Abstract
Initiation is the rate-limiting step in protein synthesis and therefore an important target for regulation. For the initiation of translation of most cellular mRNAs, the cap structure at the 5′ end is bound by the translation factor eukaryotic initiation factor 4E (eIF4E), while the poly(A) tail, at the 3′ end, is recognized by the poly(A)-binding protein (PABP). eIF4G is a scaffold protein that brings together eIF4E and PABP, causing the circularization of the mRNA that is thought to be important for an efficient initiation of translation. Early in infection, rotaviruses take over the host translation machinery, causing a severe shutoff of cell protein synthesis. Rotavirus mRNAs lack a poly(A) tail but have instead a consensus sequence at their 3′ ends that is bound by the viral nonstructural protein NSP3, which also interacts with eIF4GI, using the same region employed by PABP. It is widely believed that these interactions lead to the translation of rotaviral mRNAs, impairing at the same time the translation of cellular mRNAs. In this work, the expression of NSP3 in infected cells was knocked down using RNA interference. Unexpectedly, under these conditions the synthesis of viral proteins was not decreased, while the cellular protein synthesis was restored. Also, the yield of viral progeny increased, which correlated with an increased synthesis of viral RNA. Silencing the expression of eIF4GI further confirmed that the interaction between eIF4GI and NSP3 is not required for viral protein synthesis. These results indicate that NSP3 is neither required for the translation of viral mRNAs nor essential for virus replication in cell culture.
As obligate intracellular parasites viruses depend on the cell translation machinery to synthesize their proteins, since this is a complex process that requires numerous components that exceed by far the coding capacity of viral genomes. The successful replication of viruses depends on the ability of viral mRNAs to outcompete cellular mRNAs for the use of the host translation apparatus. To achieve this, viruses have developed remarkable strategies to ensure the efficient translation of their mRNAs while simultaneously inhibiting cellular protein synthesis (reviewed in references 2 and 30).
Initiation is the rate-limiting step in protein synthesis and thus an important target for regulation. The critical step during translation initiation is the recruitment of the small 40S ribosomal subunit to the mRNA (7, 14), a process that involves the synergistic action of the 5′-cap structure and the poly(A) tail at the 3′ end of most eukaryotic mRNAs (8, 20). The cap structure serves to recruit the eukaryotic initiation factor 4F (eIF4F), a multiproteic complex composed of eIF4E, eIF4A, and eIF4G. eIF4E is the cap binding protein and also interacts with the amino-terminal region of eIF4G; eIF4A is an ATP helicase thought to unwind RNA secondary structures present near the 5′ end of the mRNAs and associates with the middle region of eIF4G; eIF4G is a scaffolding protein that in addition to binding eIF4E and eIF4A also binds the poly(A)-binding protein (PABP), promoting the circularization of the mRNA molecule that is thought to be important for an efficient initiation of translation (15, 28, 33). There are two functional homologues of eIF4G in mammals, eIF4GI and eIF4GII, which are 46% identical at the amino acid level and are known to have similar biochemical activities and to functionally complement each other (10, 15).
Rotaviruses are the leading etiologic agent of severe diarrheal disease in infants and young children, causing an estimated 600,000 deaths each year, mostly in developing countries (24). These viruses have a genome composed of 11 segments of double-stranded RNA (dsRNA) enclosed in a capsid formed by three concentric layers of protein. During or shortly after cell entry, the infecting virus uncoats, losing the two proteins of the outer layer and yielding a double-layered particle that is transcriptionally active. The viral mRNAs contain a 5′-methylated cap structure but lack the poly(A) tail characteristic of most cellular mRNAs. Instead, rotavirus mRNAs have a consensus sequence (UGACC) at their 3′ end that is conserved in all segments of the viral genome (27). The rotavirus nonstructural protein NSP3 has been shown to bind to this consensus sequence through its amino-terminal domain. NSP3 also binds eIF4GI through its carboxy-terminal domain, in the same region used by PABP but with higher affinity; thus, it has been proposed that during rotavirus infection NSP3 evicts PABP from eIF4GI, impairing the translation of cellular mRNAs while enhancing at the same time the translation of rotaviral mRNAs (26, 27).
The expression of rotavirus genes can be efficiently and specifically silenced by RNA interference (RNAi) using small interfering RNAs (siRNAs) (1). This has proven to be a very useful tool to dissect the function of rotaviral genes in the context of virus-infected cells (1, 3, 4, 18, 19, 31). In this work we have silenced the expression of NSP3 to characterize the role of this protein in the replication cycle of the virus. We found that in the absence of NSP3 the cellular protein synthesis was not inhibited, supporting previous observations (26, 27). However, the synthesis of viral proteins was unexpectedly not affected, and even more, the yield of viral progeny increased, which correlated with an increased synthesis of dsRNA. The lack of relevance of the NSP3-eIF4GI interaction for the translation of viral transcripts was supported by the fact that silencing the expression of eIF4GI did not affect the synthesis of viral proteins. Altogether, these results indicate that NSP3 is not required for translation of viral mRNAs, as is widely accepted (2, 6, 30), and suggest that the two domains of NSP3 (eIF4GI and RNA binding) might have independent functions.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The rhesus monkey epithelial cell line MA104 was grown in advanced Eagle's minimal essential medium (MEM) (Invitrogen) supplemented with 2% fetal bovine serum and was used for all experiments carried out in this work. Rhesus rotavirus (RRV) was obtained from H. B. Greenberg, Stanford University, Stanford, CA, and was propagated in MA104 cells as described previously (23). The rabbit hyperimmune serum to NSP3 and the monoclonal antibody (MAb) HS2 directed to VP4 have been described previously (9). Polyclonal antibodies to purified RRV triple-layered particles (TLPs) and to recombinant vimentin protein were produced in rabbits as described previously (18). The antibody to eIF4GI was purchased from Santa Cruz Biotechnology (Santa Cruz, CA); goat anti-firefly luciferase antibody was obtained from Chemicon (Temecula, CA). Horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibody was from Perkin-Elmer Life Sciences (Boston, MA), and horseradish peroxidase-conjugated donkey anti-goat antibody was from Santa Cruz Biotechnology (Calif.); goat anti-mouse immunoglobulin G (IgG) coupled to Alexa 568 and goat anti-rabbit IgG coupled to Alexa 488 were from Molecular Probes (Eugene, OR).
