Abstract
The antibody responses elicited in rhesus macaques immunized with soluble human immunodeficiency virus (HIV) Env gp140 proteins derived from the R5-tropic HIV-1 SF162 virus were analyzed and compared to the broadly reactive neutralizing antibody responses elicited during chronic infection of a macaque with a simian/human immunodeficiency virus (SHIV) expressing the HIV-1 SF162 Env, SHIVSF162P4, and humans infected with heterologous HIV-1 isolates. Four gp140 immunogens were evaluated: SF162gp140, ΔV2gp140 (lacking the crown of the V2 loop), ΔV3gp140 (lacking the crown of the V3 loop), and ΔV2ΔV3gp140 (lacking both the V2 and V3 loop crowns). SF162gp140 and ΔV2gp140 have been previously evaluated by our group in a pilot study, but here, a more comprehensive analysis of their immunogenic properties was performed. All four gp140 immunogens elicited stronger anti-gp120 than anti-gp41 antibodies and potent homologous neutralizing antibodies (NAbs) that primarily targeted the first hypervariable region (V1 loop) of gp120, although SF162gp140 also elicited anti-V3 NAbs. Heterologous NAbs were elicited by SF162gp140 and ΔV2gp140 but were weak in potency and narrow in specificity. No heterologous NAbs were elicited by ΔV3gp140 or ΔV2ΔV3gp140. In contrast, the SHIVSF162P4-infected macaque and HIV-infected humans generated similar titers of anti-gp120 and anti-gp41 antibodies and NAbs of significant breadth against primary HIV-1 isolates, which did not target the V1 loop. The difference in V1 loop immunogenicity between soluble gp140 and virion-associated gp160 Env proteins derived from SF162 may be the basis for the observed difference in the breadth of neutralization in sera from the immunized and infected animals studied here.
The envelope gene of the human immunodeficiency virus type 1 (HIV-1) encodes the viral envelope glycoprotein gp160, which mediates the binding and fusion of the virus with target cells. The functional form of the HIV envelope glycoprotein (Env) on the surface of infectious virions is believed to be a trimer (12, 52, 94), although nonfunctional forms of Env are also present on the virion surface (60). HIV Env is the target of neutralizing antibodies (NAbs), and several passive antibody infusion studies have indicated that the presence of high titers of NAbs directed to the challenge virus at the time of viral exposure can protect from infection (41, 56, 57, 68, 78). Therefore, the design of HIV Env-derived immunogens capable of eliciting relevant NAb responses could greatly benefit HIV vaccine efforts.
Soluble mimics of the Env trimer comprising all of gp120 and the extracellular portion of gp41, termed gp140, have been engineered and tested as immunogens in an attempt to elicit NAbs (1, 3, 7, 13, 21-23, 26, 27, 34, 35, 46, 49, 53, 80, 92). Overall, these constructs appear to be more effective in eliciting cross-reactive NAb responses than soluble monomeric gp120 immunogens (1, 3, 23, 34, 46, 49, 92), but the breadth of neutralizing responses elicited by the currently available soluble gp140 trimers is still limited. Several groups, including ours, are attempting to engineer soluble gp140 constructs on which the immunogenicity of the most variable Env regions, those against which NAbs with narrow breadth of activity are believed to be elicited, is eliminated or greatly reduced while the immunogenicity of conserved regions is increased (1, 14, 17, 24, 28, 36, 39, 40, 43, 44, 47, 50, 51, 53-55, 59, 66, 67, 72, 77). Although it is hoped that a reduction in the immunogenicity of the more variable regions of Env will result in a concomitant increase in the immunogenicity of the more conserved regions against which cross-reactive NAbs are elicited, this has not yet been achieved. Furthermore, it is not possible to accurately predict the immunogenic properties of particular HIV Env regions by their antigenic properties (14, 50, 51, 77).
To increase our understanding of the relationship between epitope presentation and immunogenicity on HIV Env immunogens, an iterative approach in which the immunogenic properties of newly designed HIV Env immunogens are correlated with their structural and biophysical properties is required. To this end, we and others have been immunizing animals with gp140 proteins and analyzing the potency, breadth, and epitope specificities of the NAbs elicited (3, 49, 77, 80).
We previously reported on the engineering as well as the antigenic and immunogenic characterization of soluble trimeric gp140 proteins derived from the R5-tropic HIV-1 SF162 virus (1, 80). SF162 was chosen because it is highly susceptible to neutralization by broadly reactive NAbs (74), suggesting that the epitopes these NAbs recognize are efficiently exposed on SF162 Env, and immunization with SF162 Env-derived constructs may result in the generation of such NAbs. In a pilot study, SF162gp140 and a derivative lacking part of the second variable region (V2), termed ΔV2gp140, both elicited homologous NAbs in rabbits and macaques as well as NAbs against certain heterologous HIV viruses, including primary HIV-1 isolates (1). Our initial analysis indicated that a portion of the antibodies elicited by these two gp140s bound linear epitopes in the V3 loop (80). Because those pilot studies were conducted with a small number of animals and the overall heterologous NAb responses were weak and narrow in breadth, we wanted to confirm the findings in a larger number of animals as well as expand the studies by including gp140 constructs with additional modifications. We hypothesized that the elicitation of linear anti-V3 antibodies might explain the limited breadth of neutralization of those immune sera. Therefore, in the current study, we engineered two additional gp140 constructs lacking the V3 loop: one derived from SF162gp140, termed ΔV3gp140, and the second derived from ΔV2gp140, and termed ΔV2ΔV3gp140. We reevaluated the immunogenic properties of SF162gp140 and ΔV2gp140 in greater detail and compared them to the immunogenic properties of ΔV3gp140 and ΔV2ΔV3gp140.
The breadth of neutralization and epitope specificity of the antibodies elicited by these four immunogens were also compared to those of antibodies elicited in a macaque infected with the simian/human immunodeficiency virus SHIVSF162P4 (8), which expresses an Env similar to the HIV-1 SF162 Env. This animal developed broadly reactive NAbs during the course of infection, suggesting that the SF162 Env is capable of eliciting such NAbs. We also compared the epitope specificity of pooled human serum immunoglobulin G (IgG) from patients infected with heterologous HIVs, which generated cross-reactive NAb responses, to that of the above-mentioned gp140-immunized and SHIVSF162P4-infected macaque. Such a comparative analysis may assist in the design of improved gp140 immunogens.
MATERIALS AND METHODS
Cells.
Human embryonic kidney 293T cells and the U87 human astroglioma cell line expressing human CD4 and CCR5 were cultured in complete Dulbecco's minimal essential medium (DMEM) as previously described (74). The HeLa-derived TZM-bl cell line (from David Montefiori, Duke University, Durham, NC), stably expressing human CD4, CCR5, and CXCR4 and containing cassettes for beta-galactosidase and firefly luciferase (Luc), both under control of the HIV-1 long terminal repeat, was cultured in complete DMEM. The canine thymocyte cell line CF2.CD4Th.CCR5 (25) was cultured in complete DMEM supplemented with G418 (0.6 mg/ml) and hygromycin (0.15 mg/ml).
Antibodies.
Reagents recognizing the CD4-binding site (CD4BS) were CD4-IgG (PRO542), obtained from Progenics Pharmaceuticals Inc. (Tarrytown, NY); the monoclonal antibodies (MAbs) IgG1b12 and IgG1b6, both provided by Dennis Burton (The Scripps Research Institute, La Jolla, CA) (9); and F105, obtained through the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, NIH, from Marshall Posner (70, 86). MAb 17b, directed to a CD4-induced (CD4i) epitope (85), and the anti-V3 MAb LE311 were provided by James Robinson (Tulane University, New Orleans, LA). The anti-V1 MAb P3C8 was isolated from a mouse immunized with ΔV2gp140, and the anti-V3 MAb P3E1 was isolated from a mouse immunized with SF162gp140 (Derby and Stamatatos, unpublished data). The human anti-V3 MAb 447D (18, 29, 33) was provided by Susan Zolla-Pazner and Mirek Gorny (New York University, New York, NY). Fab X5 (61) was provided by Dimiter Dimitrov (National Cancer Institute, Frederick, MD). The anti-V2 MAb G3.4 was provided by Michael Fung (Tanox, Houston, TX) (38). Anti-gp41 MAbs 2F5 and 4E10 (96) were obtained through the ARRRP, as were MAb 2G12, directed to a complex glycan epitope on gp120 (73, 75), and the HIV-positive IgG pool (HIVIG). Four-domain soluble CD4 (sCD4) was from Chiron Co.
Proteins and peptides.
Purified monomeric SF162gp120, trimeric SF162gp140, and trimeric ΔV2gp140 proteins were provided by Chiron Co. The HxB2 gp41 ectodomain lacking the fusion peptide, membrane-spanning, and cytoplasmic domains was purchased from Viral Therapeutics (Ithaca, NY). Chimeric proteins (CP) containing the V3 loop of JR-CSF (V3-CP) or the V1V2 region of SF162 (V1V2-CP) on the backbone of the murine leukemia virus (MLV) envelope glycoprotein gp70 (45) were provided by Abraham Pinter (Public Health Research Institute, Newark, NJ).
Peptides derived from the V1 and V2 variable regions of the SF162 Env as well as a scrambled V3 peptide were purchased from Sigma-Genosys (The Woodlands, TX). An SF162 V3 peptide was provided by Chiron Co., and V4 and V5 peptides, derived from the SHIVSF162P3 Env, were obtained through the ARRRP. Two gp41 peptides derived from the epitopes of MAbs 2F5 and 4E10 were provided by John Mascola and Richard Wyatt (VRC/NIH, Bethesda, MD). The sequences of all above-mentioned peptides are as follows: V1 whole, representing the sequence of the V1 loop between the cysteines, TNLKNATNTKSSNWKEMDRGEIK; V2 N terminus, representing the amino-terminal side of the V2 loop, TTSIRNKMQKEYALF; V2 C terminus, representing the carboxy-terminal side of the V2 loop, YKLDVVPIDNDNTSY; V2 base, representing the five amino-terminal and five carboxy-terminal amino acids of the V2 loop linked by the GAG tripeptide as is present on the SF162ΔV2 Env (81), CSFKVGAGKLINC; V3 crown, representing the 14 central amino acids of the V3 loop, CTRKSITIGPGRAFYC; V3 scramble (used as a control), TRKSFYATPGRAITIG; seven overlapping peptides spanning the V4 loop of SHIVSF162P3, which has a sequence identical to HIV-1 SF162, CGGEFFYCNSTQLFN, FFYCNSTQLFNSTWN, NSTQLFNSTWNNTIG, LFNSTWNNTIGPNNT, TWNNTIGPNNTNGTI, TIGPNNTNGTITLPC, and NNTNGTITLPCRIKQ; six overlapping peptides spanning the V5 loop of SHIVSF162P3, IRCSSNITGLLLTRD, SNITGLLLTRDGGR(K)E, GLLLTRDGGR(K)EV(I)G(S)NT, TRDGGR(K)EV(I)G(S)NTTEIF, GR(K)EV(I)G(S)NTTEIFRPGG, and G(S)NTTEIFRPGGGDMR (these sequences differ from that of HIV-1 SF162 at the amino acids indicated in parentheses, which are those found in SF162 preceded by the amino acid found in SHIVSF162P3); peptide spanning the epitope of MAb 2F5, NEQELLELDKWASLWN; and peptide spanning the epitope of 4E10, NWFDITNWLWYIRKKK.
Generation of ΔV3gp140 and ΔV2ΔV3gp140 DNA constructs.
The generation of pEMC* DNA vectors encoding SF162gp140 and the derivative ΔV2gp140 has been described previously (1, 82), as has the construction of the gp160 versions of SF162 and ΔV2 lacking the crown of V3, termed ΔV3 and ΔV2ΔV3, respectively (74). gp140 versions of ΔV3 and ΔV2ΔV3 were generated by introducing two stop codons immediately upstream of the transmembrane region of gp41. For each construct, two versions were engineered: gp140C (used during the DNA phase of immunization; see below), in which the cleavage site between gp120 and gp41 remained intact, and gp140F (used during recombinant Env immunization; see below), in which the cleavage site was eliminated by site-directed mutagenesis to aid in protein stability for purification (79).
