, M. Bar-Josef, and T. O. Diener. 2000. Subviral agents: viroids, p. 1009-1024. In M. H. V. van Regenmortel, C. M. Fouquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy, seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 13.Gast, F. U., D. Kempe, R. L. Spieker, and H. L. Sanger. 1996. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J. Mol. Biol. 262:652-670. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, W. 1986. The RNA world. Nature 319:618. [Google Scholar]
- 15.Gora-Sochacka, A. 2004. Viroids: unusual small pathogenic RNAs. Acta Biochim. Pol. 51:587-607. [PubMed] [Google Scholar]
- 16.Gross, H. J., H. Domdey, C. Lossow, P. Jank, M. Raba, H. Alberty, and H. L. Sanger. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273:203-208. [DOI] [PubMed] [Google Scholar]
- 17.Gross, H. J., G. Krupp, H. Domdey, M. Raba, P. Jank, C. Lossow, H. Alberty, K. Ramm, and H. L. Sanger. 1982. Nucleotide sequence and secondary structure of citrus exocortis and chrysanthemum stunt viroid. Eur. J. Biochem. 121:249-257. [DOI] [PubMed] [Google Scholar]
- 18.Grundy, F. J., S. C. Lehman, and T. M. Henkin. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. USA 100:12057-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond, R. W., and R. A. Owens. 1987. Mutational analysis of potato spindle tuber viroid reveals complex relationships between structure and infectivity. Proc. Natl. Acad. Sci. USA 84:3967-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henco, K., H. L. Sanger, and D. Riesner. 1979. Fine structure melting of viroids as studied by kinetic methods. Nucleic Acids Res. 6:3041-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Y., P. A. Feldstein, P. J. Bottino, and R. A. Owens. 1996. Role of the variable domain in modulating potato spindle tuber viroid replication. Virology 219:45-56. [DOI] [PubMed] [Google Scholar]
- 22.Hu, Y., P. A. Feldstein, J. Hammond, R. W. Hammond, P. J. Bottino, and R. A. Owens. 1997. Destabilization of potato spindle tuber viroid by mutations in the left terminal loop. J. Gen. Virol. 78:1199-1206. [DOI] [PubMed] [Google Scholar]
- 23.Hull, R. 2002. Matthew's plant virology, 4 ed. Academic Press, San Diego, Calif.
- 24.Joyce, G. F. 2002. The antiquity of RNA-based evolution. Nature 418:214-221. [DOI] [PubMed] [Google Scholar]
- 25.Joyce, G. F., and L. E. Orgel. 1999. Replication of human hepatitis delta virus: recent developments, p. 49-77. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), RNA world, 2nd ed., vol. 37. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 26.Keese, P., and R. H. Symons. 1985. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 82:4582-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolonko, N., O. Bannach, K. Aschermann, K. H. Hu, M. Moors, M. Schmitz, G. Steger, and D. Riesner. 2006. Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 347:392-404. [DOI] [PubMed] [Google Scholar]
- 28.Lai, M. M. 2005. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 79:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leontis, N. B., J. Stombaugh, and E. Westhof. 2002. Motif prediction in ribosomal RNAs: lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie 84:961-973. [DOI] [PubMed] [Google Scholar]
- 30.Leontis, N. B., J. Stombaugh, and E. Westhof. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30:3497-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leontis, N. B., and E. Westhof. 1998. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA 4:1134-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leontis, N. B., and E. Westhof. 1998. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 283:571-583. [DOI] [PubMed] [Google Scholar]
- 33.Leontis, N. B., and E. Westhof. 1998. Conserved geometrical base-pairing patterns in RNA. Q. Rev. Biophys. 31:399-455. [DOI] [PubMed] [Google Scholar]
- 34.Leontis, N. B., and E. Westhof. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7:499-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lescoute, A., N. B. Leontis, C. Massire, and E. Westhof. 2005. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 33:2395-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loss, P., M. Schmitz, G. Steger, and D. Riesner. 1991. Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J. 10:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel, F., and E. Westhof. 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216:585-610. [DOI] [PubMed] [Google Scholar]
- 38.Owens, R., S. M. Khurana, D. R. Smith, M. N. Singh, and I. D. Grag. 1992. Mild strain of potato spindle tuber viroid isolated from wild Solanum spp. In India. Plant Dis. 76:527-529. [Google Scholar]
- 39.Owens, R. A., R. W. Hammond, R. C. Gardner, M. C. Kiefer, S. M. Thompsom, and D. E. Cress. 1986. Site-specific mutagenesis of potato spindle tuber viroid cDNA. Plant Mol. Biol. 6:179-192. [DOI] [PubMed] [Google Scholar]
- 40.Owens, R. A., G. Steger, Y. Hu, A. Fels, R. W. Hammond, and D. Riesner. 1996. RNA structural features responsible for potato spindle tuber viroid pathogenicity. Virology 222:144-158. [DOI] [PubMed] [Google Scholar]