siRNA transfection.
siRNANSP3 had the sequence AAUUGGAUGACUGACUCUCGA (sense) and UCGAGAGUCAGUCAUCCAAUU (antisense), corresponding to nucleotides 281 to 301 of the RRV NSP3 gene (accession number AY065842). siRNAeIF4GI had the sequence AAUUGGCUGAGGACAUGGAAA (sense) and UUUCCAUGUCCUCAGCCAAUU (antisense), corresponding to nucleotides 4245 to 4265 from the human eIF4GI gene (GenBank accession number NM_182917). As an irrelevant control, a previously reported siRNA to the green fluorescent protein (siRNAIRR) (18) was used. The siRNAs were obtained from Dharmacon Research (Lafayette, CO). Transfection of siRNAs was carried out in nearly confluent cell monolayers using Lipofectamine (Invitrogen), as described previously (18). The transfection mixture was added to cells previously washed with MEM and incubated for 8 h at 37°C. After this time the transfection mixture was removed, and the cells were washed with MEM and kept in this medium for 48 h at 37°C before virus infection.
Infection of cells and titration of viral progeny.
Transfected cell monolayers in 24- or 48-well plates were infected with RRV at a multiplicity of infection (MOI) of 3. After incubation for the indicated periods of time at 37°C, the cells were lysed by two freeze-thaw cycles, and the lysates were treated with 10 μg/ml of trypsin for 30 min at 37°C to activate the virus infectivity before titration. The virus infectious titer was obtained by an immunoperoxidase focus assay (13, 23).
Immunoassays.
Immunofluorescence was essentially carried out as previously described (4, 29), using a rabbit polyclonal antibody to NSP3 and MAb HS2 directed to VP4. The slides were analyzed with a Nikon E600 epifluorescence microscope coupled to a DXM1200 digital still camera (Nikon). Immunoblot assays were performed as described above, using antibodies to NSP3, luciferase, RRV TLPs, vimentin, or eIF4GI.
Radiolabeling of proteins.
Cells grown in 48-well plates were transfected with siRNAs and infected with RRV as described above. At the indicated times the medium was replaced by MEM without methionine, supplemented with 40 μCi/ml of Easy-tag Express-[35S] labeling mix (Dupont, NEN) and incubated for different periods of time as indicated. The cells were then lysed with Laemmli sample buffer. The samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), adjusting the amount of protein loaded in each lane by Coomassie blue staining.
Real-time reverse transcription-PCR (RT-PCR).
The level of gene 10 RNA(+), which includes both mRNA and the positive-strand RNA present in gene 10 dsRNA, was quantified by real-time PCR as described previously (19). The level of gene 10 RNA(−) was determined by initially priming the reverse transcriptase reaction with the reverse primer 5′-GAGCAATCTTCATGGTTGGAA-3′ (nucleotides 173 to 193 of RRV gene 10), which is complementary to the negative strand of gene 10. The amount of mRNA was calculated from subtracting the amount of RNA(−) (present only in, and thus equivalent to, dsRNA) from the total amount of RNA(+) obtained.
Luciferase expression.
Plasmids pTet-off, which expresses the tetracycline-controlled transactivator, and pTRE2-luc, which expresses firefly luciferase under the control of the transactivator (Clontech), were cotransfected at a ratio of 1:1 using Lipofectamine (Invitrogen). Usually, siRNA-transfected cells were infected 48 h posttransfection, and at 1 h postinfection (hpi) the cells were cotransfected with plasmids pTet-off and pTRE2-luc. At 12 hpi cells were lysed and luciferase activity was determined using the luciferase reporter gene assay kit (Roche), according to the manufacturer's instructions.
RESULTS
NSP3 is not required for the synthesis of viral proteins.
