Abstract
Genetic bottlenecks may occur in virus populations when only a few individuals are transferred horizontally from one host to another, or when a viral population moves systemically from the infection site. Genetic bottlenecks during the systemic movement of an RNA plant virus population were reported previously (H. Li and M. J. Roossinck, J. Virol. 78:10582-10587, 2004). In this study we mechanically inoculated an artificial population consisting of 12 restriction enzyme marker mutants of Cucumber mosaic virus (CMV) onto young leaves of squash plants and used two aphid species, Aphis gossypii and Myzus persicae, to transmit the virus populations from infected source plants to healthy squash plants. Horizontal transmission by aphids constituted a significant bottleneck, as the population in the aphid-inoculated plants contained far fewer mutants than the original inoculum source. Additional experiments demonstrated that genetic variation in the artificial population of CMV is not reduced during the acquisition of the virus but is significantly reduced during the inoculation period.
Plant viruses are dependent on vectors for their horizontal transmission, and aphids are the most common and important group of plant virus vectors. Aphids transmit at least 275 plant viruses, and approximately 75% of these viruses are transmitted in a nonpersistent manner, or stylet borne (14). This mode of transmission is characterized by a rapid rate of virus acquisition (<1 min) and inoculation by the aphids (8). A successful aphid transmission event is dependent upon the uptake of virus, stable retention of the acquired virions, the release of retained virions from regions within the mouth parts of the vector, and their delivery to a site of infection (10).
Nonpersistent transmission by aphids may have an important effect on the dynamics and evolution of virus populations. Infection of a host may start with a small number of virions; transmission events could constitute genetic bottlenecks, which by definition are stochastic events that can introduce random elements into the genetic structures of populations (13). A major cause of virus strain differentiation could be genetic drift as a result of population bottlenecks during aphid transmission. However, so far there has been no experimental evidence to show that genetic bottlenecks occur during the aphid transmission of plant viruses.
Cucumber mosaic virus (CMV) is efficiently transmitted in a nonpersistent manner by more than 75 species of aphids (11). The coat protein (CP) of CMV is a primary determinant of aphid transmission (1). The Fny isolate of CMV is efficiently transmitted by both Aphis gossypii (Glover) and Myzus persicae (Sulzer) (12).
In a previous study, using an artificial population of CMV consisting of 14 mutants with silent restriction enzyme marker mutations, we showed that systemic infection constituted a significant bottleneck for the CMV populations (4). To understand the role of genetic bottlenecks during aphid transmission of CMV, we mechanically inoculated 12 of the CMV mutants into squash cotyledons. A. gossypii and M. persicae were used for the transmission of CMV from inoculated source leaves to healthy squash leaves. When the newly infected squash leaves were analyzed for the presence of each of the 12 marker mutants, we found that aphid transmission induced a significant genetic bottleneck in the CMV population.
MATERIALS AND METHODS
Virus mutants and plant inoculation.
Cotyledons of squash seedlings (Cucurbita pepo cv. Elite) were mechanically inoculated with individual or mixed viral RNAs of CMV mutants (a, b, c, d, e, f, g, h, i, j, k and l) described previously (4). Three days postinoculation, the inoculated cotyledons were used as the virus source leaves for aphid transmission to healthy squash cotyledons.
Aphids and transmission assays.
A. gossypii was reared on healthy squash or cotton seedlings, and M. persicae was raised on healthy Chinese cabbage or turnips. Before acquisition, aphids were starved for 2 to 3 h on a moistened filter paper in a petri dish. Individual aphids then were transferred with a camel hair brush to the surface of the infected source leaf. The starved aphids were allowed 30- to 90-s acquisition probes on CMV-infected cotyledons. Probing was observed under a dissecting microscope, and actively feeding aphids were either transferred to healthy squash cotyledons or collected for use in reverse transcription-PCR (RT-PCR) analysis. Single aphids transferred to healthy squash cotyledons were confined in 1-cm-diameter clip cages and allowed an overnight inoculation access period. Plants were then fumigated with an insecticide, placed in a greenhouse for 7 to 10 days, and observed for symptom development. Two to three days postinoculation, tissues were collected from within the caged area where the aphids had fed. When individual aphids were directly examined for the presence of the 12 mutants, the aphids were allowed to feed on infected plants for 5 to 10 min before being removed and flash frozen in liquid nitrogen.