The templates for the ΔV3 mutagenesis were the above-mentioned “cleaved” and “fused” forms of SF162gp140 and ΔV2gp140 in pUC18 vectors. Each gp140 insert was flanked by a 5′ NheI site and a 3′ ClaI site, which were used for subsequent subcloning into the pEMC* vector. To create the 21-amino-acid V3 deletion while introducing the Gly-Ala-Gly (GAG) tripeptide linker in its place, a forward primer, ΔV3F (5′-CAATAATGGAGCTGGAGATATAAGAC AAGCAC-3′), and a reverse primer, ΔV3R (5′-TATATCTCCAGCTCCATTATTGTTAGGTCTTG-3′), which overlapped the deletion and introduced GAG (shown in the primer sequence by the underlined nucleotides), were designed. Two separate first-round PCRs were performed (reactions A and B). In reaction A, primer ΔV3R and an outer forward primer at the 5′ end of gp140, gp160F (5′-GGACCATAGTGTACATAGAATACAG-3′), were used, and in reaction B, ΔV3F and an outer reverse primer at the 3′ end of gp140, env8R (5′-CACAATCCTCGCTGCAATCAAG-3′), were used. These reactions generated separately the 5′ and 3′ halves of each gp140 construct. In the second round of PCR, the products of the two first-round PCRs were combined and amplified with primers gp160F and env8R to generate full-length gp140 constructs. These final PCR products were ligated into TOPO TA cloning vectors, digested with NheI and ClaI, and subcloned into the pEMC* expression vector (15). All of the mutagenesis reactions were performed using the Expand High Fidelity PCR system (Roche). Following mutagenesis, the entire gp140 was sequenced to verify the introduction of the desired deletions and the absence of any additional mutations. For immunization, an endotoxin-free GIGA prep kit (QIAGEN, Valencia, CA) was used to generate sterile endotoxin-free preparations of each gp140C and control plasmid.
Protein expression and Western blotting.
293T cells were transiently transfected with 5 μg of pEMC* DNA vectors expressing SF162gp140, ΔV2gp140, ΔV3gp140, or ΔV2ΔV3gp140 as previously described (82). In some cases, the cells were cotransfected with 5 μg of a plasmid encoding the cellular cleavage enzyme furin (5). Supernatants were collected 48 h posttransfection.
Transfection supernatants were subjected to electrophoresis on 6% Tris-glycine gels (Invitrogen). Proteins were transferred to Immobilon P membranes (Millipore, Billerica, MA), blocked, and probed with the mouse anti-V1 MAb P3C8 (1 μg/ml) or the human anti-gp41 MAb 2F5 (2 μg/ml) overnight at 4°C. Membranes were incubated with goat anti-mouse IgG (heavy plus light chains)-horseradish peroxidase (1:5,000; Bio-Rad, Hercules, CA) or goat anti-human IgG (heavy plus light chains)-horseradish peroxidase (1:3,000; Bio-Rad) antibodies and washed, and protein bands were visualized by enhanced chemiluminescence (Amersham, Piscataway, NJ).
Production and purification of gp140 proteins for immunization purposes.
Purification of SF162gp140F and ΔV2gp140F trimeric proteins has been described previously (79). Similar methods were used to purify the ΔV3gp140F and ΔV2ΔV3gp140F proteins. Briefly, suspension-adapted 293T cells were transfected with pEMC* vectors encoding ΔV3gp140F or ΔV2ΔV3gp140F. gp140 proteins in the supernatants were purified first on Galanthus nivalis lectin-agarose and then on DEAE resin. Eluted trimeric gp140-containing fractions were pooled, concentrated, and size fractionated on a Superdex-200 column (Pfizer Pharmacia, New York, NY) (79).
Antigenicity of purified gp140Fs by ELISA.
Epitope exposure on the purified trimeric gp140 proteins was determined by enzyme-linked immunosorbent assay (ELISA) according to the previously described protocol (80).
Macaques.
All animals in this study were Indian rhesus macaques. Those immunized with ΔV3gp140 (n = 3) or ΔV2ΔV3gp140 (n = 3) or mock immunized (n = 3) were housed at the Tulane National Primate Research Center (Covington, LA). Those immunized with SF162gp140 (n = 6) and ΔV2gp140 (n = 6) were part of a separate challenge study (91) and were housed at the Washington National Primate Research Center (Seattle, WA). The macaque that was chronically infected with SHIVSF162P4 (animal C640) was previously described by our group (8).
Immunization protocol.
All macaques were immunized by the DNA prime plus protein boost vaccination method as previously described (1, 8, 91). Animals received three monthly immunizations with 2 mg of DNA encoding the appropriate gp140C (or the empty vector for mock immunizations) in 1 ml of endotoxin-free water. The DNA was delivered both intramuscularly (0.8 mg at two sites in the quadriceps muscle) and intradermally (0.2 mg at two sites). Following the third DNA immunization (7 months for the SF162gp140 and ΔV2gp140 groups and 12 months for the ΔV3gp140, ΔV2ΔV3gp140, and mock groups), the animals were immunized intramuscularly in the deltoids with 0.1 mg of the corresponding recombinant purified soluble gp140F proteins emulsified in MF-59 adjuvant (Chiron) in a final volume of 0.5 ml per animal. During recombinant protein boost, the SF162gp140- and ΔV2gp140-immunized animals were also immunized a fourth time with DNA vectors. The ΔV3gp140 and ΔV2ΔV3gp140 animals were immunized with the recombinant protein alone. The mock-immunized group received adjuvant alone. At the time of each immunization as well as intermittently in the time between the DNA and protein phases of immunization, the animals were bled, and the plasma was isolated and stored at −80°C. Before use, plasma was heat inactivated for 1 h at 56°C and centrifuged at 16,000 × g for 10 min. At 2, 4, and 6 weeks after protein boost, serum rather than plasma was similarly isolated and stored at −80°C until use.
Purification of IgG from macaque serum.
Heat-inactivated sera (preimmunization and from 2 weeks after recombinant protein boost) from macaques in each group were pooled, and the serum IgG was isolated by affinity purification on protein A/G agarose (Pierce) columns according to the manufacturer's instructions. IgG was eluted using imidazole, pH 2.0 (Pierce). The binding of each fraction to SF162gp140 was determined by ELISA, and the anti-SF162gp140 IgG-containing fractions were pooled, diluted 2,000-fold in phosphate-buffered saline (PBS), and concentrated by centrifugation through an Amicon Ultra-15 50-kDa-cutoff filter (Millipore). The concentration of the final product was determined by UV absorbance at 280 nm in comparison with a bovine gamma globulin standard (Bio-Rad).
Determination of anti-Env binding antibody titers.
Relative endpoint serum antibody titers were determined by ELISA as previously described (1, 80, 91) on plates coated overnight with SF162gp120, HxB2 gp41, V1V2SF162-CP, or V3JR-CSF-CP proteins (50 ng/well in 100 mM NaHCO3) at room temperature or with peptides derived from the gp120 variable regions or the gp41 epitopes for 2F5 and 4E10 (100 ng/well in 200 mM NaHCO3) at 37°C. Relative endpoint antibody titers were determined by subtracting twice the preimmunization or preinfection optical density at 490 nm (OD490) from the postimmunization or postinfection OD490 at each serum dilution and determining the serum dilution at which the resulting value was zero.
Production of single-round-competent viruses.
Luciferase reporter viruses capable of only a single round of replication were generated as previously described with a few minor modifications (74). 293T cell transfections were performed using the Gene Juice transfection reagent (Invitrogen). In most cases, viruses were concentrated through an Amicon Ultra-15 100-kDa-cutoff filter (Millipore) to enhance the infectious titer per milliliter. Pseudovirus p24 content was assayed using a kit from the NIH ARRRP.
Single-round-competent virions that expressed the following Env proteins were generated: SF162, 89.6, ADA, JRFL, and YU2 (all isolates from chronic infection) and 6535, SS1196, REJO, and RHPA (primary isolates collected within 3 months following HIV infection [48]). Pseudoviruses expressing an SF162 Env with mutations in the 2F5 and 4E10 epitopes (Δ2F5.4E10 Env, provided by John Mascola and Richard Wyatt) that abrogate the binding of the corresponding MAbs were also generated. Single-round-competent virions expressing the unrelated amphotropic MLV Env were also generated and used as negative controls in neutralization assays. Finally, pseudoviruses that did not express an Env protein (Δenv) were generated and used as negative controls during virus-cell entry assays.
Neutralization assays.
TZM-bl cells (3 × 103 cells in 100 μl of medium) or U87 CD4 CCR5 cells (7 × 103 cells in 100 μl of medium) were seeded in 96-well flat-bottomed tissue culture plates (Corning) and allowed to adhere overnight. Macaque sera collected prior to the initiation of immunization or infection (prebleeds) and sera collected at 2 and 4 weeks following immunization with recombinant gp140 or at various times after SHIVSF162P4 infection (immune sera) were titrated and incubated in triplicate with an equal volume of pseudovirus for 1 h at 37°C in 96-well U-bottomed tissue culture plates (Corning). The amount of single-round-competent virus used was predetermined to give a cell-associated Luc reading of 105 to 106 relative luciferase units (RLU) per well. The sera-virus mixture was added to Polybrene-treated cells for 72 h at 37°C. The cells were lysed in 1× cell culture lysis buffer (Promega, Madison, WI) per the manufacturer's instructions and frozen at −80°C. The cell lysate was thawed and transferred to a black 96-well plate (for TZM-bl cells) or a white 96-well plate (for U87 cells). Luc substrate (Promega) was added, and the RLU per well were recorded on a Fluoroskan Ascent fluorimeter (Thermo electron) according to the manufacturer's instructions. Percent neutralization at each serum dilution was calculated from the reduction in single-round-competent virus entry in the presence of immune sera relative to the entry in the presence of the prebleed sera from that animal as described mathematically here: [(RLUprebleed − RLUimmune)/RLUprebleed] × 100.
When purified macaque serum IgG or human serum IgG (HIVIG) was used in the neutralization assay, the following changes were made to the above protocol: the virus-IgG mixture was incubated with cells for only 3 h, after which the cells were washed and received fresh medium. Following 48 h of incubation, the cells were lysed as described above.
Epitope mapping of neutralizing antibodies. (i) Detection of serum neutralizing antibodies to gp120 epitopes.
For detection of serum NAbs to linear epitopes in gp120 defined by the peptides described above, serial dilutions of sera (20 μl) were incubated in triplicate with an equal volume of peptide (10 μg/ml during this incubation step) for 1 h at 37°C prior to incubation with 20 μl of single-round-competent virus for 1 h at 37°C. In other experiments, we used the above-mentioned chimeric proteins (CP) expressing the V1V2 or V3 HIV-1 Env regions on the MLV background. Fifty microliters of the sera-peptide (or CP)-virus mixture was then incubated with Polybrene-treated U87 CD4 CCR5 cells for 72 h at 37°C, the cells were lysed, and Luc was read as described above. The percent reduction in neutralization in the presence of the competing agent was determined at the serum dilution that resulted in 70% inhibition of infection in the absence of peptide or CP. As a control for a potential effect of peptide or CP on neutralization, we evaluated the effect of the V3 peptide and the V3-CP on SF162 pseudovirus infectivity in the absence of immune sera and found that at 10 μg/ml, neither the peptide nor the CP had an effect.
(ii) Detection of 2F5- and 4E10-like neutralizing antibodies in sera.
To determine whether 2F5- or 4E10-like NAbs were elicited during gp140 immunization or during SHIVSF162P4 or HIV-1 infection, we preabsorbed sera with 2F5 or 4E10 peptides, as described above. We also used a second approach, during which the neutralizing potential of serum was evaluated against a virus expressing the Δ2F5.4E10 Env. This virus is resistant to 2F5- or 4E10-mediated neutralization but is susceptible to other MAbs (J. R. Mascola, personal communications and data presented below).
(iii) Post-CD4 neutralization assay.
A modified version of a recently described assay for detection of antibodies that bind to CD4-induced epitopes (20) was employed. Briefly, single-round-competent SF162 virus (20 μl) was first incubated without or with an equal volume of sCD4 for 1 h at 37°C at an sCD4 concentration (0.3 μg/ml) that results in less than 50% inhibition of infection. Serially diluted immune sera (20 μl) were then added to the sCD4-virus mixture for another hour at 37°C, and 50 μl of the mixture was added to Polybrene-treated TZM-bl cells for 72 h at 37°C. The percent neutralization in the presence of sCD4 was determined at the 50% inhibitory concentration (IC50) in the absence of sCD4.
(iv) Virus capture competition.
Virus capture by MAbs has been described previously (10, 11, 31, 58, 60, 62, 64, 69). The epitope reactivity profiles of sera were determined by their ability to inhibit capture of virus-like particles (VLPs) expressing the JRFL Env by a panel of MAbs. Microwells were coated with MAbs representing major epitope clusters overnight at 5 μg/ml. The wells were then washed and blocked with 3% bovine serum albumin in PBS. Graded dilutions of macaque sera were added to the virus, and the virus-serum mixtures were then added to MAb-coated ELISA wells for 3 h followed by washing with PBS. CF2.CD4.CCR5 cells were overlaid, and 2 days later infection was measured by assaying Luc as described above. The reciprocal serum dilution that inhibited MAb-mediated virus capture by 50% was recorded. Because the JRFL VLPs may be neutralized by the competing antibody, VLPs also expressed on their surfaces vesicular stomatitis virus G (an amphotropic and highly fusogenic molecule) to provide a readout of captured virus irrespective of neutralization, as previously described (60).