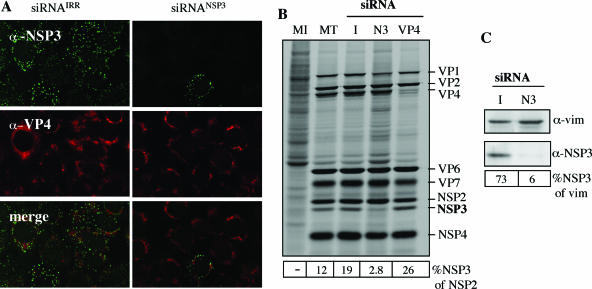
To study the role of the viral nonstructural protein NSP3 in rotavirus-infected cells, the expression of this protein was silenced by RNAi. For this, a chemically synthesized siRNA that targets RRV gene 8 was tested for its effect on the synthesis of NSP3 and on total viral protein synthesis. MA104 cells were transfected with siRNAs to either NSP3 (siRNANSP3), to the green fluorescent protein as an irrelevant sequence (siRNAIRR), or to VP4 (siRNAVP4) as a positive control (4). Forty-eight hours posttransfection the cells were infected with RRV and were either metabolically labeled for 8 h at 4 hpi or fixed and immunostained at 8 hpi. By immunofluorescence only about 20% of the infected cells transfected with siRNANSP3 were positive to an anti-NSP3 antibody, compared to infected control cells transfected with the irrelevant siRNA, in all of which NSP3 could be clearly detected (Fig. 1A). Accordingly, by SDS-PAGE and autoradiography, the synthesis of NSP3 was found to be severely decreased in cells transfected with siRNANSP3, compared to cells transfected with siRNAVP4 or siRNAIRR (Fig. 1B). This observation was confirmed by Western blot analysis of NSP3 (Fig. 1C). A densitometric analysis of NSP3 and NSP2 in the autoradiogram and of NSP3 and vimentin (which was used as a loading control) in the Western blot showed that at 12 hpi the relative amount of NSP3 with respect to vimentin (percentage) in cells transfected with the control siRNA or with siRNANSP3 was 73% and 6%, respectively (Fig. 1C), indicating that the amount of NSP3 that accumulates in the presence of siRNANSP3 is about 10 times lower than that accumulated in cells treated with the irrelevant siRNA. The amount of NSP3 relative to NSP2 was also calculated in the autoradiogram, where it was found that NSP3 represented about 16% of NSP2 in the control-transfected cells, while this number dropped to 2.8% in the cells where NSP3 was silenced, indicating that the amount of NSP3 under these conditions decreased about six times (Fig. 1B). These results indicate that siRNANSP3 specifically and effectively silenced the expression of the RRV NSP3 gene. Surprisingly, given the central role proposed for NSP3 during translation of rotaviral mRNAs (26, 27), the level of all other viral proteins remained largely unaffected (Fig. 1B). On the other hand, the background of cellular proteins appeared more intense when NSP3 was silenced than when no siRNA or the siRNAIRR was used, indicating that in the absence of NSP3 the shutoff of cellular proteins observed in rotavirus-infected cells is less pronounced, as previously suggested (26, 27).
FIG. 1.
NSP3 is not required for the synthesis of viral proteins. MA104 cells were transfected with the indicated siRNA to either NSP3 (N3), VP4, or an irrelevant (I) sequence and infected with RRV at an MOI of 3, as described in Materials and Methods. (A) At 8 hpi the cells were fixed and immunostained with a rabbit polyclonal antibody to NSP3 and MAb HS2 to VP4 as primary antibodies, as indicated, followed by incubation with goat anti-mouse IgG coupled to Alexa 568 and goat anti-rabbit IgG coupled to Alexa 488 as secondary antibodies. (B) At 4 hpi the cells were metabolically labeled with 40 μCi/ml of Easy-tag Express-[35S] for 8 h and then lysed in Laemmli sample buffer. The labeled proteins were resolved by SDS-10% PAGE and detected by autoradiography. MT, mock-transfected cells, infected with RRV; MI, mock-infected cells. The position of the viral proteins is indicated. (C) Immunoblot analysis of RRV NSP3. The transferred proteins were incubated with hyperimmune sera to NSP3 and vimentin (vim) as indicated, and the bound antibodies were detected by incubation with a peroxidase-labeled anti-rabbit immunoglobulin antibody. The amount of loaded protein was previously adjusted by visual inspection of Coomassie blue-stained gels. A densitometric analysis of NSP3 and NSP2 in the autoradiogram and of NSP3 and vimentin (which was used as a loading control) in the Western blot was performed. The numbers below the gels represent the relative amount (percentage) of NSP3 with respect to NSP2 or vimentin.
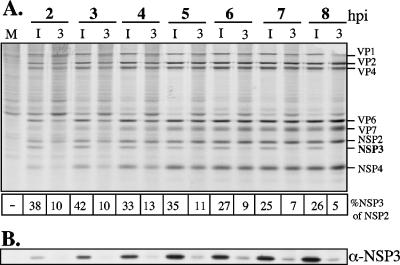
To determine if NSP3 was required for the synthesis of viral proteins at some point during the infection cycle, infected cells transfected with siRNANSP3 or the irrelevant siRNA were metabolically labeled for 1 h at different times postinfection, and the pattern of viral proteins was analyzed by SDS-PAGE and autoradiography (Fig. 2A); the presence of NSP3 at each time point was also detected by Western blotting (Fig. 2B). At all times evaluated (from 2 to 8 hpi), the synthesis of all viral proteins, with the exception of NSP3, was found to be very similar in cells transfected with either siRNANSP3 or siRNAIRR, suggesting that the synthesis of viral proteins is independent of NSP3 throughout the entire virus replication cycle. In general, the relative amount of NSP3 with respect to NSP2 ranged from 25 to 42% throughout the different time points in control-transfected cells, while in the cells where NSP3 was knocked down, it decreased to 5 to 13%, NSP2 being taken as 100% (Fig. 2A).
FIG. 2.
Viral protein synthesis is independent of NSP3 throughout the virus replication cycle. MA104 cells in 48-well plates were transfected with either an irrelevant siRNA (I) or siRNANSP3 (N3) and were infected with RRV. (A) At 30 min before the indicated times postinfection (hours) the cells were pulse-labeled for 30 min with Easy-tag Express-[35S]. The cells were then lysed in Laemmli sample buffer and the proteins separated by SDS-10% PAGE and detected by autoradiography. (B) Immunoblot analysis of RRV NSP3, carried out as described in the legend for Fig. 1. The amount of loaded protein was previously adjusted by visual inspection of Coomassie blue-stained gels. A densitometric analysis of NSP3 and NSP2 in the autoradiogram was performed. The numbers below the gel in panel A represent the relative amount (percentage) of NSP3 with respect to NSP2. Lane M, mock-infected cells.