Extraction of viral RNA from plants and aphids.
Total RNA from source and test plants was extracted as described previously (4). For analysis of individual aphids that had probed virus-infected source tissue, whole single aphids were placed in 1.5-ml microcentrifuge tubes, along with about 15 mg of healthy plant tissue as a carrier, and flash frozen in liquid nitrogen. The whole frozen aphids were ground with a sterile pestle and suspended in 200 μl of NTS buffer (0.1 M NaCl, 0.01 M Tris [pH 8.0], 1 mM EDTA, and 1% sodium dodecyl sulfate). An equal volume of phenol-chloroform (1:1; saturated with Tris-EDTA) was added, and the sample was vortexed for 2 min and centrifuged for 2 min at full speed in a microcentrifuge. The supernatant was removed to a fresh tube and precipitated with 2 volumes of ethanol by standard methods. The precipitated RNA was resuspended in 50 μl of sterile water.
CMV detection in source tissue and individual aphids.
Total RNA from source tissue and individual aphids was used in a real-time fluorescent RT-PCR assay to quantify the amount of virus, as previously described (16). The primers for CMV (forward primer, nt 260 to 279, 5′GCGCGCTGATAATGCTATTT3′, and reverse primer, nt 322 to 301, 5′GCAATACGACCGTGGGTTACTT3′) that amplified a portion of the 3a gene and the probe (nt 283 to 300, TCCGGCCCCTCGTTCCCG) used in the real-time RT-PCR assay were kindly provided by Keith Perry, Cornell University. Fluorescence from the 6-carboxyfluorescein reporter was detected at a 505- to 537-nm wavelength. The cycle threshold (CT) values for each reaction were calculated automatically width Smart Cycler (Cepheid) detection software by determining the point in time (PCR cycle number) at which the reporter fluorescence exceeded 10 times the computer-determined standard deviation for background.
Analysis of populations.
Total RNA from source plants, test plants, or aphids was used as a template for RT-PCR and subsequent enzyme digestion of PCR products as described previously (4).
Data analysis.
The transmission efficiency of each virus mutant was calculated as a ratio of the number of plants infected to the number of plants infested with the aphids that had been allowed to acquire the virus. An analysis of variance was used to test the significance of the mean number of mutants recovered from test plants infected by either A. gossypii or M. persicae. A test of least significant difference was used to compare mean transmission efficiencies among mutants and aphid vectors (17).
RESULTS
Detection of bottlenecks as a result of aphid transmission.
Five independent experiments determined the transmissibility of each of the 12 mutants when acquired from a source leaf inoculated with all 12 mutants (Table 1) . The 12 mutants were always found in the source leaves used in each experiment. Fifty-two and 26 squash seedlings inoculated by single A. gossypii or M. persicae aphids, respectively, developed systemic symptoms. Tissues were analyzed only from those test plants that later developed systemic symptoms. Probing behavior of both aphid species was similar on the virus-infected squash leaves, indicating that the different host plants that the aphid species were reared on did not contribute to any differences in transmission efficiency.
TABLE 1.