SHIVSF162P4 challenge.
Six weeks following immunization with the recombinant HIV Env gp140, the macaques in the ΔV3gp140, ΔV2ΔV3gp140, and mock groups were challenged intravenously with 100 50% tissue culture infective doses of cell-free SHIVSF162P4. Blood was collected every week for the first month postchallenge and monthly thereafter. The viral RNA copy number in plasma was determined by branched-chain DNA amplification assay (Bayer Reference Testing Laboratory, Tarrytown, NY) in which the lower limit of detection is 125 RNA copies per ml. Statistical analysis of peak viremia was performed using a two-tailed t test on the log-transformed values for peak virus RNA.
RESULTS
Expression and processing of SF162gp140, ΔV2gp140, ΔV3gp140, and ΔV2ΔV3gp140.
The expression and fusogenic potentials of SF162-derived gp160 proteins lacking parts of the V2 and/or the V3 loops have been reported previously (74, 83), as has the expression of soluble gp140 forms of these proteins, SF162gp140 and ΔV2gp140 (82). Here we generated ΔV3gp140 and ΔV2ΔV3gp140 constructs and compared their expression and recognition by MAbs to those of SF162gp140 and ΔV2gp140. All gp140C and gp140F proteins were expressed and secreted into the culture medium (see Fig. S1A and B in the supplemental material), indicating that neither the deletion of the V2 and/or V3 loops nor the elimination of the gp120-gp41 cleavage site abrogated protein expression or processing, which is in agreement with previous reports (72, 82, 89).
As expected, in the supernatants of gp140F-expressing cells, a single HIV Env protein species was present (see Fig. S1A in the supplemental material). In the supernatants of gp140C-expressing cells, however, two species were present (see Fig. S1B in the supplemental material): one species of higher molecular weight (MW) that was recognized by both the anti-gp120 MAb P3C8 and the anti-gp41 MAb 2F5 and a second species of a lower MW that was recognized only by P3C8. Coexpression of the cellular protease furin during transfection with the gp140Cs resulted in the presence of only the lower-MW species, recognized by only P3C8, in the supernatant (see Fig. S1B in the supplemental material). Therefore, the higher-MW protein species most likely corresponds to gp140 that is not properly cleaved due to its overexpression and saturation of the cellular cleavage machinery (4, 5). The coexpression of furin, which, as expected, had no effect on the noncleavable gp140F proteins (see Fig. S1A in the supplemental material), resulted in complete processing of gp140C (see Fig. S1B in the supplemental material). Under the denaturing conditions of sodium dodecyl sulfate-polyacrylamide gel electrophoresis, gp140C dissociated into gp120 (see Fig. S1B in the supplemental material) and the extracellular part of gp41 (4, 5; not shown).
Antigenic characterization of purified trimeric gp140 proteins.
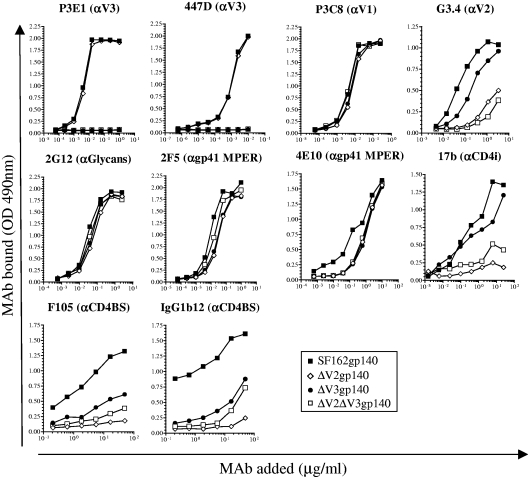
Several groups have shown that monomers, dimers, trimers, and other oligomeric species are present in supernatants of cells transfected with DNA vectors expressing gp140 proteins (3, 60, 76, 79). We purified the trimers from the other gp140 Env forms as previously described (79). Purified trimeric SF162gp140F, ΔV2gp140F, ΔV3gp140F, and ΔV2ΔV3gp140F proteins (containing less than 5% of other gp140 Env forms [data not shown]) were evaluated by ELISA to determine whether and how the modifications affected the exposure of various gp120 and gp41 epitopes (Fig. 1). The mouse anti-V3 MAb P3E1 (which neutralizes SF162 efficiently; see below) and the well-characterized neutralizing anti-V3 MAb 447D recognized SF162gp140 and ΔV2gp140 equally and, as expected, failed to recognize ΔV3gp140 or ΔV2ΔV3gp140. The anti-V1 MAb P3C8 (which also neutralizes SF162 efficiently; see below) recognized all four proteins equally. The epitope for MAb 2G12, a conformational array of closely spaced high-mannose sugars on gp120, was also equally exposed on all gp140 proteins. Another MAb that recognized all four proteins equally and with high affinity was the anti-gp41 MAb 2F5, although the anti-gp41 MAb 4E10, which recognizes an epitope directly C-terminal to the epitope of 2F5, was recognized less efficiently on the V2- and V3-deleted proteins than on SF162gp140. Deletion of the crown of the V2 loop reduced the binding of MAb G3.4 to its epitope at the base of V2 (84). It appears that deletion of the V3 loop itself had an effect on the binding of G3.4, since this antibody recognized ΔV3gp140 less efficiently than SF162gp140 and recognized ΔV2ΔV3gp140 less efficiently than ΔV2gp140. The negative effect of V3 loop deletion on G3.4 binding observed in this study is in contrast to results from a previous study (95) in which the binding of G3.4 to modified monomeric gp120s derived from JRFL was examined. It is possible that the V2 loops of SF162 and JRFL are differentially positioned. It is also possible that the observed differences are due to the use of gp140 and not gp120 as a binding target for G3.4 in our studies.
FIG. 1.
Recognition of gp140s by MAbs. The binding of MAbs directed to linear and conformational gp120 and gp41 epitopes on SF162gp140F (filled squares), ΔV2gp140F (open diamonds), ΔV3gp140F (filled circles), and ΔV2ΔV3gp140F (open squares) was determined as discussed in Materials and Methods.
The 17b MAb, which preferentially recognizes its epitope on gp120 in the CD4-bound configuration, bound to SF162gp140 and ΔV3gp140 more efficiently than ΔV2gp140 and ΔV2ΔV3gp140. Two anti-CD4BS MAbs, b12 and F105, which recognize overlapping epitopes in the CD4BS (65, 87), bound their epitopes on SF162gp140, but deletion of the V2 or V3 loops reduced their binding. These results are consistent with previous data that have implicated the V2 and V3 loops in the binding of certain anti-CD4BS MAbs (65, 71, 95).
Taken together, these antigenicity results are consistent with previous reports indicating that the exposure of distinct neutralization epitopes on HIV Env is differentially affected by the V2 and/or V3 loops. In the case of the SF162-derived Env gp140s examined here, the removal of the V2 and/or V3 loops reduced the exposure of certain epitopes in the CD4-binding site and epitopes that become preferentially exposed upon Env-CD4 binding. To examine whether and how these modifications and the absence of the V2 and/or V3 loops affect the ability of these gp140 proteins to elicit anti-HIV NAbs, we immunized macaques and compared the neutralization potentials and epitope specificities of the elicited antibodies.
Antibodies elicited during immunization or infection with homologous SHIVSF162P4 and heterologous HIV-1.
Relative endpoint titers for serum antibodies specific for gp120 and gp41 throughout the immunization course were determined for all groups (Table 1). Both the individual titers for each animal and the median titer for each group are reported. Little to no binding antibody was detectable by ELISA during the DNA phase of the immunization protocol (data not shown), similar to what we have described in our previous studies (1, 8). Following recombinant HIV Env gp140F protein boosting, all animals developed anti-HIV Env binding antibodies against both gp120 and gp41. These titers peaked at 2 weeks after protein boost and subsequently declined in most animals over the next several weeks (Table 1 and data not shown).
TABLE 1.
Antibody titers and epitope specificities of antibodies present in immune sera
| Group | Animal | Antibody titer fora:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gp120b
|
gp41b
|
V1V2
|
V3
|
V4V5
|
Control (V3 scramble) | ||||||||
| 2 wpb | 4 wpb | 2 wpb | 4 wpb | V1 | V2 | V1V2-CP | V3 | V3-CP | V4 | V5 | |||
| SF162 | A02177 | 60,000 | 24,500 ± 3,535 | 8,000 | 6,500 ± 707 | 1,700 ± 1,838 | — | 7,000 | 250 ± 212 | 3,700 | — | — | — |
| A02179 | 18,000 | 2,300 ± 1,273 | 3,500 | —f | 550 ± 212 | — | 1,000 | — | — | — | — | — | |
| A02186 | 19,000 | 7,500 ± 3,536 | 2,900 | 1,500 ± 141 | 350 ± 71 | — | 1,300 | 400 ± 141 | 260 | — | — | — | |
| A02188 | 33,000 | 21,500 ± 707 | 6,500 | 3,500 ± 2,121 | 5,500 ± 707 | — | 13,500 | 260 ± 198 | 1,500 | — | — | — | |
| 02193 | 12,000 | 7,500 ± 2,121 | 2,000 | 1,500 ± 0 | 1,000 ± 849 | — | 3,000 | 100 ± 100 | 700 | — | — | — | |
| Medianc | 19,000 | 7,500 | 3,500 | 1,500 | 1,000 | — | 3,000 | 250 | 700 | — | — | — | |
| ΔV2 | 02187 | 15,000 | 8,000 ± 2,828 | 2,600 | 1,400 ± 141 | 2,000 ± 1,414 | — | 4,000 | — | — | — | — | — |
| A02183 | 3,000 | 6,750 ± 2,475 | 1,500 | — | 110 ± 14 | — | 1,300 | — | — | — | — | — | |
| 02184 | 12,000 | 8,500 ± 2,121 | 11,000 | 7,500 ± 707 | 750 ± 71 | — | 4,000 | 100 ± 0 | 400 | — | — | — | |
| A02178 | 19,000 | 23,000 ± 4,243 | 12,000 | 9,000 ± 1,414 | 2,400 ± 2,263 | — | 7,000 | 140 ± 57 | 340 | — | — | — | |
| Medianc | 11,000 | 8,250 | 6,800 | 4,450 | 1,375 | — | 4,000 | 75 | 195 | — | — | — | |
| ΔV3 | AV28 | 18,000 | 20,000 | 3,400 | 1,600 | 18,500 ± 16,264 | — | 26,000 | — | — | — | — | — |
| BD59 | 10,000 | 21,000 ± 1,414 | 10,000 | 5,500 ± 707 | 4,000 ± 2,828 | — | 7,000 | — | — | — | — | — | |
| CD15 | 7,200 | 6,750 ± 1,061 | 3,300 | 2,250 ± 354 | 4,850 ± 4,455 | — | 4,600 | — | — | — | — | — | |
| Medianc | 10,000 | 20,000 | 3,400 | 2,250 | 4,850 | — | 7,000 | — | — | — | — | — | |
| ΔV2ΔV3 | CN63 | 31,000 | 30,000 ± 0 | 2,000 | — | 3,100 ± 2,687 | — | 4,200 | — | — | — | — | — |
| M733 | 2,000 | 2,350 ± 919 | 900 | 1,200 ± 283 | 2,950 ± 2,899 | — | 1,100 | — | — | — | — | — | |
| N422 | 20,000 | 21,000 ± 1,414 | 3,300 | 2,500 ± 707 | 15,500 ± 7,778 | — | 1,450 | — | — | — | — | — | |
| Medianc | 20,000 | 21,000 | 2,000 | 1,200 | 3,100 | — | 1,450 | — | — | — | — | — | |
| SHIVSF162P4) | C640, day 178d | 200,000 | 200,000 | 11,000 ± 5,657 | — | 29,000 | 2,250 ± 354 | 80,000 | 350 ± 70 | 250 ± 70 | — | ||
| C640, day 304 | 300,000 | 300,000 | 7,500 ± 707 | — | 100,000 | 3,250 ± 1,061 | 117,000 | — | — | — | |||
| C640, day 643 | 100,000 | 200,000 | 1,550 ± 71 | — | 100,000 | 500 ± 0 | 47,000 | 550 ± 70 | 30 ± 10 | — | |||
| HIVIGe | 0.4 ± 0.1 | 0.04 ± 0.02 | 39.5 ± 14.8 | — | 3.4 | 13.5 ± 7.8 | 0.7 | 5.9 ± 3.7 | 10.5 ± 5.9 | — | |||
Values indicate relative endpoint ELISA titer against the indicated proteins or peptides.
Sera were collected 2 and 4 weeks following the gp140 immunization (wpb). Serum from animal C640 was collected at the indicated days post-SHIVSF162P4 infection.