- 41.Owens, R. A., S. M. Thompson, and G. Steger. 1991. Effects of random mutagenesis upon potato spindle tuber viroid replication and symptom expression. Virology 185:18-31. [DOI] [PubMed] [Google Scholar]
- 42.Qi, Y., and B. Ding. 2003. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15:1360-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi, Y., and B. Ding. 2002. Replication of potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: the role of specific nucleotides in determining replication levels for host adaptation. Virology 302:445-456. [DOI] [PubMed] [Google Scholar]
- 44.Qi, Y., X. Zhong, A. Itaya, and B. Ding. 2004. Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 32:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu, F., C. Heinrich, P. Loss, G. Steger, P. Tien, and D. Riesner. 1993. Multiple pathways of reversion in viroids for conservation of structural elements. EMBO J. 12:2129-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riesner, D., K. Henco, U. Rokohl, G. Klotz, A. K. Kleinschmidt, H. Domdey, P. Jank, H. J. Gross, and H. L. Sanger. 1979. Structure and structure formation of viroids. J. Mol. Biol. 133:85-115. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, M. W., and T. W. Okita. 1991. Accurate in vitro transcription of plant promoters with nuclear extracts prepared from cultured plant cells. Plant Mol. Biol. 16:771-786. [DOI] [PubMed] [Google Scholar]
- 48.Sanger, H. L., G. Klotz, D. Riesner, H. J. Gross, and A. K. Kleinschmidt. 1976. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 73:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrader, O., T. Baumstark, and D. Riesner. 2003. A mini-RNA containing the tetraloop, wobble-pair and loop E motifs of the central conserved region of potato spindle tuber viroid is processed into a minicircle. Nucleic Acids Res. 31:988-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroder, A. R., and D. Riesner. 2002. Detection and analysis of hairpin II, an essential metastable structural element in viroid replication intermediates. Nucleic Acids Res. 30:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Semancik, J. S., J. A. Szychowski, A. G. Rakowski, and R. H. Symons. 1993. Isolates of citrus exocortis viroid recovered by host and tissue selection. J. Gen. Virol. 74:2427-2436. [DOI] [PubMed] [Google Scholar]
- 52.Simons, R. W., and M. Grunberg-Manago. 1998. RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Singh, R. P., K. F. M. Ready, and X. Nie. 2003. Viroids of solanaceous species, p. 125-133. In A. Hadidi, R. Flores, J. W. Randles, and J. S. Semancik (ed.), Viroids. CSIRO, Collingwood, Australia.
- 54.Skoric, D., M. Conerly, J. A. Szychowski, and J. S. Semancik. 2001. CEVd-induced symptom modification as a response to a host-specific temperature-sensitive reaction. Virology 280:115-123. [DOI] [PubMed] [Google Scholar]
- 55.Steger, G., and D. Riesner. 2003. Molecular characteristics, p. 15-29. In A. Hadidi, R. Flores, J. W. Randles, and J. S. Semancik (ed.), Viroids. CSIRO, Collingwood, Australia.
- 56.Sudarsan, N., J. K. Wickiser, S. Nakamura, M. S. Ebert, and R. R. Breaker. 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 17:2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunter, G., and D. M. Bisaro. 2003. Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305:452-462. [DOI] [PubMed] [Google Scholar]
- 58.Szewczak, A. A., and P. B. Moore. 1995. The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 247:81-98. [DOI] [PubMed] [Google Scholar]
- 59.Tabler, M., and M. Tsagris. 2004. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 9:339-348. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, J. M. 2003. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 11:185-190. [DOI] [PubMed] [Google Scholar]
- 61.Visvader, J., and R. H. Symons. 1985. Eleven new sequence variants of citrus exocortis viroid and the correlation of sequence with pathogenicity. Nucleic Acids Res. 13:2907-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wassenegger, M., S. Heimes, and H. L. Sanger. 1994. An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J. 13:6172-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wassenegger, M., R. L. Spieker, S. Thalmeir, F. U. Gast, L. Riedel, and H. L. Sanger. 1996. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226:191-197. [DOI] [PubMed] [Google Scholar]
- 64.Wimberly, B., G. Varani, and I. Tinoco, Jr. 1993. The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry 32:1078-1087. [DOI] [PubMed] [Google Scholar]
- 65.Zhong, X., A. Itaya, and B. Ding. 2005. Transfecting protoplasts by electroporation to study viroid replication, p. 16D.4.1-16D.4. 11. In R. Coico, T. Kowalik, J. M. Quarles, B. Stevenson, and R. K. Taylor (ed.), Current protocols in microbiology, vol. 1. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, Y., Y. Qi, Y. Xun, R. Owens, and B. Ding. 2002. Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]