NSP3 is required to shut off the synthesis of cellular proteins.
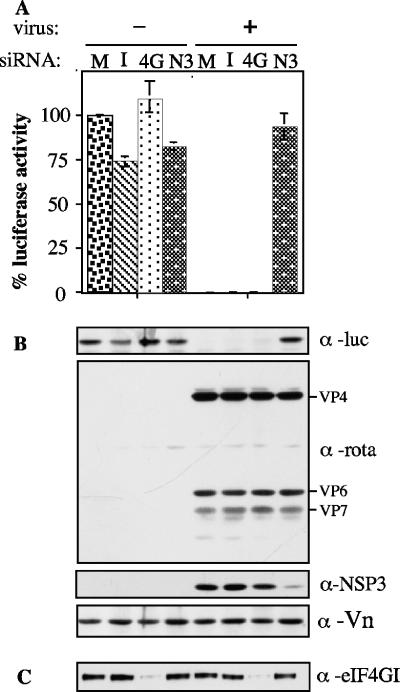
To investigate whether the synthesis of cellular proteins in rotavirus-infected cells was indeed less inhibited in the absence of NSP3, the activity of a reporter protein was determined. Cells were transfected with the different siRNAs, and 48 h posttransfection the cells were infected with rotavirus and cotransfected with plasmids pTet-off and pTRE2-luc to direct the synthesis of luciferase (see Materials and Methods). At 12 hpi the level of the reporter protein was assayed both by activity and by Western blotting (Fig. 3A). The levels of expression of luciferase were not significantly modified in mock-infected cells transfected with the different siRNAs. In contrast, luciferase was not detected when the cells were infected with rotavirus, with the exception of those cells in which the expression of NSP3 was knocked down; in that case the level of the reporter protein was comparable to those found in noninfected cells both by Western blotting and by the activity assay (Fig. 3A). The synthesis of NSP3, as well as that of the viral structural proteins, was also monitored by Western blotting in these same experiments. As described above for metabolically labeled proteins (Fig. 1B), the amount of viral structural proteins synthesized did not depend on the presence of NSP3 (Fig. 3B). The complete inhibition of the translation of the luciferase mRNA, which is capped and polyadenylated, in virus-infected cells and its restoration by silencing the expression of NSP3 strongly support the role of this protein in shutting off the translation of cellular mRNAs. The all-or-nothing synthesis effect observed for the luciferase in the above-described conditions, compared to the observation that the cell protein synthesis was not completely restored when NSP3 was knocked down, might be due to the fact that the luciferase reporter system used in these assays involves the translation of two proteins, including the activator protein from the pTet-off plasmid, as well as the translation of the luciferase mRNA, which might result in an exacerbated inhibition and restoration of luciferase translation.
FIG. 3.
Silencing the expression of NSP3 prevents the shutoff of cellular proteins. (A) MA104 cells in 24-well plates were transfected with the indicated siRNAs to either NSP3 (N3), eIF4GI (4G), or an irrelevant sequence (I); 48 h posttransfection the cells were either infected (+) or mock infected (−) with RRV and then cotransfected with plasmids pTRE2-luc and pTet-off. At 12 hpi the cells were lysed and the luciferase activity was determined as indicated in Materials and Methods. The data are expressed as percentages of the luciferase activity present in noninfected cells that were mock transfected. (B) Immunoblot analysis (SDS-10% PAGE) of the same samples shown in panel A; the same membrane was probed with antibodies to either luciferase (α-luc), RRV TLPs (α-rota), NSP3 (α-NSP3), or vimentin (α-Vn). The viral proteins VP4, VP6, and VP7 are indicated. (C) Immunoblot analysis (SDS-7% PAGE) of the same samples shown in panel A, probed with an antibody to eIF4GI.
RNA synthesis is increased in the absence of NSP3.
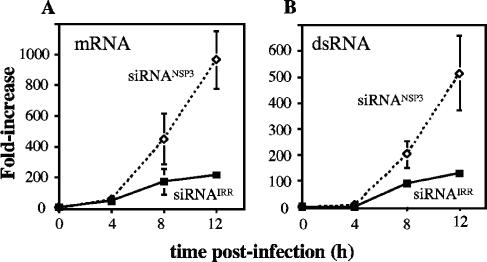
To evaluate if the level of viral RNA was affected in the absence of NSP3, the relative amounts of viral mRNA and dsRNA produced in cells transfected with either siRNANSP3 or siRNAIRR were determined by semiquantitative RT-PCR. The accumulation at different times postinfection of gene 10-positive (RNA+) and -negative (RNA−) RNA strands was used to estimate the presence of viral mRNA and dsRNA, respectively. Gene 10 was selected as a marker for the general synthesis of viral RNA, since the quantitative RT-PCR conditions for its determination were already set up in the laboratory (C. Ayala-Breton et al., unpublished data). The relative amounts of both viral mRNA (Fig. 4A) and genomic dsRNA (Fig. 4B) were increased in cells transfected with siRNANSP3 compared to cells transfected with the irrelevant siRNA. At 12 hpi there was about four times more viral mRNA and approximately three times more dsRNA in cells transfected with siRNANSP3, suggesting an enhanced replication of viral RNA in the absence of NSP3.
FIG. 4.