Median antibody titer of each group.
The number of days after infection with SHIVSF162P4 is indicated.
Titers recorded for HIVIG are in μg/ml.
—, not detected.
Within each group as a whole (as well as in most individual animals), the median titer of anti-gp120 antibodies was greater (2- to 20-fold) than the median titer of anti-gp41 antibodies, suggesting that gp120 is more immunogenic than gp41 on the gp140 proteins. The apparent lower titers of anti-gp41 than anti-gp120 antibodies could also be related to the use of the homologous SF162gp120 and heterologous HxB2 gp41 proteins (91% amino acid homology with the corresponding region from SF162gp41).
The ΔV2gp140 immunogen elicited the highest anti-gp41 antibody titers of all the gp140 immunogens tested here (Table 1). It appears, therefore, that deletion of V2 alters the immunogenicity of certain gp41 epitopes. Such an increase in immunogenicity could be related to an increase in exposure of certain epitopes, but these epitopes do not include the 2F5 epitope, which was equally exposed on all gp140 proteins, or the 4E10 epitope, which was less exposed on the modified gp140s than on SF162gp140 (Fig. 1 and additional data presented below). In general, within each group, the animals with the highest response to gp120 also had the highest response to gp41, suggesting that some animals responded better to immunization than others, leading to the development in these animals of a higher total concentration of HIV Env-specific IgG.
In addition to the gp140-immunized macaques, we also investigated the antibody responses elicited during infection of macaque C640 with SHIVSF162P4, which expresses the homologous (SF162) Env, and in HIVIG, which is serum IgG pooled from several humans infected with heterologous HIV-1 isolates. These macaque and human sera were chosen for comparison because they contained broadly reactive NAbs (see below). Overall, sera from the infected animal and from humans had much higher anti-Env antibody titers than the gp140 immune sera (Table 1). Also, sera from the SHIVSF162P4-infected animal, C640, collected between 6 months and 2 years postinfection had either roughly equal titers of anti-gp120 and anti-gp41 antibodies or higher titers to gp41 than gp120. HIVIG contained approximately 10-fold-higher titers of anti-gp41 than anti-gp120 antibodies.
Serum antibody reactivity with SF162gp140, ΔV2gp140, ΔV3gp140, and ΔV2ΔV3gp140.
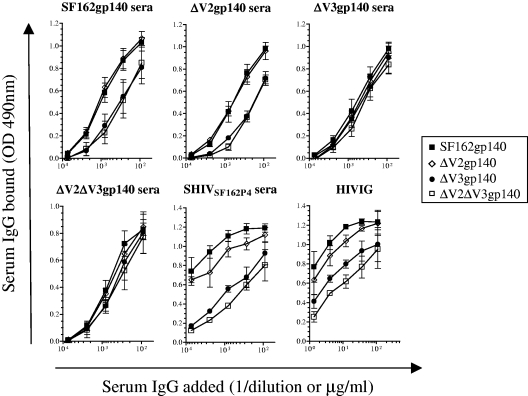
To determine whether deletion of V2 and/or V3 from SF162gp140 created new immunogenic epitopes (neoepitopes), we investigated how immune sera collected from the animals immunized with the four gp140 immunogens recognized these proteins (Fig. 2). For these experiments, we pooled the sera from animals within each immunized group. The relative recognition of these proteins by SHIVSF162P4 sera and HIVIG was also determined. The SF162gp140 and ΔV2gp140 proteins were similarly recognized by the SF162gp140- and ΔV2gp140-elicited serum antibodies. However, the gp140 proteins with deletions in the V3 loop were less efficiently recognized by these sera. This most likely suggests that SF162gp140 did not elicit antibodies to the V2 loop and that a fraction of the antibodies elicited by both SF162gp140 and ΔV2gp140 were directed to the V3 loop. Sera from animals immunized with either ΔV3gp140 or ΔV2ΔV3gp140 recognized all four gp140 proteins similarly, suggesting that deletion of the V3 loop from SF162gp140 or ΔV2gp140 did not create neoepitopes on these proteins.
FIG. 2.
Recognition of gp140s by serum antibodies. Sera collected 4 weeks following the recombinant gp140 immunization step from the immunized animals, from the SHIVSF162P4-infected macaque, C640, at day 643 postinfection and HIVIG were added to ELISA wells coated with the indicated gp140s, and the binding of serum IgG was determined. Sera from each immunized group were pooled. Preimmunization or preinfection sera were used as a control for nonspecific binding, and the means with standard deviation of results from three separate experiments are shown.
In contrast, serum antibodies from the SHIVSF162P4-infected animal and HIVIG recognized the SF162gp140 protein more efficiently than ΔV2gp140 (Fig. 2). The gp140s containing deletions in the V3 loop were less efficiently recognized. The decreased recognition of the V3-deleted proteins might be explained in part by the presence of V3-directed antibodies in these sera. The somewhat weaker binding to V2-deleted proteins may be due to the presence of V2-directed antibodies in the sera or to antibodies directed to non-V2 epitopes which become masked or eliminated by V2 deletion, for example, CD4BS epitopes (Fig. 1). SHIVSF162P4 serum antibodies and HIVIG recognized the gp140 proteins with a similar pattern.
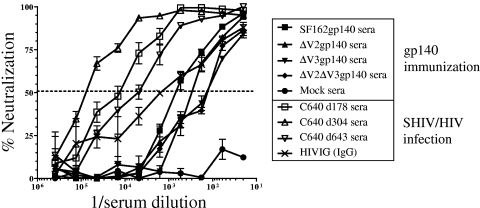
Neutralization of SF162 virus by immune sera.
The neutralizing potential of serum antibodies from the immunized animals was first evaluated against the homologous SF162 virus. Neutralization of individual or pooled sera (Table 2 and Fig. 3) was determined. All four gp140 immunogens elicited homologous anti-SF162 NAbs. The titers of anti-SF162 NAbs declined in all animals from week 2 to week 4 after recombinant gp140 immunization (Table 2), which is similar to what we observed with the binding antibody titers (Table 1). Immunization with V2- or V3-deleted gp140 proteins elicited lower titers of homologous anti-SF162 NAbs (Table 2 and Fig. 3). Sera from the SHIVSF162P4-infected macaque, C640, contained higher titers of anti-SF162 NAbs than the sera collected from the gp140-immunized animals, consistent with the higher overall anti-Env antibody titers in sera from the SHIVSF162P4-infected animal (Table 1). HIVIG also contained anti-SF162 NAbs but at lower titers than the SHIVSF162P4 sera (assuming 10 mg/ml for serum IgG) (42) (Table 2 and Fig. 3).
TABLE 2.
Epitope specificity of serum neutralizing antibodies
| Group | Animal | 50% Neutralization titer (IC50)a
|
% Reduction in neutralization at the IC70
|
Percent increase in neutralization at the IC50 with sCD4d | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 wpb | 4 wpb | V1 | V1V2-CP | V3 | V3-CP | V3 scr.b | 2F5c | 4E10c | |||
| SF162 | A02177 | 4,050 ± 354 | 1,917 ± 567 | 24% ± 6% | 41% ± 11% | 7% ± 6% | — | — | — | — | |
| A02179 | 4,050 ± 354 | 733 ± 231 | 61% ± 24% | 52% ± 4% | 9% ± 8% | — | — | — | — | ||
| A02186 | 1,180 ± 113 | 260 ± 14 | —e | 33% ± 23% | 16% ± 7% | 14% | — | — | — | ||
| A02188 | 4,200 ± 1,414 | 3,500 ± 707 | 55% ± 1% | 54% ± 6% | 9% ± 2% | — | — | — | — | ||
| 02193 | 390 ± 113 | 275 ± 35 | 41% ± 5% | 36% ± 18% | 8% ± 2% | 9% | — | — | — | ||
| Median | 4,050 | 733 | 41% | 41% | 9% | — | — | — | — | 30% ± 5% | |
| ΔV2 | 02187 | 687 ± 103 | 237 ± 97 | 63% ± 4% | 46% ± 5% | 20% ± 1% | — | — | — | — | |
| A02183 | 823 ± 282 | 695 ± 148 | 55% ± 2% | 48% ± 8% | 7% ± 5% | — | — | — | — | ||
| 02184 | 640 ± 53 | 190 ± 14 | 53% ± 0% | 44% ± 3% | 5% ± 2% | — | — | — | — | ||
| A02178 | 1,047 ± 81 | 505 ± 219 | 26% ± 8% | 32% ± 15% | — | — | — | — | — | ||
| Median | 755 | 371 | 54% | 45% | 6% | — | — | — | — | 28% ± 15% | |
| ΔV3 | AV28 | 850 ± 212 | 225 ± 92 | 67% ± 4% | 70% ± 4% | — | — | — | — | — | |
| BD59 | 413 ± 4 | 193 ± 116 | 72% ± 11% | 70% ± 4% | — | — | — | — | — | ||
| CD15 | 280 ± 113 | 140 ± 98 | 79% ± 0% | 71% ± 1% | — | — | — | — | — | ||
| Median | 413 | 193 | 72% | 70% | — | — | — | — | — | 27% ± 7% | |
| ΔV2ΔV3 | CN63 | 142 ± 40 | 106 ± 48 | 88% ± 2% | 62% ± 17% | — | — | — | — | — | |
| M733 | 120 ± 21 | 93 ± 13 | 64% ± 0% | 55% ± 7% | — | — | — | — | — | ||
| N422 | 1,335 ± 474 | 633 ± 350 | 49% ± 6% | 50% ± 0% | — | — | — | — | — | ||
| Median | 142 | 106 | 64% | 55% | — | — | — | — | — | 33% ± 7% | |
| Mock | AM10 | — | — | NDf | ND | ND | ND | ND | ND | ND | ND |
| AM94 | — | — | ND | ND | ND | ND | ND | ND | ND | ND | |
| AP18 | — | — | ND | ND | ND | ND | ND | ND | ND | ND | |
| Median | — | — | ND | ND | ND | ND | ND | ND | ND | ND | |
| SHIVSF162P4 | C640, day 178 | 17,500 ± 3,535 | — | — | 36% | — | — | — | ND | ND | |
| C640, day 304 | 40,000 ± 20,000 | — | 36% | 10% | — | — | — | ND | ND | ||
| C640, day 643 | 5,000 ± 50 | — | — | 18% | 19% | — | — | — | 25% ± 0% | ||
| HIVIGg | 13.5 ± 8 | — | — | 33% | 29% | — | — | — | 29% ± 1% | ||
Sera were collected 2 and 4 weeks following the gp140 immunization (wpb). Serum from animal C640 was collected at the indicated days post-SHIVSF162P4 infection.
V3 scr., V3 scrambled peptide.
Peptides recognized by MAbs 2F5 and 4E10.
This indicates detection of NAbs following incubation of pseudovirus with soluble CD4.
—, <5% reduction in the presence of peptide/chimeric protein.
ND denotes not done.
Titers recorded for HIVIG are in μg/ml.
FIG. 3.
Neutralization of SF162 by immune sera and HIVIG. Pooled sera collected from gp140-imunized animals 4 weeks after protein boost or from mock-immunized animals, sera collected from the SHIVSF162P4-infected macaque, C640, at days 178, 304, and 643 postinfection, and HIVIG were tested for neutralization against SF162. The percent neutralization was calculated using preimmunization or preinfection sera as described in Materials and Methods. The lowest serum dilution tested was 1:20. The highest HIVIG concentration was 0.5 mg/ml, which is roughly equivalent to a 1:20 serum dilution.
Heterologous neutralization by purified serum IgG.
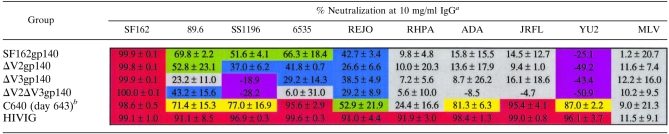
We previously reported that immunization with SF162gp140 and ΔV2gp140 elicited antibodies capable of neutralizing certain heterologous isolates although the cross-reactive NAb responses were narrow in breadth and weak in potency (1). Here we determined whether elimination of the V3 loop from these immunogens improved the elicitation of cross-reactive NAbs (Table 3). Initial experiments with high concentrations of sera from the ΔV3gp140- and ΔV2ΔV3gp140-immunized animals indicated that these sera had unusually high cell-killing activity and this resulted in an artificial enhancement of their neutralizing potential. We therefore decided to purify IgG from the sera of all immunized animals and directly test their neutralizing potential against SF162 and heterologous isolates. Sera from the animals of each group were pooled prior to IgG purification. Pooled IgG from the immunized animals as well as the HIV-positive IgG pool, HIVIG, were used at a concentration of 10 mg/ml, which is approximately the concentration of IgG in normal human serum (42). Due to the limited quantity of serum from the SHIVSF162P4-infected animal, C640, serum from this animal was used at a 1:20 dilution, which is equivalent to 0.5 mg/ml. In these neutralization experiments, the unrelated virus, MLV, was used as a control for nonspecific IgG-associated neutralizing activity. A serum IgG was considered to have anti-HIV neutralizing activity against a particular isolate when the percent neutralization obtained against that isolate was greater than 1 standard deviation above the percent neutralization recorded with that serum IgG against MLV.