Changes in CMV populations during horizontal transmission by Aphis gossypii and Myzus persicae
| Expt. |
A. gossypii
|
M. persicae
|
||||
|---|---|---|---|---|---|---|
| Planta | Mutants recovered
|
Planta | Mutants recovered
|
|||
| No. | Composition | No. | Composition | |||
| 1 | Source leaf | 12 | abcdefghijkl | Source leaf | 12 | abcdefghijkl |
| Ag1-1 | 1 | c | Mp1-1 | 2 | cj | |
| Ag1-2 | 1 | j | Mp1-2 | 4 | abhj | |
| Ag1-3 | 3 | egj | Mp1-3 | 3 | cdj | |
| Ag1-4 | 1 | e | Mp1-4 | 1 | c | |
| Ag1-5 | 3 | adl | Mp1-5 | 2 | ce | |
| Ag1-6 | 1 | l | Mp1-6 | 4 | cdeh | |
| Ag1-7 | 3 | ehl | ||||
| Ag1-8 | 3 | cdj | ||||
| Ag1-9 | 4 | acel | ||||
| Ag1-10 | 3 | chl | ||||
| Ag1-11 | 2 | dj | ||||
| Ag1-12 | 4 | dhjl | ||||
| Ag1-13 | 3 | ajl | ||||
| Ag1-14 | 2 | cl | ||||
| Ag1-15 | 2 | al | ||||
| Ag1-16 | 3 | cej | ||||
| 2 | Source leaf | 12 | abcdefghijkl | Source leaf | 12 | abcdefghijkl |
| Ag2-1 | 2 | jl | Mp2-1 | 2 | ce | |
| Ag2-2 | 2 | dj | Mp2-2 | 2 | bc | |
| Ag2-3 | 5 | abchl | Mp2-3 | 1 | c | |
| Ag2-4 | 1 | c | ||||
| Ag2-5 | 3 | bgk | ||||
| Ag2-6 | 2 | jl | ||||
| Ag2-7 | 5 | dehjl | ||||
| Ag2-8 | 4 | chil | ||||
| Ag2-9 | 5 | aceil | ||||
| Ag2-10 | 3 | del | ||||
| Ag2-11 | 4 | cijl | ||||
| Ag2-12 | 1 | e | ||||
| Ag2-13 | 2 | el | ||||
| Ag2-14 | 3 | dek | ||||
| 3 | Source leaf | 12 | abcdefghijkl | Source leaf | 12 | abcdefghijkl |
| Ag3-1 | 3 | chl | Mp3-1 | 1 | c | |
| Ag3-2 | 3 | cde | Mp3-2 | 2 | cj | |
| Ag3-3 | 4 | ehjk | Mp3-3 | 3 | cjk | |
| Ag3-4 | 3 | chl | Mp3-4 | 3 | cej | |
| Ag3-5 | 2 | hl | Mp3-5 | 4 | chjk | |
| Ag3-6 | 3 | cej | Mp3-6 | 2 | ch | |
| Ag3-7 | 3 | aeh | Mp3-7 | 5 | bdghi | |
| Ag3-8 | 1 | c | Mp3-8 | 3 | dgi | |
| Ag3-9 | 3 | ceh | Mp3-9 | 5 | cdhjl | |
| 4 | Source leaf | 12 | abcdefghijkl | Source leaf | 12 | abcdefghijkl |
| Ag4-1 | 2 | ce | Mp4-1 | 7 | bdefghj | |
| Ag4-2 | 4 | achj | Mp4-2 | 1 | h | |
| Ag4-3 | 3 | agh | Mp4-3 | 1 | e | |
| Ag4-4 | 3 | aef | ||||
| Ag4-5 | 3 | deg | ||||
| Ag4-6 | 4 | efgl | ||||
| Ag4-7 | 6 | bcdehl | ||||
| Ag4-8 | 2 | hi | ||||
| Ag4-9 | 6 | bcehjl | ||||
| 5 | Source leaf | 12 | abcdefghijkl | Source leaf | 12 | abcdefghijkl |
| Ag5-1 | 2 | ce | Mp5-1 | 2 | cl | |
| Ag5-2 | 2 | ei | Mp5-2 | 1 | c | |
| Ag5-3 | 2 | al | Mp5-3 | 2 | ce | |
| Ag5-4 | 4 | acgh | Mp5-4 | 3 | cdl | |
| Mp5-5 | 2 | kl | ||||
Cotyledons of squash seedlings were mechanically inoculated with 12 CMV mutants and used as source leaves for aphid feeding. Numbers identify individual plants that were successfully infected by aphids in that particular experiment.
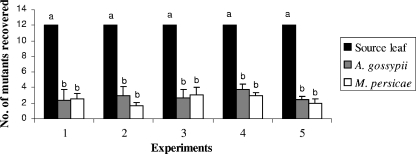
Results from five independent experiments showed that the number of mutant viruses decreased significantly (P < 0.001) during the transmission of the population from the source plants to the test plants by both A. gossypii and M. persicae (Fig. 1). The number of mutants recovered from the aphid-inoculated leaves of test plants ranged from one to six with an average of three for A. gossypii and from one to seven with an average of three for M. persicae. Reduction in the number of mutants was statistically significant (P < 0.05) (Fig. 1) in all five experiments and did not differ with aphid species (Table 1). The compositions of the mutant virus populations recovered from individual test plants infected by the two aphid vectors were different from each other (Table 1). Transmission of individual mutants was largely stochastic, indicating that a bottleneck existed during aphid transmission of CMV.