The synthesis of the viral mRNA and dsRNA is increased in NSP3-silenced cells. Cells transfected with either an irrelevant siRNAIRR or siRNANSP3 were infected with RRV, and at the indicated times total RNA was extracted with Trizol, and the levels of gene 10 RNA(+) and RNA(−) strands were determined by real-time RT-PCR. The results are expressed as an increase (n-fold) over the levels detected at time zero infection (immediately after the end of the virus adsorption period). The amount of mRNA shown in panel A was calculated from subtracting the amount of RNA(−) (present only in, and thus equivalent to, dsRNA), shown in panel B, from the total amount of RNA(+) obtained. The arithmetic mean ± standard deviation of two independent experiments, performed in triplicate, is shown.
The yield of viral progeny is increased in the absence or with reduced amounts of NSP3.
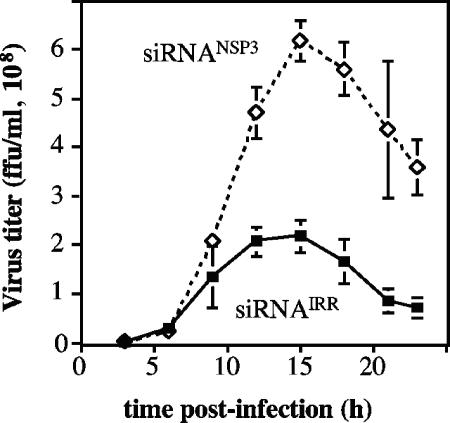
To determine if the enhanced replication of viral RNA observed in NSP3-silenced cells results in an increase in the production of infectious viral progeny, cells transfected with either siRNANSP3 or siRNAIRR were infected with RRV, and 12 hpi the cells were harvested and the titer of the virus present in the cell lysates was determined by an immunoperoxidase focus-forming unit assay (23). In the absence of NSP3 the virus yield was consistently found to be 2 to 2.5 times higher than that obtained in control-transfected cells (not shown). The production of increased levels of infectious viral progeny in NSP3-silenced cells was also clearly observed in one-step growth curve experiments (Fig. 5). At 9 hpi there was already a difference in the amount of infectious virus particles produced, and at 15 hpi the infectious virus recovered from cells transfected with siRNANSP3 was three times as much as that obtained from control-transfected cells.
FIG. 5.
The yield of progeny virus increases when NSP3 is silenced. MA104 cells in 48-well plates were transfected with siRNANSP3 or with the siRNAIRR, and 48 h posttransfection the cells were infected with RRV at an MOI of 3. The cells were harvested at the indicated times postinfection, and the progeny virus produced was determined by an immunoperoxidase assay, as described in Materials and Methods. Virus titer is expressed as the number of focus-forming units per ml. Data shown represent the arithmetic means ± standard deviations of two independent experiments.
The cytopathic effect caused by rotavirus is delayed when NSP3 is knocked down.
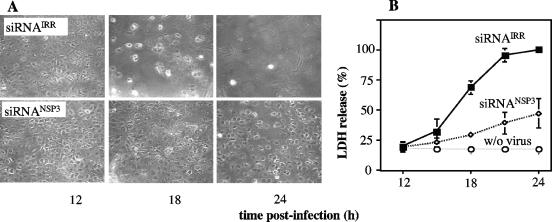
During the course of the experiments carried out in this work, it was observed that the cell monolayers were less damaged when the cells were transfected with the siRNANSP3 than when they were transfected with an siRNAIRR. To further characterize this observation, MA104 monolayers transfected with either siRNANSP3 or siRNAIRR were infected with RRV and at various times postinfection the cytopathic effect was assessed by contrast microscopy and by measuring the release of the cytoplasmic enzyme lactate dehydrogenase (LDH). While the infected cell monolayer transfected with the siRNA control showed evident damage at 12 hpi, the cell monolayer in which NSP3 was knocked down appeared healthy, with few cells detached (Fig. 6A). This effect was more pronounced at 24 hpi, when the control-transfected cell monolayer was already destroyed, while the cytopathic effect was barely noticeable in the monolayer transfected with siRNANSP3 (Fig. 6A). Similarly, there was about 50% less LDH activity released into the culture medium from the infected cells transfected with siRNANSP3 compared to siRNAIRR-transfected cells (Fig. 6B). These results indicate that in the absence, or in the presence of low levels, of NSP3, the cytopathic effect caused by rotavirus infection is less pronounced, which could explain the increased production of viral progeny in NSP3-silenced cells. Also, these results suggest that the inhibition of cell protein synthesis caused by NSP3 might result in the loss of cell viability.
FIG. 6.
The cytopathic effect caused by rotavirus is delayed when NSP3 is knocked down. MA104 cells in 48-well plates were transfected with siRNANSP3 or siRNAIRR, and 48 h posttransfection the cells were infected or not with RRV at an MOI of 3. The cells were harvested at the indicated times postinfection and photographed using phase-contrast microscopy (A), and the activity of lactate dehydrogenase was determined in the cell culture medium at each time point (B), using a commercial kit (Sigma) following the manufacturer's instructions. The LDH activity is expressed as a percentage of the activity released at 24 hpi, which was taken as 100%. “w/o virus” indicates the LDH activity released by mock-infected cells. Data shown represent the arithmetic means ± standard deviations of two independent experiments.
The synthesis of viral proteins is independent of eIF4GI.