TABLE 3.
Neutralization of heterologous isolates by purified serum IgG
The values are the average with standard deviation of results from two independent experiments performed in duplicate. The C640 sera were tested at a 1:20 serum dilution and not 10 mg/ml. Red, >90% neutralization; yellow, 70.0 to 89.9% neutralization; green, 50.0 to 69.9% neutralization; blue, <50% neutralization but above background; gray, background; purple, enhancement of infection.
Sera from the SHIVSF162P4-infected animal at 643 days postinfection.
Purified serum IgG from all immunized groups neutralized the homologous SF162 virus very efficiently (Table 3), which indicates that the neutralizing activity of the sera (Table 2 and Fig. 3) was IgG mediated. HIVIG (at 10 mg/ml) and SHIVSF162P4-derived sera (at a 1:20 serum dilution, which corresponds to approximately 0.5 mg/ml IgG) also potently neutralized SF162. At that IgG concentration, HIVIG potently neutralized all the heterologous HIV-1 isolates tested here, while SHIVSF162P4 sera neutralized (to various degrees) all isolates except RHPA (Table 3). The differential abilities of SHIVSF162P4 sera and HIVIG to neutralize RHPA is most likely due to the presence of certain antibodies in HIVIG, but not in SHIVSF162P4 sera, that recognize a particular Env epitope. The greater potency with which HIVIG neutralized the remaining heterologous isolates is most likely related to the fact that the concentration of IgG used in these assays was 20-fold greater for HIVIG than for the SHIVSF162P4 sera. In contrast, serum IgG from any of the four gp140-immunized groups failed to neutralize RHPA, ADA, JRFL, and YU2. Serum IgG elicited by SF162gp140 and ΔV2gp140 did neutralize 89.6, SS1196, 6535, and REJO, with the potency of neutralization by the ΔV2gp140-elicited serum IgG being lower. In contrast, serum IgG from the groups immunized with ΔV3gp140 and ΔV2ΔV3gp140 showed only weak neutralizing activity against 89.6 and REJO, and 6535 was weakly neutralized by only the ΔV3gp140-elicited serum IgG. Finally, infection of YU2 was slightly enhanced by all of the gp140-elicted IgGs, although it was neutralized efficiently by both SHIVSF162P4 sera and HIVIG.
Epitope mapping.
The difference in neutralization breadth between the gp140-immunized and SHIVSF162P4-infected animals could be due to the observed differences in the total anti-Env antibodies present in those sera or to differences in the epitope specificity of antibodies present in the sera (Fig. 2). To address this, we first performed epitope mapping studies in an ELISA format using SF162 Env-derived peptides (Table 1). A scrambled V3 peptide, used as a control for nonspecific binding by sera, was not recognized by any of the sera tested or by the MAb P3E1 directed to the V3 loop crown (Table 1 and data not shown). Antibodies to the V4 or V5 regions were not detectable in any of the sera from the gp140-immunized animals, irrespective of the gp140 immunogen, although they were detectable at a very low level in the sera from the SHIVSF162P4-infected animal. The absence of significant titers of V4- and V5-specific responses is not surprising, since the V4 and V5 regions of the HIV Env are highly glycosylated and are not expected to be very immunogenic (88). HIVIG also contained antibodies capable of binding to these regions even though the peptides used here were derived from HIV-1 SF162 while HIVIG was isolated from humans infected with heterologous HIV-1 isolates. Antibodies against the V2 loop were undetectable in the SF162gp140 sera, in accordance with our previous observations (80), or in ΔV3gp140 sera. Similarly, the V2 loop was not immunogenic on the surface of SF162 Env-expressing infectious SHIVSF162P4 virions, and HIVIG contained no antibodies capable of recognizing peptides derived from the SF162 V2 loop.
In contrast to the low immunogenicity of the V2, V4, and V5 regions, both the V1 and V3 loops elicited antibodies during immunization and during infection (Table 1). In fact, the V1 loop was immunogenic on all gp140 proteins, and V1-specific antibodies accounted for approximately 10 to 25% of the total anti-gp120 antibodies present in sera from the gp140-immunized animals (based on ELISA assays). While deletion of V2 did not affect the elicitation of anti-V1 antibodies, deletion of V3 enhanced both the actual titer of anti-V1 antibodies and their relative proportion in the total gp120-specific pool. The crown of the V3 loop was immunogenic on both SF162gp140 and ΔV2gp140 although much less so than the V1 loop. These results are in accordance with the reduced binding of sera from SF162gp140- and ΔV2gp140-immunized animals to the V3-deleted proteins (Fig. 2). As expected, antibodies against the crown of the V3 loop were absent from the ΔV3gp140 and ΔV2ΔV3gp140 sera.
Similar to the gp140-immunized animals, the SHIVSF162P4-infected animal elicited higher anti-V1 than anti-V3 antibody responses, especially during the early stages of infection (Table 1). However, the relative ratio of anti-V1 to anti-V3 antibodies was lower in the SHIVSF162P4-infected animal than in the gp140-immunized animals. The antibodies present in HIVIG had a different ratio of epitope specificity than those observed in either gp140-immunized or SHIVSF162P4-infected animal sera, since antibodies specific for the V3 region were present at a threefold higher titer than those specific for the V1 region. This may in part explain the greater breadth of neutralization recorded with HIVIG than SHIVSF162P4 sera (Table 3). However, as seen in the SHIVSF162P4-infected animal, V1- and V3-specific antibodies represented only a small fraction of the total anti-gp120 antibodies in HIVIG (Table 1). In part, these results obtained with HIVIG may be related to the use of SF162-derived peptides for these experiments.
The above results were obtained with peptides as targets for serum antibodies, and most likely they report on the relative titers of antibodies recognizing linear epitopes on HIV Env. To examine whether the gp140 proteins elicited antibodies that recognize conformational epitopes within the V1, V2, and V3 regions, we used CPs which, on the background of MLV Env, express the V1V2 region of SF162 or the V3 region of JR-CSF. The relative antibody titers to V1V2-CP were higher than those to the V1 peptide for the SF162gp140-, ΔV2gp140-, and ΔV3gp140-immunized animals (Table 1). This is not surprising, since the CPs are recognized by antibodies to linear and conformational epitopes, while the V1 peptide is most likely recognized only by antibodies to linear epitopes. Alternatively, antibodies to conformational epitopes on V2 may be present in those sera and recognize V1V2-CP but not the V2 peptides. Interestingly, the dual deletion of the V2 and V3 loops resulted in higher antibody titers to the V1 peptide than the V1V2-CP, suggesting that the animals immunized with ΔV2ΔV3gp140 made predominantly antibodies to linear V1 epitopes that are not efficiently exposed on the V1V2-CP.
Higher relative antibody titers were also recorded against V3-CP than the V3 peptide in the SF162gp140 and ΔV2gp140 sera (Table 1), again most likely because the sera also contain antibodies to conformational V3 epitopes.
The SHIVSF162P4-infected animal also had higher titers of antibodies to the V1V2-CP and V3-CP than the corresponding peptides (Table 1). The relative ratio of antibodies recognizing the CPs over the peptides also increased over the course of infection. Early (day 178) during infection, the ratio of anti-V1V2-CP to anti-V1 peptide antibodies was approximately 3, while later during infection (days 304 and 643), this ratio increased to 100. The ratio of anti-V3-CP to anti-V3 peptide antibodies was high (around 30 to 40) at 178 and 304 days postinfection and increased (close to 100) at 643 days postinfection. The above results suggest that during the course of SHIVSF162P4 infection, more and more antibodies to conformational epitopes on the V1V2 and V3 regions were generated. Although immunization with our gp140 constructs also resulted in the generation of higher titers of antibodies to conformational than linear V1V2 or V3 epitopes, the relative ratio of antibodies to conformational versus linear epitopes was lower than that recorded during SHIVSF162P4 infection.
Similar to the results with the SHIVSF162P4-derived sera, a higher proportion of antibodies in HIVIG recognized the chimeric proteins than the corresponding linear peptides (Table 1). This is not a surprise, since HIVIG was isolated from patients infected with viruses other than SF162 and therefore any cross-reactive antibodies present in these sera would most likely recognize conformational epitopes rather than linear amino acid sequences.
Mapping the epitopes recognized by NAbs in immunized and infected animals or humans.
Antibodies can bind to their epitopes on Env-derived peptides or CPs in ELISA assays without recognizing these epitopes on the virion-associated Env. Thus, although the epitope mapping studies described above provide information on the immunogenic properties of specific epitopes on gp140 proteins, they do not inform us about whether antibodies to a given epitope actually neutralize HIV. To address this directly, we performed “epitope interference” neutralization experiments during which the neutralizing potency of sera was determined in the absence or presence of the peptides and CPs discussed above. In these experiments, reductions in serum neutralizing potency were recorded at the serum dilution that resulted in 70% inhibition of infection of SF162 in the absence of peptide (IC70). The mean reduction in serum neutralizing potency at that serum dilution from two to three independent experiments is recorded in Table 2 as the percent reduction in neutralization, and the medians of those means for each group are also shown.
(i) V1V2 and V3 epitopes.
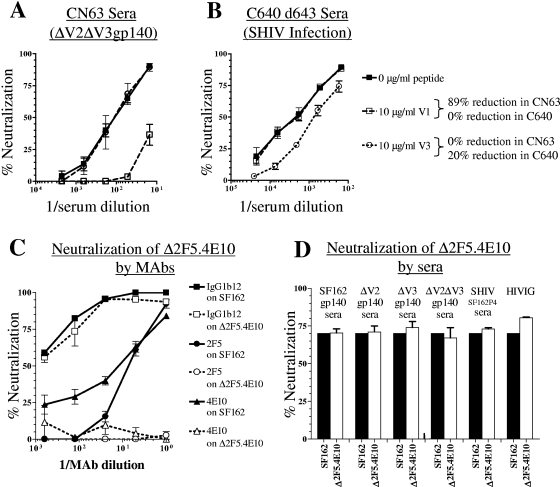
We first examined the contribution of V1- and V3-directed antibodies to neutralization of SF162 (Table 2), since epitopes in these regions were immunogenic on the gp140 proteins (Table 1). In control experiments, we showed that the neutralization potentials of P3C8 (anti-V1) and P3E1 (anti-V3) against SF162 were reduced by 81% and 93%, respectively, in the presence of the corresponding V1 and V3 peptides (10 μg/ml) (see Fig. S2A in the supplemental material). As expected, the neutralizing activity of the anti-gp41 MAb 4E10 was unaffected by the presence of the V3 peptide (see Fig. S2A in the supplemental material), and a scrambled V3 peptide did not significantly affect the neutralizing potency of any of the MAbs or sera tested (Table 2), although it resulted in a 6% reduction in neutralization by the anti-V3 MAb P3E1 (see Fig. S2A in the supplemental material). As a result, during these experiments any reduction in serum neutralizing activity of less than 5% was considered to be nonspecific. Preincubation of sera from the gp140-immunized animals with the V1 peptide significantly reduced the serum neutralizing activity. The group medians were between 41% (in the case of SF162gp140 sera) and 72% (in the case of ΔV3gp140 sera) (Table 2). The highest median reduction was recorded with the ΔV3gp140 sera, and the highest reduction in an individual animal was recorded with animal CN63 (88% reduction in neutralizing potency) in the ΔV2ΔV3gp140 group (Fig. 4A and Table 2). These results indicate that a large fraction of the NAbs present in sera from gp140-immunized animals, regardless of the gp140 immunogen used, was directed to the V1 loop, and this was particularly evident in sera from animals immunized with V3-deleted immunogens and consistent with the high titers of binding antibodies to V1 in these animals (Table 1). In contrast, the neutralizing activity of serum antibodies from the SHIVSF162P4-infected animal was unaffected by preincubation with the V1 peptide (Fig. 4B and Table 2), irrespective of the time the sera were collected during infection. Similarly, preincubation of HIVIG with the V1 peptide did not reduce its neutralizing activity (Table 2). These results, obtained by preincubation of sera with the V1 peptide, are supported by the finding that the SF162 virus from which the V1 loop had been deleted (ΔV1) was resistant to neutralization by sera from the gp140-immunized animals but not by sera from the SHIVSF162P4-infected animal (see Table S1 in the supplemental material).
FIG. 4.