FIG. 1.
Reduction in CMV populations during aphid transmission from mechanically inoculated leaves to healthy plants. Experiments 1 to 5 are independent experiments. All values are means plus standard errors. Bars with different letters (a and b) are significantly different from each other (P < 0.05), as determined by the significant difference test.
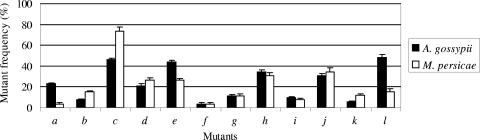
As shown in Fig. 2, each of the 12 mutant viruses from five separate experiments was transmissible by either species of aphid. Pairwise comparison showed that the mean transmission efficiency of three of the mutants (a, c, and l) varied significantly (P < 0.05) between transmission by A. gossypii and that by M. persicae. For example, mutant a was transmitted 23% of the time and mutant l 48% of the time by A. gossypii, while the efficiency of transmission by M. persicae was reduced to 3.8% for mutant a and 15% for mutant l. Similarly, mutant c was transmitted 73% of the time by M. persicae but only 46% of the time by A. gossypii. The transmission efficiencies of the remaining nine mutants (b, d, e, f, g h i, j, and k) were not significantly different between the two aphid species, although mutant f was poorly transmitted (3.8%) by both aphid species. Hence, the mutants fell into three classes: those that were randomly transmitted (the majority), those that were poorly transmitted, and those that were transmitted with different efficiencies by the different aphid species. For the poorly transmitted and differentially transmitted mutants, there appear to be selective forces in play, even though all of the mutations were synonymous.
FIG. 2.
Frequency of recovery of mutants from tissues of test plants infected by A. gossypii and M. persicae. The recovery percentage of each mutant in test tissue was calculated for each experiment as for Fig. 1. Values are means plus 1 standard error.
Virus population complexity affects transmission efficiency.
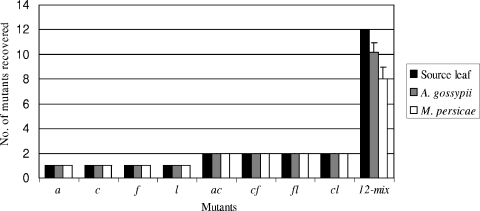
To further understand the nonstochastic transmission of some of the CMV mutants by both aphid species, we selected mutant f, which was transmitted poorly by both aphid species, mutants a and l, which were transmitted more efficiently by A. gossypii than by M. persicae, and mutant c, which was transmitted more efficiently by M. persicae than A. gossypii (Fig. 2). Cotyledons of squash seedlings were mechanically inoculated with viral RNA of individual mutants a, c, f, and l or combinations of two mutants (ac, cf, cl, and fl) and used as source tissue for aphid feeding 3 days postinoculation. At least 10 single aphids of each species were observed to probe each of the source tissues. Immediately following, RNA was extracted from single whole aphids and from their respective source tissues and used in real-time RT-PCR assays to compare the titers of the virus in source tissues.
Virus was detected in 80 to 100% of A. gossypii and 90 to 100% of M. persicae aphids tested, and CT values for the respective source tissues in real-time RT-PCR were not significantly different (P > 0.05) (Table 2), indicating that viral loads were similar in all source tissues for individual mutants or combinations of mutants. Analysis of the RT-PCR products from individual aphids by restriction enzyme digestion indicated that in all cases, individual mutants or combinations of mutants were present in each aphid and in the mechanically inoculated source tissues (Fig. 3).
TABLE 2.