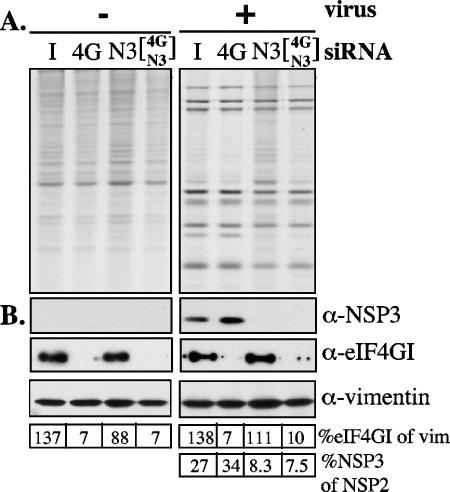
It has been shown that NSP3 interacts directly with the initiation translation factor eIF4GI, and it has been proposed that this interaction is needed to ensure the efficient translation of the viral mRNAs (12, 26, 27). Since in the absence or with reduced levels of NSP3 the synthesis of viral proteins was not affected, we investigated whether eIF4GI was nonetheless important for translation of viral mRNAs. For this, the expression of eIF4GI was silenced with a specific siRNA (siRNAeIF4GI). This siRNA was effective in blocking the expression of the target gene, since 48 h after transfection the level of eIF4GI, as detected by Western blot analysis, was decreased by about 90% compared to cells transfected with either siRNAIRR or siRNANSP3 (Fig. 7B). In the absence or with reduced levels of eIF4GI, a slight decrease in the synthesis of cellular proteins was detected by SDS-PAGE and autoradiography of 35S-labeled proteins (Fig. 7A). This decrease corresponded to 20 to 25% of total protein synthesis, as determined by incorporation of 35S-labeled amino acids into trichloroacetic acid-precipitable material. The same level of reduction in the incorporation of 35S was observed in infected as well as noninfected cells. The effect of silencing the expression of eIF4GI on the synthesis of luciferase was also characterized. The synthesis of the reporter protein, as determined both by activity and by Western blotting, was not significantly altered in cells where the levels of eIF4GI had been markedly reduced by siRNAeIF4GI (Fig. 3A and B); the knockdown of eIF4GI in this assay was verified by Western blotting (Fig. 3C). Of note, when the siRNAeIF4GI-transfected cells were infected with rotavirus 48 h posttransfection, the synthesis of viral proteins was not significantly altered at 12 hpi, compared to control cells transfected with either siRNAIRR or siRNANSP3 (Fig. 3A). Furthermore, the yield of infectious viral progeny was also unaffected in eIF4GI-silenced cells (119% ± 18.6% of infectious virus produced, where 100% is the amount of virus obtained from cells transfected with siRNAIRR). Altogether, these results suggest that while the overall protein synthesis of MA104 cells is very little affected by silencing the expression of eIF4GI, the synthesis of the rotavirus proteins does not depend on the presence of this translation factor.
FIG. 7.
Synthesis of cellular and viral proteins in eIF4GI-silenced cells. (A) MA104 cells in 48-well plates were transfected with an irrelevant siRNA (I), siRNAeIF4GI (4G), siRNANSP3 (N3), or a mixture of both 4G and NSP3, and 48 h posttransfection the cells were infected (+) or not (−) with RRV. At 4 hpi the cells were labeled with 40 μCi/ml of Easy-tag Express-[35S] for 8 h. The cells were then lysed in Laemmli sample buffer and the proteins separated by SDS-10% PAGE and detected by autoradiography. (B) Immunoblot analysis of NSP3 (in SDS-10% PAGE), eIF4GI protein (in SDS-7% PAGE), or vimentin synthesized in cells transfected with the indicated siRNAs. The transferred proteins were incubated with antibodies to NSP3, eIF4GI, and vimentin, and the bound antibodies were developed by incubation with a peroxidase-labeled anti-rabbit immunoglobulin antibody. The amount of loaded protein was previously adjusted by visual inspection of Coomassie blue-stained gels. A densitometric analysis of NSP3 and NSP2 in the autoradiogram and of eIF4G1 and vimentin (which was used as an internal control) in the Western blots was performed. The numbers below the gels represent the relative amount (percentage) of eIF4G1 with respect to vimentin or the relative amount of NSP3 with respect to NSP2, as indicated.
The effect of knocking down eIF4GI and NSP3 at the same time was also studied. Cells were transfected with a mixture of siRNAeIF4GI and siRNANSP3 and then infected with rotavirus 48 h posttransfection. By Western blotting the level of NSP3 with respect to NSP2 decreased about fourfold when silenced alone or in combination with eIF4GI (Fig. 7A and B); similarly the level of eIF4GI was reduced about 15 times compared to vimentin. Interestingly, when both proteins were silenced, the viral protein synthesis was not significantly affected while the cellular protein synthesis decreased compared to the profile obtained when NSP3 was silenced alone, suggesting that the cellular protein synthesis that takes place when NSP3 is silenced depends on eIF4GI. Silencing both proteins resulted in a cellular protein pattern similar to that observed when both proteins were present, i.e., the pattern observed in a regular infection, confirming that NSP3 prevents the synthesis of cellular proteins by interacting with eIF4GI.
DISCUSSION
The role of NSP3 as a substitute factor for PABP for translation of rotaviral mRNAs has been widely used and accepted as an argument to support the relevance of circularization during translation of cellular mRNAs (2, 6, 30). The experimental evidence that supports the model that proposes that NSP3 is responsible for shutting off cellular protein synthesis, while ensuring at the same time the translation of viral mRNAs, has been generated by experiments in vitro or by expression of the NSP3 gene in uninfected cells. These observations include the following: (i) the N-terminal domain of NSP3 binds with high affinity and specificity to the 3′-end consensus sequence of rotaviral mRNAs (5, 26, 27, 34); (ii) the carboxy-terminal domain of NSP3 binds eIF4GI with high affinity (12, 25, 26), evicting PABP from its interaction with this translation factor (26); (iii) swine testicular cells stably transfected to express NSP3 showed a reduced protein synthesis of capped and polyadenylated RNAs up to onefold in vivo, and in an in vitro system the synthesis of polyadenylated RNAs was inhibited up to 10-fold (34); (iv) the expression of NSP3 from a recombinant vaccinia virus decreased the level of cellular protein synthesis, and this inhibition was correlated with the concentration of NSP3 produced in the cell (22).