Mapping the epitopes of anti-SF162 neutralizing antibodies. (A) Peptide-mediated interference of SF162 neutralization by sera from the ΔV2ΔV3gp140-immunized macaque CN63. Neutralization was performed in the absence (filled symbols and solid lines) and presence (open symbols and dashed lines) of V1 (squares) or V3 (circles) peptides. The percent reduction in neutralization at the IC70 is indicated. (B) Peptide-mediated interference of SF162 neutralization by sera from the SHIVSF162P4-infected macaque C640. The symbols are the same as in panel A. (C) Neutralization of SF162 and Δ2F5.4E10 by MAbs. IgG1b12, squares; 2F5, circles; 4E10, triangles; SF162, filled symbols; and Δ2F5.4E10, open symbols. (D) Neutralization of SF162 and Δ2F5.4E10 by pooled sera from the immunized animals and from the SHIVSF162P4-infected animal. The IC70 is shown for each serum pool on SF162 (black bars), and neutralization at the IC70 for SF162 is shown for each serum pool on Δ2F5.4E10 (white bars).
As expected, the neutralizing potency of sera from the ΔV3gp140- and ΔV2ΔV3gp140-immunized animals was unaffected by the V3 peptide (Fig. 4A and Table 2). Consistent with the low but detectable titer of binding antibodies to the V3 peptide in sera from SF162gp140- and ΔV2gp140-immunized animals, the neutralizing activity of SF162gp140-elicted serum antibodies was reduced by 9% and that of ΔV2gp140-elicited antibodies was reduced by 6% following preincubation with the V3 peptide (Table 2). Interestingly, the SF162gp140-immunized animal (A02186) with the highest percentage (16%) of V3-specific NAbs had no detectable V1-specific NAbs. Preabsorption with the V3 peptide of sera collected early during SHIVSF162P4 infection (day 178) resulted in a greater reduction in serum neutralizing activity than that recorded with sera collected later during infection (Fig. 4B and Table 2). Approximately 33% of the anti-SF162 neutralizing activity of HIVIG was also blocked by preabsorption with the SF162-derived V3 peptide (Table 2).
Although the SF162-derived V3 peptide was capable of absorbing NAbs present in sera from all of the SF162gp140-immunized animals, this was not the case when the JR-CSF-derived V3-CP was used (Table 2). In addition, although the neutralizing activity of serum antibodies from the SHIVSF162P4-infected macaque was reduced by preincubation with the V3 peptide at all times tested postinfection, absorption with the V3-CP resulted in a reduction in neutralizing activity only at the latest time point tested (643 days postinfection) (Table 2), in keeping with the preponderance of conformation-dependent V3 binding antibodies on that day (Table 1). Thus, it appears that during SHIVSF162P4 infection, NAbs to conformational V3 epitopes are generated, while the gp140 immunogens used here do not elicit such antibodies, even though low titers of conformational V3 binding antibodies were present in the sera from some of the SF162gp140- and ΔV2gp140-immunized animals (Table 1). Most likely, these antibodies are not present at high titers, and they may not bind to the virion surface, although they bind to the V3-CP (JR-CSF). Despite the higher titer of binding antibodies to V3-CP than to the V3 peptide in HIVIG, the neutralizing activity of HIVIG was reduced to similar degrees by the V3 peptide and the V3-CP (Table 2).
Preabsorption of gp140-elicted serum antibodies with the V1V2-CP resulted in a reduction of neutralizing activity similar to that recorded with the V1 peptide (Table 2), indicating that, as suggested by the ELISA epitope mapping data (Table 1), the V1-specific NAbs elicited by immunization were directed to linear epitopes. In contrast, the neutralizing activity of HIVIG was not affected by absorption with V1V2-CP (Table 2). Interestingly, the neutralizing activity of serum antibodies from the SHIVSF162P4-infected animal was reduced by absorption with the V1V2-CP only at 304 days after SHIVSF162P4 infection. This suggests that conformational NAbs against the V1V2 region of the SF162 Env were transiently elicited in that animal.
(ii) gp41 membrane-proximal external region epitopes.
The highly conserved epitopes for the broadly neutralizing MAbs 2F5 and 4E10 are present and exposed on all four gp140 immunogens tested here (Fig. 1). We therefore examined whether or not our gp140 immunogens elicited 2F5- or 4E10-like NAbs and whether HIVIG or SHIVSF162P4 sera contained such NAbs.
Two methodologies were used to detect the presence of 2F5- and 4E10-like NAbs in macaque sera and HIVIG. First, sera were absorbed with peptides spanning the 2F5 and 4E10 epitopes. Control experiments indicated that, indeed, these peptides absorbed the neutralizing activity of the corresponding MAbs, while no effect was observed on the neutralization of the anti-CD4BS MAb IgG1b12 (see Fig. S2A in the supplemental material). However, no effect on the neutralizing potency of any of the sera tested was evident (Table 2). Second, we used an SF162 virus that expresses a modified Env with mutations in the 2F5 and 4E10 epitopes, termed Δ2F5.4E10. These mutations render the virus resistant to neutralization by 2F5 and 4E10 while having no effect on the susceptibility of the virus to other unrelated MAbs, such as IgG1b12 (Fig. 4C). It is anticipated that if NAbs directed to the 2F5 or 4E10 epitopes were present in the immune sera, the sera would neutralize Δ2F5.4E10 less efficiently than SF162. If such NAbs were not present in the sera, then the two viruses would be equally susceptible to neutralization by the tested sera. The SF162 and Δ2F5.4E10 viruses were similarly susceptible to neutralization by all sera tested (Fig. 4D), indicating that 2F5- or 4E10-like NAbs were not present (at least to a level that was detectable by the assays used here) in sera from the gp140-immunized animals or the SHIVSF162P4-infected animal or in HIVIG. These results indicate that the neutralizing activity associated with those sera was 2F5 and 4E10 independent.
(iii) Post-CD4-binding epitopes.
Epitopes induced upon the binding of gp120 to CD4 have recently been recognized as highly conserved across lentiviruses and highly immunogenic during infection (20). To determine whether our gp140 immunogens elicit CD4-induced (CD4i) antibodies and whether such antibodies are present in sera from the SHIVSF162P4-infected animals that display broad neutralizing activity, we preincubated SF162 without or with sCD4 (at concentrations that do not induce more than 50% inhibition of infection in the absence of serum) and then added the virus-sCD4 mixture to serially diluted pooled sera from each gp140-immunized group of animals. Neutralization of the sCD4-treated and untreated viruses was compared, and the increase in neutralization in the presence of sCD4 was determined at the serum dilution that resulted in 50% inhibition of infection in the absence of sCD4. This increase in neutralizing potency at the IC50 is recorded in Table 2 as the percent increase in neutralization. As a control in this experiment, we demonstrated that neutralization of SF162 by the CD4i MAb 17b was enhanced in a concentration-dependent manner by preincubation with sCD4 (see Fig. S2B in the supplemental material). The preincubation of SF162 with sCD4 resulted in an increase in the neutralizing activity of sera from all gp140-immunized groups and from the SHIVSF162P4-infected animal and the HIV-1-infected humans (HIVIG) (Table 2). Here we need to emphasize that incubation of virions with sCD4 exposes diverse epitopes, including epitopes interacting with coreceptor molecules (such as the 17b epitope), V3 epitopes, and others (19). We were unable to detect any differences in the titers of these various types of antibodies between the different gp140 sera because the assay reports on the binding of antibodies to different types of epitopes that become exposed upon sCD4 binding. Furthermore, we could not detect significant differences in the frequency of CD4i antibodies between the gp140 sera and SHIVSF162P4 sera or HIVIG.
(iv) Characterization of serum antibody epitopes by competition with MAbs.
We next determined the ability of serially diluted sera to inhibit virus capture by specific MAbs (60). In these experiments, single-round-competent virions expressing the JRFL Env were used in conjunction with pooled sera from each immunized group collected 4 weeks following the recombinant gp140 immunization step and sera from the SHIVSF162P4-infected animal (Table 4). In certain instances, the virus was incubated with sCD4 prior to its incubation with sera. As a positive control, virus was preincubated with the same MAb used for capture (self competition). As a negative control, a pool of preimmunization sera from all of the animals was shown to have no effect on virus binding to the MAbs tested. In these experiments, we did not use HIVIG.
TABLE 4.
Epitope specificities of serum antibodies determined by MAb binding competition
| Group | Binding to JRFL gp120a | Neutralization of JRFLb
|
Competition titerc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4BS
|
Glycan (2G12) | V3
|
CD4i (X5/sCD4) | Cluster I (7B2) | |||||||
| Standard | After CD4 | b6 | 15e | b12 | LE311 | LE311/sCD4 | |||||
| SF162gp140 | 5,000 | <100 | 6,000 | <100 | <100 | <100 | <100 | 125 | 250 | <100 | <100 |
| ΔV2gp140 | 4,000 | <100 | 3,000 | 100 | 225 | <100 | <100 | 100 | 500 | <100 | <100 |
| ΔV3gp140 | 10,000 | <100 | 120 | <100 | 100 | <100 | <100 | 100 | 500 | 150 | <100 |
| ΔV2ΔV3gp140 | 17,000 | <100 | <100 | 100 | 125 | <100 | <100 | <100 | 150 | <100 | <100 |
| SHIVSF162P4d | 150,000 | 250 | >30,000 | 300,000 | 500 | <100 | 500 | 17,000 | 70,000 | 24,000 | >25,000 |
| Prebleede | <1,000 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 |
| Selff | 0.01 | 0.08 | 0.02 | 0.2 | 0.008 | 0.008 | 0.005 | 0.005 | |||
Values indicate the reciprocal serum dilution resulting in half-maximal binding.
Values indicate reciprocal serum dilution resulting in 50% neutralization in the absence of sCD4 (standard) or following preincubation of viruses with sCD4 (after CD4).
Values indicate the reciprocal serum dilution at which MAb-mediated virus capture was competed 50%.
For SHIVSF162P4, the sera tested were collected 643 days postinfection.
For prebleed, preimmunization sera pooled from all animals were included in all groups.
Self denotes competition with the same MAb used for capture recorded in μg/ml.
Immunization with the four gp140 immunogens resulted in the generation of binding antibodies to JRFL gp120 (Table 4), suggesting that antibodies to epitopes which are conserved between SF162 and JRFL were elicited; however, these antibodies did not neutralize the JRFL virus. Their titers were lower than those elicited during SHIVSF162P4 infection (similar to what we observed with the homologous SF162gp120 [Table 1]). Although neutralization of JRFL was not recorded with any of the gp140 sera, it was observed with the SHIVSF162P4 sera. Interestingly, however, when JRFL was preincubated with sCD4, it became neutralizable by the gp140 sera (the exception being the ΔV2ΔV3gp140 sera). This indicates that the gp140 immunogens used here (with the exception of ΔV2ΔV3gp140) elicited low titers of antibodies to conserved Env regions, but these epitopes were not accessible on the virus during virus-cell fusion.
Concerning the presence of antibodies that bind to epitopes overlapping the CD4BS on the HIV Env, b12-like antibodies were not detected in any of the gp140 sera or in sera from the SHIVSF162P4-infected animal (Table 4), suggesting that such antibodies did not contribute to the neutralizing potential of these sera. However, 15e- and b6-like antibodies that are able to neutralize only the most sensitive HIV isolates (6) were generated during SHIVSF162P4 infection and may have been present at very low titers in sera from macaques immunized with the V2- and/or V3-deleted gp140 immunogens although none were detected in sera from the SF162gp140-immunized animals. Interestingly, low titers of antibodies capable of blocking the binding of 2G12 were present in the SHIVSF162P4 sera but not in any of the gp140 sera. As expected from the neutralization results discussed above, antibodies that bind to neutralization epitopes that preferentially become exposed following the interaction of Env with CD4 were present in low titers in sera from the gp140-immunized animals and in higher titers in the SHIVSF162P4-infected animal. Such antibodies comprised mostly anti-V3 antibodies (as indicated by the presence of LE311-like antibodies). CD4i antibodies (as indicated by the presence of X5-like antibodies) were elicited at low titers by ΔV3gp140 but not by the other immunogens. Of note, although antibodies like LE311 captured JRFL in the absence of sCD4 preincubation, they did so most effectively once the virus was preincubated with sCD4. In contrast, X5 does not capture JRFL in the absence of sCD4 pretreatment (19).
The fact that sera from the ΔV3gp140- and ΔV2ΔV3gp140-immunized animals competed the binding of JRFL to the anti-V3 MAb LE311 (Table 4) is most likely due to the presence of antibodies in those sera that sterically prevent the binding of virions to certain anti-V3 antibodies, such as LE311.