Real-time RT-PCR analysis of individual A. gossypii and M. persicae aphids allowed to feed on squash cotyledons inoculated with different CMV mutants
| Squash inoculum |
A. gossypii
|
M. persicae
|
||||
|---|---|---|---|---|---|---|
| % Positive (no. positive/no analyzed)a |
CT value forb:
|
% Positive (no. positive/no analyzed)a |
CT value forb:
|
|||
| Single aphids | Source tissuec | Single aphids | Source tissuec | |||
| a | 90 (9/10) | 34.7 ± 3.7 | 11.75 | 100 (10/10) | 24.8 ± 4.6 | 12.09 |
| c | 100 (10/10) | 38.5 ± 1.4 | 15.2 | 100 (10/10) | 24.9 ± 6.0 | 12.3 |
| f | 100 (10/10) | 32.3 ± 2.7 | 11.63 | 90 (9/10) | 26.9 ± 5.7 | 13.06 |
| l | 80 (8/10) | 36.1 ± 5.1 | 11.55 | 100 (10/10) | 26.4 ± 4.9 | 12.01 |
| ac | 80 (8/10) | 37.2 ± 2.8 | 11.76 | 100 (10/10) | 27.2 ± 4.4 | 12.29 |
| cf | 100 (10/10) | 36.9 ± 4.3 | 13.63 | 100 (10/10) | 27.8 ± 5.6 | 12.56 |
| cl | 80 (8/10) | 34.5 ± 3.4 | 11.76 | 90 (9/10) | 25.3 ± 3.4 | 12.53 |
| fl | 100 (10/10) | 38.1 ± 1.5 | 11.35 | 90 (9/10) | 27.3 ± 3.9 | 12.43 |
| 12-mix | 90 (9/10) | 34.2 ± 3.7 | 11.49 | 80 (8/10) | 21.3 ± 6.0 | 12.33 |
Percent virus (not the number of mutants) detected in 9 aphids out of 10.
CT value represents total virus, not individual mutants. CT values for A. gossypii and M. persicae are expressed as means ± standard deviations.
Source tissue represents a single cotyledon inoculated with single, double, or all 12 mutants (12-mix).
FIG. 3.
Acquisition efficiency of CMV populations by single A. gossypii or M. persicae aphids. Source represents the squash cotyledon tissue mechanically inoculated with single mutants, double mutants, or all 12 mutants (12-mix). Values are means plus 1 standard error.
The above experiments indicated that aphids could acquire multiple virus mutants without preference, but it was not clear if all 12 mutants could be acquired simultaneously by a single aphid. In a final experiment, cotyledons of squash seedlings were mechanically inoculated with an equal mixture of all 12 mutant viruses. At least 10 single aphids of each species were allowed to feed on the inoculated source leaves, and each aphid was analyzed individually for the presence of mutants. From real-time RT-PCR analysis, 9 of 10 A. gossypii aphids had acquired virus while 8 of 10 M. persicae aphids were positive (Table 2). For A. gossypii, the number of mutants recovered from a single aphid ranged from 8 to 12 with an average of 10, while for M. persicae it ranged from 6 to 11 with an average of 8 (Fig. 3). The populations of mutants recovered from aphids of both species frequently contained mutants a, c, f, and l (data not shown), which were either poorly or efficiently transmitted from mixed infections by aphids (Fig. 2).
DISCUSSION
Here we report the transmission of a population of genetically marked mutants of CMV from source plants to test plants using two aphid species, A. gossypii and M. persicae. Our results showed that the 12 CMV mutants were readily acquired from the source plants by both aphid species but the number of mutants decreased significantly when the aphids transmitted the population to test plants. The reduction in the recovery of CMV mutants from the test plant was stochastic except for a few mutants that were either poorly or highly transmitted by two aphid species (Fig. 2). A. gossypii is generally a more efficient vector of CMV than M. persicae, and the difference in the rate of transmission of the mutants may be the result of a combination of a reduction in the stability of virions and intrinsic differences between the two aphid species (9). Our data indicated that the bottleneck event occurred during the inoculation period (or infection event) rather than the acquisition access period. This finding indicates that nonpersistent aphid transmission plays an important role in the genetic structures of RNA virus populations in infected plants.
The real-time RT-PCR analysis of the source tissue inoculated with single, double, and all 12 mutants showed that CMV individuals had no effect on each other, as shown by the CT values (Table 2), and all of them replicated in the inoculated host plant and were efficiently acquired by single aphids. Similarly, both A. gossypii and M. persicae acquired the population without any selection at the acquisition access period from the source tissue, supporting the previous findings that virus acquisition occurs via ingestion from virus-infected plants to their retention sites within the food canal of the maxillary stylets (14).