Even though the interactions of NSP3 with eIF4GI and the 3′ end of viral mRNAs have been clearly established, there is no direct evidence that NSP3 engages simultaneously in these two interactions to promote the circularization of viral mRNAs, and although it is generally accepted, there is also no evidence that these interactions favor the translation of viral mRNAs. Using an in vitro system, Vende et al. (34) observed that a recombinant NSP3 protein enhanced the translation of a reporter gene containing the 3′ untranslated region of rotaviral genes; however, in their system the recombinant protein also stimulated the translation of a poly(A)-containing reporter. Furthermore, an NSP3 mutant in which the eIF4GI binding domain was deleted failed to stimulate the translation of the reporter mRNA containing the rotaviral 3′ untranslated region, but it also seemed to inhibit the translation of a reporter containing a poly(A) tail at its 3′ end; unfortunately these results were not discussed. In this work we found that the synthesis of viral proteins was not affected by knocking down the expression of NSP3, suggesting that binding of this protein to the 3′ end of the viral mRNAs is not necessary for their translation. It is not possible to rule out the possibility that very little NSP3 (not detectable in our assays) might be sufficient to allow the synthesis of viral proteins. However, this observation is supported by the fact that in the earlier experiments performed by Piron et al. (26), the viral protein synthesis was already apparent at 2 hpi, and NSP3 was not detected in association with eIF4GI until 3.5 to 4 hpi, suggesting that the interaction of NSP3 with eIF4GI is not necessary for viral translation. Furthermore, an increased level of viral RNA synthesis (both single stranded and double stranded) was detected in cells where NSP3 was silenced, suggesting that rather than promoting the translation of viral mRNAs, the interaction of NSP3 with the 3′ end of viral mRNAs might prevent them from being selected for replication, thus ensuring a pool of viral transcripts available for translation. Alternatively, the binding of NSP3 to the viral mRNAs might protect them from degradation. If this was the case, the fact that we did not find a decrease in the amount of mRNA when NSP3 was silenced could be explained by the increased viral RNA replication observed under these conditions, associated with an enhanced secondary transcription that could compensate for the putative lower stability of the viral mRNAs in the absence of NSP3. Interestingly, when the structural and biophysical characteristics of the NSP3-RNA complex were determined, it was found that the N-terminal domain of NSP3 had high-affinity RNA binding (Kd, 79 nM) and low dissociation (half-life of 8 h) rates. Based on these observations Deo et al. (5) suggested that the binding of NSP3 to the viral RNA might even interfere with rotavirus genome replication.
The fact that in NSP3-silenced cells the virus yield, as well as the amount of viral single-stranded RNA and dsRNA, was increased indicates that NSP3 is not required for the replication of the virus in cell culture (MA104, CV1, A549, and MDCK cells were tested, with the same results [not shown]). However, this protein might still be relevant in an in vivo infection, where either protection of viral mRNAs from degradation, rescuing mRNA from entering the replication pathway, or even helping mRNAs to be translated might be required. Further experiments are required to evaluate these possibilities.
To further demonstrate that the interaction of NSP3 with eIF4GI is not essential for the translation of viral proteins, we silenced the expression of this translation factor. In agreement with our previous results we found that the synthesis of viral proteins, as well as the production of viral progeny, was not modified when this factor was silenced, while the synthesis of cellular proteins was reduced by about 25%. Even though the reduction of the total protein synthesis seems low in view of the central role that eIF4GI plays in translation, similar findings were reported when this factor was cleaved by the 2Apro protease of several picornaviruses, which preferentially cleave the factor early after cell infection (11, 32); thus, protein synthesis decreased by 35% in Xenopus laevis oocytes injected with 2Apro from coxsackievirus B4, even though eIF4GI was completely cleaved (17). These results have been explained by the fact that eIF4GII can functionally complement eIF4GI (10, 15); thus, the small reduction of total protein synthesis observed in this work when the expression of eIF4GI was silenced could result from complementation of eIF4GII under these conditions. The fact that in standard rotavirus-infected cells (where NSP3 is expressed at normal levels) a more severe shutdown of cellular protein synthesis is observed, compared to noninfected cells in which eIF4GI was silenced, suggests that NSP3 binds to both eIF4GI and eIF4GII. Indeed, the region of eIF4GI that interacts with NSP3 is very similar, if not identical, in eIF4GII (12); thus, although not formally proven, it might be expected that NSP3 could bind both factors, displacing PABP from both eIF4GI and eIF4GII, resulting in the severe shutoff of cell protein synthesis induced by rotavirus. It cannot be ruled out, however, that rotaviruses might use more than one mechanism to control the translation machinery of the cell. Silencing other rotavirus proteins, and assessing their effect on cellular protein synthesis, will show if this is the case.