We also examined whether antibodies to the gp41 cluster I region were generated, because this region is immunodominant in gp41 and has been demonstrated to be highly immunogenic during HIV-1 infection of humans (63, 90). To assess whether this region was similarly immunogenic on our gp140 immunogens and on the surface of SHIVSF162P4 virions, we competed capture of virus to the cluster I-specific MAb 7B2 with sera (Table 4). Only serum antibodies from the SHIVSF162P4-infected animal were able to compete out capture by the MAb 7B2, indicating that during SHIVSF162P4 infection, as during HIV infection, the gp41 cluster I region is highly immunogenic, while it is poorly immunogenic on our gp140 immunogens. Therefore, the greater immunogenicity of the extracellular gp41 region during SHIVSF162P4 infection compared to gp140 immunization (Table 1) is most likely due to the generation of anti-cluster I antibodies during infection.
Previous immunization reduced peak viremia following homologous challenge.
We have previously shown that immunization with SF162gp140 and ΔV2gp140 results in the generation of homologous NAbs that reduce SHIVSF162P4 replication in macaques challenged intravenously, even in the absence of antiviral CD8+-T-cell-mediated immune responses (8). Here we examined whether the antibodies elicited by ΔV3gp140 and ΔV2ΔV3gp140 also affect SHIVSF162P4 replication in vivo. As mentioned above, the SF162gp140- and ΔV2gp140-immunized animals were part of a different challenge study during which the animals were challenged with the heterologous SHIV 89.6P virus (91). Six weeks following immunization with recombinant ΔV3gp140 or ΔV2ΔV3gp140 proteins, two animals in the ΔV3gp140 group, all three animals in the ΔV2ΔV3gp140 group, as well as three mock-immunized animals were intravenously challenged with SHIVSF162P4. Although none of the animals was protected from infection (sterilizing immunity), peak viremia levels (at 2 weeks postchallenge) were lower in the immunized animals than in controls (see Table S2 in the supplemental material). The number of animals used per group was insufficient to perform a statistical analysis on each group of immunized macaques separately compared to the controls. However, a statistically significant reduction in peak viremia was reached (P = 0.05) when the ΔV3gp140- and ΔV2ΔV3gp140-immunized animals were grouped. Although the number of animals used here is very small to draw solid conclusions, the results suggest that NAbs that target regions other than the crown of the V3 loop that were present in the ΔV3gp140 and ΔV2ΔV3gp140 sera were capable of blunting homologous in vivo replication. Since these two immunogens elicited robust anti-V1 NAb responses (Table 2), it is possible that the dampening of viral replication recorded here was due to such antibodies. Additional studies are required to define the relative contributions of the various types of NAbs elicited during immunization (or infection) in controlling viral replication in vivo.
DISCUSSION
The neutralizing antibody response elicited during immunization with our four SF162-derived gp140 immunogens was primarily due to V1-directed antibodies. Immunization with gp140 proteins lacking the V3 loop individually or in combination with the V2 loop resulted in increased titers of anti-V1 NAbs. This may be a consequence of a loss in the interaction between the V1 and V3 loops within the Env trimer resulting in more effective exposure of certain V1 loop epitopes (16), which do not include the epitope recognized by MAb P3C8, which recognized the V3 loop-deleted gp140s as efficiently as SF162gp140. Alternatively, the relative immunogenicity of V1 may be boosted by any modification that reduces the immunogenicity of V3, such as has been shown on the HxB2 gp120 Env by the addition of a glycosylation site to the V3 crown (28). This focusing of the immune response to the V1 region upon gp140 immunization in part explains the narrow breadth of neutralizing activities elicited by our immunogens. The V1 region is highly variable, and thus it is expected that anti-V1 antibodies elicited by SF162-derived gp140 Env will not bind to the virion-associated V1 loops of heterologous isolates. During SHIVSF162P4 infection, anti-V1 binding antibodies were generated, but their titers decreased over time. This decrease in titer could be due to a gradual decrease in V1 immunogenicity due to amino acid changes within V1 or due to changes in other Env regions that occur during SHIVSF162P4 replication and which could affect the presentation of V1. Alternatively, it could be due to the use of a V1 peptide derived from the SF162/SHIVSF162P4 Env which does not have the same amino acid composition as the V1 sequence of the viral variants that gradually emerged during SHIVSF162P4 infection and against which variant V1 sequence antibodies may have been elicited late in infection. However, even with sera collected early during infection (when limited V1 variation was observed [data not shown]), we did not record a significant contribution of anti-V1 antibodies to the neutralizing potential of sera from the SHIVSF162P4-infected animal. Thus, although the neutralizing activity of gp140 sera was primarily focused on the V1 loop, that of SHIVSF162P4 sera targeted other Env regions. Since the V1 peptide used here to detect the presence of anti-V1 antibodies spans almost the entire V1 loop of SF162 Env, we do not know at this point whether the anti-V1 antibody response targeted multiple epitopes or one specific immunodominant epitope, as has been recently shown by others using YU2-derived Env immunogens (49). The fact that significant titers of anti-V1 antibodies are elicited by immunization with SF162, YU2, HxB2, and W61D-derived Env immunogens (2, 28, 37, 49) suggests that the V1 loop will be immunogenic on soluble HIV Env constructs in general. Of course, the relative immunogenicity of the V1 loop and of other Env regions will depend on the Env background and on the oligomeric structure of the immunogen. For example, immunization with group M consensus Env results in the generation of primarily anti-V3 NAbs (27). Nevertheless, based on previous results obtained with monomeric gp120 (37) and those presented here obtained with trimeric gp140 constructs, it appears that the V1 loop is highly immunogenic on the SF162 Env.
In part, the immunogenic properties of the V1 loop are related to its exposure on the SF162 Env. Our antigenicity studies with the recently isolated mouse anti-V1 antibody, P3C8, indicate that the epitope recognized by this MAb is well exposed on all four gp140 constructs examined here. However, relative epitope exposure may not necessarily predict epitope immunogenicity. The epitope of b12 is also present on SF162gp140, and b12 can efficiently neutralize SHIVSF162P4, yet b12-like antibodies were not detectable in the gp140-immunized or SHIVSF162P4-infected animals. It remains unclear why anti-V1 antibodies were readily elicited during immunization with this gp140 immunogen but b12-like antibodies were not. However, our results are in general agreement with those previously reported by others that b12 epitope exposure does not correlate well with b12 epitope immunogenicity (77). Similarly, we did not detect the generation of 2F5- or 4E10-like antibodies during immunization or during SHIVSF162P4 infection, even though these two antibodies recognize their epitopes on the gp140 immunogens used here and neutralize SF162 (74).
Our SF162gp140 immunogen elicited anti-V3 antibodies, and deletion of the V2 and/or V3 loops reduced their titers. This most likely is due to alterations in the positioning of the V3 loop with respect to other Env regions when the V2 loop is eliminated that may alter its immunogenic properties. The anti-V3 NAbs elicited by SF162gp140 targeted mostly linear epitopes (although two animals in this group, A02186 and 02193, appeared to also generate low titers of conformational anti-V3 antibodies). Since sera elicited by gp140 constructs lacking the V3 loop showed significantly reduced heterologous neutralizing activity compared with sera elicited by SF162gp140, we believe that the limited cross-neutralizing activity recorded with the SF162gp140 immunogen was due to the presence of such anti-V3 NAbs. In contrast, the SHIVSF162P4-infected macaque gradually developed anti-V3 NAbs that recognized both linear and conformational epitopes on V3 (similar to what we observed with HIVIG). Anti-V3 antibodies that recognize conformational epitopes have a broader neutralizing potential than anti-V3 antibodies that recognize linear epitopes (30, 32). Thus, the conformational anti-V3 antibodies present in the SHIVSF162P4 sera collected on the later days of infection most likely contributed to the broad neutralizing activity associated with those sera. In fact, the breadth of neutralization associated with the SHIVSF162P4 sera increased over time (data not shown). Of note, conformational anti-V3 antibodies were detected with the use of a chimeric protein expressing the JR-CSF V3 loop and not that of SF162. This supports the proposal that the conformational V3 antibodies present in SHIVSF162P4 sera recognize a common V3 loop structure.
Differences between the binding titer of certain antibodies and their neutralizing activity were noted. For example, animal A02177 had a higher titer of anti-V1 antibodies than animal A02179, but the anti-V1 antibodies from A02179 had a higher neutralizing activity than those of A02177. It is possible that different animals developed antibodies against different regions of the V1 loop, some of which may be more exposed on the virion surface than others. A more detailed mapping of the epitope specificities of the V1 antibodies elicited during immunization might provide information that would explain this observation. Alternatively, differences between binding and neutralization could be due to differences in the presentation of the V1 loop on virions, on gp140 proteins, and as peptides on ELISA plates.
All four gp140 immunogens elicited homologous NAbs despite the fact that some had major deletions in the V2 and V3 regions. This is most likely due to the fact that SF162 is susceptible to neutralization by antibodies that bind to diverse epitopes on gp120 (6, 74). This contrasts with results obtained during immunizations with modified Env immunogens derived from either the DH12 or HxB2 Env's (47, 53). However, deletion of the V3 loop from our gp140 immunogens reduced the titer of homologous NAbs, indicating that V3 loop-directed antibodies are significant contributors to the homologous neutralization recorded with the sera from the SF162gp140-immunized animals. It is also possible that the differences in immunization protocol between the SF162gp140 and ΔV2gp140 groups compared with the ΔV3gp140 and ΔV2ΔV3gp140 groups (lack of a fourth DNA immunization and longer rest period between the DNA and protein phases of immunization) contributed to the observed differences in titer. However, differences in binding antibody titers between the SF162gp140 and ΔV2gp140 groups (as well as between the ΔV3gp140 and ΔV2ΔV3gp140 groups) were evident even though the animals were immunized with the same protocol. In contrast to our previous observations (1), here the ΔV2gp140-immunized animals did not elicit broader NAb responses than the SF162gp140-immunized animals. Individual animals respond to immunization or infection differently, and even though in the previous study the two animals immunized with ΔV2gp140 showed an enhancement modest in neutralization breadth (and weak in potency) compared to SF162gp140-immunized animals, this most likely was not related to differences in the immunogenic properties of the SF162gp140 and ΔV2gp140 proteins.
The four gp140 immunogens elicited low titers of antibodies that bind to CD4-induced V3 loop epitopes. The SHIVSF162P4-infected animal elicited higher titers of such antibodies and also elicited high titers of CD4i antibodies. Such antibodies may have contributed to the cross-neutralizing activity associated with the SHIVSF162P4 sera. Further studies are required to assess in more detail the participation of such antibodies during heterologous neutralization by these sera.
The development of broader NAb responses during infection with a SHIVSF162P4 that expresses the SF162 Env than during immunization with SF162-derived gp140 immunogens could be explained in different ways. The gp140 immunogens tested here do not perfectly mimic the Env structure present on infectious virions, and as a result, they may not present key neutralization epitopes as efficiently to the immune system. For example, during the DNA phase of immunization the animals received the “cleaved” versions of our gp140 immunogens (gp140C). Based on our in vitro expression results, we expect that various forms of gp140C, some of which were properly processed and some of which were not, were expressed in vivo. These distinct forms may differentially prime for the development of relevant NAb responses. During the recombinant protein immunization phase, we use the “fused” version of our gp140 immunogens (gp140F), and such structures may also not present relevant neutralization epitopes in the most effective way, as has been suggested by others (4, 76). A second possibility is that broader NAb responses may have developed in the SHIVSF162P4-infected animal due to continuous viral replication which provided a sustained Env presentation to B cells during infection. A third possibility is that broader neutralizing responses may have developed due to the evolution of the viral Env in these animals. Such evolution may gradually focus the immune response to conserved epitopes on the viral Env or, alternatively, to successive variants of the variable regions of Env. HIVIG also contained broadly reactive NAbs, but our studies indicate that their epitopes may be different from those elicited during SHIVSF162P4 infection.
Overall, our studies suggest that in order for gp140 immunogens to elicit broad neutralizing anti-HIV antibody responses, efforts should be made to reduce the immunogenicity of the V1 region on such constructs. Potentially, this could be achieved by deleting the V1 loop from those immunogens. However, even the deletion of the V1 loop may not result in the enhancement of the immunogenicity of more conserved Env epitopes (93). Immunization with Env proteins lacking the V1 loop, either alone or in combination with other loops, has not yet elicited high titers of broadly reactive NAbs (93), and in fact in certain cases, V1 deletion abrogated homologous NAb responses (47, 53). In the case of SF162, deletion of the V1 loop renders this virus less susceptible to CD4BS NAbs (74), suggesting that the modification may in fact reduce the exposure, and potentially the immunogenicity, of CD4BS epitopes. If this is the case, alternative approaches need to be developed to deal with the high immunogenicity of the V1 loop on gp140 constructs.
Supplementary Material
Acknowledgments
These studies were supported by NIH grants AI47708 (L.S.), AI51217 (L.S.), and AI58763 (J.M.B.); the M. J. Murdock Charitable Trust; the J. B. Pendleton Charitable Trust; and the AIDS and Infectious Diseases Science Center of the Torrey Pines Institute for Molecular Studies. N.R.D. was also supported by NIH training grant AI007509.