Virus destined for inoculation is retained at the distal tips of the stylet bundle within the aphid vector. The stylet bundle of aphids comprises a pair of inner maxillary stylets forming the food and salivary canals. These canals are fine channels with a diameter of approximately 0.7 μm (food canal) and 0.3 μm (salivary canal) that remain separate from each other (15) but converge 2 to 4 μm from the tips to form a common duct where mixing of food and salivary canal contents may occur (14). Ingested virions adhering to the cuticular lining of the common duct may therefore be flushed out during the intracellular secretion of watery saliva. The binding of the virion within the vector must be readily reversible, and salivation may function to enhance the release of bound virions and their delivery into plant cells (7). It appears that salivation plays an essential role in the release of virions. An understanding of virus entry may ultimately require a better understanding of salivation. Little is known about the mechanism of binding and the release of virions in the aphid vector.
The ability of a virus to be transmitted by aphids in a nonpersistent manner is dependent not only on the acquisition but also on the stable retention of virions in the mouthparts (18). Little is known about the ligands to which viruses bind in the aphid mouthparts or the stability of virus particles in aphids (5). The coat protein (CP) of CMV is the sole viral determinant of aphid transmission (1), and the specificity and efficiency of transmission have been mapped to several domains in the amino acid sequence of the CP (5, 9, 10). In our artificial CMV populations, all the mutations were silent and the CP amino acid sequence was not altered in any mutant. No changes in aphid-virus interactions due to mutation were expected (4). However, CMV virions are dependent on RNA-protein interactions for their integrity (11), and modest changes in pH can also affect stability (Adam Zlotnick, personal communication). The effect of silent point mutations on the virion stability of the CMV particle within the aphid vectors is not known. It is also possible that A. gossypii retains more virions at the distal tip while M. persicae takes up the vast majority of virions into the food canal.
Probing plays a major role in the spread of nonpersistently transmitted viruses in the field. When an aphid lands on a plant, it initially probes the epidermal cells to determine host suitability. It is these brief probes (usually less than 30 s) that are optimal for acquisition or inoculation of nonpersistently transmitted viruses, and the virions are easily lost during more extended probing periods (3, 19). The inoculation of CMV by A. gossypii occurs during the first phase of the intracellular stylet puncture in superficial plant tissues (7).
RNA viruses can generate highly polymorphic populations known as quasispecies. A quasispecies is a population at an equilibrium of mutation and selection and varies in a cloud around one or a few central master sequences (2). The mutation frequency of a population can be reduced specifically by selection or randomly by genetic bottlenecks. Selection is a directional process by which the fittest variants in a specific environment will increase their frequency in the population (positive selection) while less fit variants will decrease their frequency in the population (negative selection), and as a consequence the most fit variants will have more progeny in the next generation than less fit variants.
In contrast, genetic bottlenecks are stochastic events that reduce genetic variation of a population and result in founding populations that can lead to genetic drift. When a few genomes or even a single genome of a virus population is randomly chosen to generate a new population, there is a high probability that it carries a mutation relative to more fit genomes of the parental population. Genetic drift within small populations could minimize the effect of differences in fitness by reducing the relative effect of selection and thus increase the rates of genetic fixation and extinction. Genetic drift depends on the effective size of population (the number of individuals that pass their genes to the next generation) and not on the census size (the total number of individual in the population). Genetic drift has an important role in determining the frequency and fate of mutations in the effective size of the population.
Genetic drift and natural selection are always at play in a population. However, the degree to which mutants are affected by drift and selection varies according to circumstances. In a large population, where genetic drift occurs very slowly, even weak selection on a mutant will push its frequency upwards or downwards (depending on whether the mutation is beneficial or harmful). However, if the population is very small, drift will predominate. In this case, weak selective effects may not be seen at all as the small changes in frequency they would produce are overshadowed by drift.
Our data constitute the first report of a genetic bottleneck during experimental transmission of CMV by two aphid species in a nonpersistent manner. We observed that the founder numbers of mutants in the test plants were small due to severe bottlenecks, and the effective numbers could be much smaller than census number. Thus, genetic drift could reshape the population of mutants in the test plants. This study extends our understanding of the dynamics and mechanisms of the long-term evolution of viruses in nature. However, field studies of CMV populations will be required to determine whether genetic drift remains a factor in natural population dynamics or whether the presumed high error rate of the viral polymerase is sufficient to overcome the negative effects of bottlenecks by rapidly regenerating the most fit variant, as predicted by quasispecies theory (6).