Altogether, the data obtained in this work suggest that the eIF4G and RNA binding domains of NSP3 function independently. The eIF4G binding domain seems to be responsible, at least partially, for the shutoff of cell protein synthesis, as has been previously suggested (26), by competing off the binding of PABP needed for the translation of many cellular mRNAs (16). The relative abundance of viral capped mRNAs in an infected cell and the inability of PABP-bound polyadenylated mRNAs to bind to eIF4G (sequestered by NSP3) might explain the successful and efficient translation of the viral mRNAs. On the other hand, the RNA binding domain of NSP3 might function to protect the viral mRNAs from degradation and/or to keep a pool of viral mRNAs in the cytosol, available for translation. This latter function could be achieved either by preventing the binding of the viral RNA polymerase VP1 to the 3′ ends of mRNAs or by taking out recently synthesized mRNAs from viroplasms, their site of synthesis (31), to ensure their availability for translation.
Viruses have evolved different strategies to shut down the synthesis of cellular proteins in order to ensure the translation of their own proteins, avoiding competition with cellular mRNAs (reviewed in references 2 and 30). If NSP3 is not needed for translation of viral mRNAs, and in NSP3-silenced cells the protein synthesis machinery can efficiently translate both cellular and viral mRNAs without any apparent detriment in the synthesis of viral proteins, why have rotaviruses evolved such a sophisticated method for shutting off cell protein synthesis? One possibility could be that the virus needs to shut off the synthesis of a particular set of cellular proteins that could interfere with the replication cycle and/or propagation of the virus in vivo. The inhibition of protein synthesis could also be required to impair the structural integrity of the cell, facilitating cell lysis and the release of progeny viruses, as has been reported for adenoviruses; in this case, when the shutoff of protein synthesis was inhibited in adenovirus-infected cells, the yield of viral progeny was not affected, while the adenovirus-mediated cytopathic effect was decreased (35). This would seem to be the case for rotaviruses, since in NSP3-silenced cells the characteristic rotavirus-induced cytopathic effect is delayed. Interestingly, Mossel and Ramig found that the gene that codes for NSP3 is responsible for the extraintestinal spread of rotavirus in mice (21). It would be interesting to determine the identity of the cellular protein(s) whose absence results in the lysis of rotavirus-infected cells.
Acknowledgments
We are grateful to Rafaela Espinosa for her excellent technical assistance with cell culture.
This work was partially supported by grants 55003662 and 55000613 from the Howard Hughes Medical Institute and G37621N from the National Council for Science and Technology-Mexico. H.M. is a recipient of a scholarship from DGEP/UNAM.
REFERENCES
- 1.Arias, C. F., M. A. Dector, L. Segovia, T. Lopez, M. Camacho, P. Isa, R. Espinosa, and S. Lopez. 2004. RNA silencing of rotavirus gene expression. Virus Res. 102:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campagna, M., C. Eichwald, F. Vascotto, and O. R. Burrone. 2005. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J. Gen. Virol. 86:1481-1487. [DOI] [PubMed] [Google Scholar]
- 4.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deo, R. C., C. M. Groft, K. R. Rajashankar, and S. K. Burley. 2002. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell 108:71-81. [DOI] [PubMed] [Google Scholar]
- 6.Flint, S. J., L. W. Enquist, V. R. Racaniello, and A. M. Skalka. 2004. Principles of virology: molecular biology, pathogenesis, and control of animal viruses, 2nd ed., p. 378-410. ASM Press, Washington, D.C.
- 7.Gebauer, F., and M. W. Hentze. 2004. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5:827-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez, R. A., R. Espinosa, P. Romero, S. Lopez, and C. F. Arias. 2000. Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch. Virol. 145:1963-1973. [DOI] [PubMed] [Google Scholar]
- 10.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groft, C. M., and S. K. Burley. 2002. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell 9:1273-1283. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero, C. A., S. Zarate, G. Corkidi, S. Lopez, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahvejian, A., Y. V. Svitkin, R. Sukarieh, M. N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keiper, B. D., and R. E. Rhoads. 1997. Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res. 25:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez, T., M. Camacho, M. Zayas, R. Najera, R. Sanchez, C. F. Arias, and S. Lopez. 2005. Silencing the morphogenesis of rotavirus. J. Virol. 79:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez, T., M. Rojas, C. Ayala-Breton, S. Lopez, and C. F. Arias. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 86:1609-1617. [DOI] [PubMed] [Google Scholar]
- 20.Michel, Y. M., D. Poncet, M. Piron, K. M. Kean, and A. M. Borman. 2000. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275:32268-32276. [DOI] [PubMed] [Google Scholar]
- 21.Mossel, E. C., and R. F. Ramig. 2002. Rotavirus genome segment 7 (NSP3) is a determinant of extraintestinal spread in the neonatal mouse. J. Virol. 76:6502-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padilla-Noriega, L., O. Paniagua, and S. Guzman-Leon. 2002. Rotavirus protein NSP3 shuts off host cell protein synthesis. Virology 298:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Pando, V., P. Isa, C. F. Arias, and S. Lopez. 2002. Influence of calcium on the early steps of rotavirus infection. Virology 295:190-200. [DOI] [PubMed] [Google Scholar]
- 24.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piron, M., T. Delaunay, J. Grosclaude, and D. Poncet. 1999. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 73:5411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poncet, D. 2003. Translation of rotavirus mRNAs in the infected cell, p. 185-205. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis. Elsevier Science, Amsterdam, The Netherlands.
- 28.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516-520. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-San Martin, C., T. Lopez, C. F. Arias, and S. Lopez. 2004. Characterization of rotavirus cell entry. J. Virol. 78:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 31.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarun, S. Z., Jr., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 34.Vende, P., M. Piron, N. Castagne, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 74:7064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y., and R. J. Schneider. 1994. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J. Virol. 68:2544-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]