We thank all those who contributed materials used in these studies. We also thank Ruth McCaffrey and Cheryl Saunders for their assistance in the construction of the V3-deleted immunogens described here.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beddows, S., S. Lister, R. Cheingsong, C. Bruck, and J. Weber. 1999. Comparison of the antibody repertoire generated in healthy volunteers following immunization with a monomeric recombinant gp120 construct derived from a CCR5/CXCR4-using human immunodeficiency virus type 1 isolate with sera from naturally infected individuals. J. Virol. 73:1740-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beddows, S., N. Schulke, M. Kirschner, K. Barnes, M. Franti, E. Michael, T. Ketas, R. W. Sanders, P. J. Maddon, W. C. Olson, and J. P. Moore. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 79:8812-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S. L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower, J. F., X. Yang, J. Sodroski, and T. M. Ross. 2004. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 78:4710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckner, C., L. G. Gines, C. J. Saunders, L. Vojtech, I. Srivastava, A. Gettie, R. Bohm, J. Blanchard, S. W. Barnett, J. T. Safrit, and L. Stamatatos. 2004. Priming B cell-mediated anti-HIV envelope responses by vaccination allows for the long-term control of infection in macaques exposed to a R5-tropic SHIV. Virology 320:167-180. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. I. Barbas. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 10.Cavacini, L. A., M. Duval, J. Robinson, and M. R. Posner. 2002. Interactions of human antibodies, epitope exposure, antibody binding and neutralization of primary isolate HIV-1 virions. AIDS 16:2409-2417. [DOI] [PubMed] [Google Scholar]
- 11.Cavacini, L. A., J. E. Peterson, E. Nappi, M. Duval, R. Goldstein, K. Mayer, and M. R. Posner. 1999. Minimal incidence of serum antibodies reactive with intact primary isolate virions in human immunodeficiency virus type 1-infected individuals. J. Virol. 73:9638-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Center, R. J., R. D. Leapman, J. Lebowitz, L. O. Arthur, P. L. Earl, and B. Moss. 2002. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J. Virol. 76:7863-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti, B. K., W. P. Kong, B. Y. Wu, Z. Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakrabarti, B. K., X. Ling, Z. Y. Yang, D. C. Montefiori, A. Panet, W. P. Kong, B. Welcher, M. K. Louder, J. R. Mascola, and G. J. Nabel. 2005. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine 23:3434-3445. [DOI] [PubMed] [Google Scholar]
- 15.Chapman, B. S., R. M. Thayer, K. A. Vincent, and N. L. Haigwood. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 19:3979-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 17.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 18.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crooks, E. T., P. L. Moore, D. Richman, J. Robinson, J. A. Crooks, M. Franti, N. Schulke, and J. M. Binley. 2006. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:1-13. [PMC free article] [PubMed] [Google Scholar]
- 20.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong, M., P. F. Zhang, F. Grieder, J. Lee, G. Krishnamurthy, T. VanCott, C. Broder, V. R. Polonis, X. F. Yu, Y. Shao, D. Faix, P. Valente, and G. V. Quinnan, Jr. 2003. Induction of primary virus-cross-reactive human immunodeficiency virus type 1-neutralizing antibodies in small animals by using an alphavirus-derived in vivo expression system. J. Virol. 77:3119-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria-Rose, N. A., G. H. Learn, A. G. Rodrigo, D. C. Nickle, F. Li, M. Mahalanabis, M. T. Hensel, S. McLaughlin, P. F. Edmonson, D. Montefiori, S. W. Barnett, N. L. Haigwood, and J. I. Mullins. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 79:11214-11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 25.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsell, M. N., Y. Li, M. Sundback, K. Svehla, P. Liljestrom, J. R. Mascola, R. Wyatt, and G. B. Karlsson Hedestam. 2005. Biochemical and immunogenic characterization of soluble human immunodeficiency virus type 1 envelope glycoprotein trimers expressed by Semliki Forest virus. J. Virol. 79:10902-10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, F., E. A. Weaver, Z. Lu, Y. Li, H.-X. Liao, B. Ma, S. M. Alam, R. M. Scearce, L. L. Sutherland, J.-S. Yu, J. M. Decker, G. M. Shaw, D. C. Montefiori, B. T. Korber, B. H. Hahn, and B. F. Haynes. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J. Virol. 79:1154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrity, R. R., G. Rimmelzwaan, A. Minassian, W. P. Tsai, G. Lin, J. J. de Jong, J. Goudsmit, and P. L. Nara. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279-289. [PubMed] [Google Scholar]
- 29.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorny, M. K., L. Stamatatos, B. Volsky, K. Revesz, C. Williams, X. H. Wang, S. Cohen, R. Staudinger, and S. Zolla-Pazner. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorny, M. K., J. Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 34.Grundner, C., Y. Li, M. Louder, J. Mascola, X. Yang, J. Sodroski, and R. Wyatt. 2005. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology 331:33-46. [DOI] [PubMed] [Google Scholar]
- 35.Gzyl, J., E. Bolesta, A. Wierzbicki, D. Kmieciak, T. Naito, M. Honda, K. Komuro, Y. Kaneko, and D. Kozbor. 2004. Effect of partial and complete variable loop deletions of the human immunodeficiency virus type 1 envelope glycoprotein on the breadth of gp160-specific immune responses. Virology 318:493-506. [DOI] [PubMed] [Google Scholar]
- 36.Haigwood, N. L., J. R. Shuster, G. K. Moore, H. Lee, P. V. Skiles, K. W. Higgins, P. J. Barr, C. George-Nascimento, and K. S. Steimer. 1990. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res. Hum. Retrovir. 6:855-869. [DOI] [PubMed] [Google Scholar]
- 37.He, Y., W. J. Honnen, C. P. Krachmarov, M. Burkhart, S. C. Kayman, J. Corvalan, and A. Pinter. 2002. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J. Immunol. 169:595-605. [DOI] [PubMed] [Google Scholar]
- 38.Ho, D. D., M. S. Fung, Y. Z. Cao, X. L. Li, C. Sun, T. W. Chang, and N. C. Sun. 1991. Another discontinuous epitope on glycoprotein gp120 that is important in human immunodeficiency virus type 1 neutralization is identified by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 88:8949-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho, J., K. S. MacDonald, and B. H. Barber. 2002. Construction of recombinant targeting immunogens incorporating an HIV-1 neutralizing epitope into sites of differing conformational constraint. Vaccine 20:1169-1180. [DOI] [PubMed] [Google Scholar]
- 40.Ho, J., R. A. Uger, M. B. Zwick, M. A. Luscher, B. H. Barber, and K. S. MacDonald. 2005. Conformational constraints imposed on a pan-neutralizing HIV-1 antibody epitope result in increased antigenicity but not neutralizing response. Vaccine 23:1559-1573. [DOI] [PubMed] [Google Scholar]
- 41.Hofmann-Lehmann, R., J. Vlasak, R. A. Rasmussen, B. A. Smith, T. W. Baba, V. Liska, F. Ferrantelli, D. C. Montefiori, H. M. McClure, D. C. Anderson, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, H. Katinger, G. Stiegler, L. A. Cavacini, M. R. Posner, T. C. Chou, J. Andersen, and R. M. Ruprecht. 2001. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J. Virol. 75:7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janeway, C. 2004. Immunobiology: the immune system in health and disease, 6th ed. Garland Science, New York, N.Y.
- 43.Joyce, J. G., W. M. Hurni, M. J. Bogusky, V. M. Garsky, X. Liang, M. P. Citron, R. C. Danzeisen, M. D. Miller, J. W. Shiver, and P. M. Keller. 2002. Enhancement of alpha-helicity in the HIV-1 inhibitory peptide DP178 leads to an increased affinity for human monoclonal antibody 2F5 but does not elicit neutralizing responses in vitro. Implications for vaccine design. J. Biol. Chem. 277:45811-45820. [DOI] [PubMed] [Google Scholar]
- 44.Kang, S. M., F. S. Quan, C. Huang, L. Guo, L. Ye, C. Yang, and R. W. Compans. 2005. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology 331:20-32. [DOI] [PubMed] [Google Scholar]
- 45.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, M., Z. S. Qiao, D. C. Montefiori, B. F. Haynes, E. L. Reinherz, and H. X. Liao. 2005. Comparison of HIV type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res. Hum. Retrovir. 21:58-67. [DOI] [PubMed] [Google Scholar]
- 47.Kim, Y. B., D. P. Han, C. Cao, and M. W. Cho. 2003. Immunogenicity and ability of variable loop-deleted human immunodeficiency virus type 1 envelope glycoproteins to elicit neutralizing antibodies. Virology 305:124-137. [DOI] [PubMed] [Google Scholar]
- 48.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 80:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 51.Lorin, C., L. Mollet, F. Delebecque, C. Combredet, B. Hurtrel, P. Charneau, M. Brahic, and F. Tangy. 2004. A single injection of recombinant measles virus vaccines expressing human immunodeficiency virus (HIV) type 1 clade B envelope glycoproteins induces neutralizing antibodies and cellular immune responses to HIV. J. Virol. 78:146-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 53.Lu, S., R. Wyatt, J. F. Richmond, F. Mustafa, S. Wang, J. Weng, D. C. Montefiori, J. Sodroski, and H. L. Robinson. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retrovir. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 54.Luo, M., F. Yuan, Y. Liu, S. Jiang, X. Song, P. Jiang, X. Yin, M. Ding, and H. Deng. 2006. Induction of neutralizing antibody against human immunodeficiency virus type 1 (HIV-1) by immunization with gp41 membrane-proximal external region (MPER) fused with porcine endogenous retrovirus (PERV) p15E fragment. Vaccine 24:435-442. [DOI] [PubMed] [Google Scholar]
- 55.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 58.Mbah, H. A., S. Burda, M. K. Gorny, C. Williams, K. Revesz, S. Zolla-Pazner, and P. N. Nyambi. 2001. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 75:7785-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKeating, J. A., C. Shotton, S. Jeffs, C. Palmer, A. Hammond, J. Lewis, K. Oliver, J. May, and P. Balfe. 1996. Immunogenicity of full length and truncated forms of the human immunodeficiency virus type I envelope glycoprotein. Immunol. Lett. 51:101-105. [DOI] [PubMed] [Google Scholar]
- 60.Moore, P. L., E. T. Crooks, L. Porter, P. Zhu, C. S. Cayanan, H. Grise, P. Corcoran, M. B. Zwick, M. Franti, L. Morris, K. H. Roux, D. R. Burton, and J. M. Binley. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nyambi, P. N., A. Nadas, H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, and S. Zolla-Pazner. 2000. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J. Virol. 74:10670-10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2004. Improved design of an antigen with enhanced specificity for the broadly HIV-neutralizing antibody b12. Protein Eng. Des. Sel. 17:749-758. [DOI] [PubMed] [Google Scholar]
- 68.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 71.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selvarajah, S., B. Puffer, R. Pantophlet, M. Law, R. W. Doms, and D. R. Burton. 2005. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 79:12148-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 79.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, D. C. Montefiori, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 77:11244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Srivastava, I. K., K. VanDorsten, L. Vojtech, S. W. Barnett, and L. Stamatatos. 2003. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J. Virol. 77:2310-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stamatatos, L., M. Lim, and C. Cheng-Mayer. 2000. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 16:981-994. [DOI] [PubMed] [Google Scholar]
- 83.Stamatatos, L., M. Wiskerchen, and C. Cheng-Mayer. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV type 1 isolate on viral envelope structure, cell entry, and replication. AIDS Res. Hum. Retrovir. 14:1129-1139. [DOI] [PubMed] [Google Scholar]
- 84.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilkinson, R. A., C. Piscitelli, M. Teintze, L. A. Cavacini, M. R. Posner, and C. M. Lawrence. 2005. Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120. J. Virol. 79:13060-13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 89.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu, J.-Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu, R., I. K. Srivastava, L. Kuller, I. Zarkikh, Z. Kraft, Z. Fagrouch, N. L. Letvin, J. L. Heeney, S. W. Barnett, and L. Stamatatos. 2006. Immunization with HIV-1 SF162-derived envelope gp140 proteins does not protect macaques from heterologous simian-human immunodeficiency virus SHIV89.6P infection. Virology 349:276-289. [DOI] [PubMed] [Google Scholar]
- 92.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang, Z. Y., B. K. Chakrabarti, L. Xu, B. Welcher, W. P. Kong, K. Leung, A. Panet, J. R. Mascola, and G. J. Nabel. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 78:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu, P., E. Chertova, J. Bess, Jr., J. D. Lifson, L. O. Arthur, J. Liu, K. A. Taylor, and K. H. Roux. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA 100:15812-15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.