Acknowledgments
We thank Keith Perry for providing CMV real-time PCR primers and probe. We also thank Phillip Harries and Michael Udvardi for careful reviews of the manuscript.
This work was supported by The Samuel Roberts Noble Foundation.
REFERENCES
- 1.Chen, B., and R. I. B. Francki. 1990. Cucumovirus transmission by the aphid Myzus persicae is determined solely by the coat protein. J. Gen. Virol. 71:939-944. [Google Scholar]
- 2.Domingo, E. 2002. Quasispecies theory in virology. J. Virol. 76:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray, S. M., and N. Banerjee. 1999. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, H., and M. J. Roossinck. 2004. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 78:10582-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, S., X. He, G. Park, C. Josefsson, and K. L. Perry. 2002. A conserved capsid protein surface domain of Cucumber mosaic virus is essential for efficient aphid vector transmission. J. Virol. 76:9756-9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manrubia, S. C., C. Escarmís, E. Domingo, and E. Lázaro. 2005. High mutation rates, bottlenecks, and robustness of RNA viral quasispecies. Gene 347:273-282. [DOI] [PubMed] [Google Scholar]
- 7.Martín, B., J. L. Collar, W. F. Tjallingii, and A. Fereres. 1997. Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J. Gen. Virol. 78:2701-2705. [DOI] [PubMed] [Google Scholar]
- 8.Ng, J. C. K., C. Joseffsson, A. J. Clark, A. W. E. Franz, and K. Perry. 2005. Virion stability and aphid vector transmissibility of Cucumber mosaic virus mutants. Virology 332:397-405. [DOI] [PubMed] [Google Scholar]
- 9.Ng, J. C. K., S. Liu, and K. L. Perry. 2000. Cucumber mosaic virus mutants with altered physical properties and defective in aphid vector transmission. Virology 276:395-403. [DOI] [PubMed] [Google Scholar]
- 10.Ng, J. C. K., and K. L. Perry. 2004. Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5:505-511. [DOI] [PubMed] [Google Scholar]
- 11.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281-348 [DOI] [PubMed] [Google Scholar]
- 12.Perry, K. L., L. Zhang, and P. Palukaitis. 1998. Amino acid changes in the coat protein of cucumber mosaic virus differentially affect transmission by the aphids Myzus persicae and Aphis gossypii. Virology 242:204-210. [DOI] [PubMed] [Google Scholar]
- 13.Pirone, T. P., and S. Blanc. 1996. Helper-dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34:227-247. [DOI] [PubMed] [Google Scholar]
- 14.Powell, G. 2005. Intracellular salivation is the aphid activity associated with inoculation of non-persistently transmitted viruses. J. Gen. Virol. 86:469-472. [DOI] [PubMed] [Google Scholar]
- 15.Powell, G., C. R. Tosh, and J. Hardie. 2006. Host plant selection by aphids: behavioral, evolutionary, and applied perspectives. Annu. Rev. Entomol. 51:309-330. [DOI] [PubMed] [Google Scholar]
- 16.Schneider, W. L., D. J. Sherman, A. L. Stone, V. D. Damsteegt, and R. D. Frederick. 2004. Specific detection and quantification of Plum pox virus by real-time fluorescent reverse transcription-PCR. J. Virol. Methods 120:97-105. [DOI] [PubMed] [Google Scholar]
- 17.Sokal, R. R., and F. J. Rohlf. 1995. Biometry: the principles and practices of statistics in biological research, 3rd ed. W. H. Freeman and Company, New York, N.Y.
- 18.Wang, R. Y., E. D. Ammar, D. W. Thornbury, J. J. Lopez-Moya, and T. P. Pirone. 1996. Loss of potyvirus transmissibility and helper-component activity correlate with non-retention of virions in aphid stylets. J. Gen. Virol. 77:861-867. [DOI] [PubMed] [Google Scholar]
- 19.Wang, R. Y., and S. A. Ghabrial. 2002. Effect of aphid behavior on efficiency of transmission of Soybean mosaic virus by the soybean-colonizing aphid, Aphis glycines. Plant Dis. 86:1260-1264. [DOI] [PubMed] [Google Scholar]