Abstract
Hantaviruses constitute a genus in the family Bunyaviridae. They are enveloped negative-strand RNA viruses with a tripartite genome encoding the nucleocapsid (N) protein, the two surface glycoproteins Gn and Gc, and an RNA-dependent RNA polymerase. The N protein is the most abundant component of the virion; it encapsidates genomic RNA segments forming ribonucleoproteins and participates in genome transcription and replication as well as virus assembly. In the course of RNA encapsidation, N protein forms intermediate trimers via head-to-head and tail-to-tail interactions. We analyzed the amino-terminal trimerization domain (amino acid residues 1 to 77) of Tula hantavirus using computer modeling, mammalian two-hybrid assay, and immunofluorescence assay. The results obtained were consistent with the existence of an antiparallel coiled-coil stabilized by interactions between hydrophobic residues. Residues L44, V51, and L58 were important for the N-N interaction; other residues, e.g., L25 and V32, also made a contribution, albeit a modest one. Our alignments of the N-terminal domain of the hantaviral N proteins suggest the coiled-coil structure, and hence the mode of N-protein oligomerization, is conserved among hantaviruses.
Hantaviruses constitute a distinct genus, Hantavirus, in the family Bunyaviridae. They are enveloped viruses with a tripartite negative-sense RNA genome. The three viral RNA segments L, M, and S encode an RNA-dependent RNA polymerase (L protein), the two envelope glycoproteins Gn and Gc, and nucleocapsid (N) protein, respectively (11). Hantaviruses are carried by rodents and can cause hemorrhagic fever with renal syndrome or hantavirus (cardio)pulmonary syndrome when transmitted to humans (40).
The hantaviral N protein contains 429 to 433 amino acid (aa) residues and has a molecular mass of approximately 50 kDa. It is the most abundant viral component in both virions and infected cells (19). Its main function is to encapsidate both minus-strand viral RNA and plus-strand cRNA forming the viral ribonucleoprotein (RNP). The RNA-binding domain has been located in the central, highly conserved part of the N protein that spans aa residues 175 to 217 (41). Lysine residues dispersed between positions 175 and 429 and also three residues, E192, Y206, and S217, located in the RNA-binding domain were shown to be important for RNA binding (36). Recently, N protein was shown to act as an RNA chaperone (31). In the course of transcription and replication, N protein interacts with the L protein (21); it probably also reacts with the cytoplasmic tail of Gn protein during assembly of new virions. In addition, N protein performs some ambassadorial duties by interacting with actin filaments (34) and also Daxx and SUMO-1 pathway components in infected cells (17, 22, 23, 27).
Hantaviral N protein can form stable trimers, which are thought to serve as intermediates in the process of oligomerization and RNP formation (2, 3, 15, 16, 18, 30). Trimeric N protein seems to bind specifically to a panhandle structure of the RNPs formed by the base-paired 3′- and 5′-genome RNA termini (30). Both C- and N-terminal domains of the N protein contribute to the trimerization. The current data are consistent with the “head-to-head, tail-to-tail” model for hantavirus N-protein trimerization (3, 16, 18). This model suggests that trimerization is a two-step process that involves an initial interaction between the N-terminal domains followed by a consolidating interaction between the C-terminal domains.
The N-terminal trimerization domain of the hantaviral N protein most likely folds into a coiled-coil structure (2, 3). Generally, coiled-coils consist of two or more alpha-helices that wrap around each other in a highly organized manner. Coiled-coils are characterized by a seven-residue periodicity (heptad repeat), with the occurrence of hydrophobic residues preferentially in the first, and usually also the fourth, position of the heptad (26). In the formation of the coiled-coil, these hydrophobic residues of two or more helices often interact in a “knobs-into-holes” manner to form a stable hydrophobic seam (10). In a coiled-coil, the hydrophobic seam twists slowly around the helix as two helices try to bury their hydrophobic surfaces. Coiled-coils of right-handed helices are generally left-handed. Thus, the number of residues per turn in each helix can be reduced from 3.6 to 3.5, which leads to a periodicity of seven residues, i.e., two complete turns (10). Coiled-coils are known to be a versatile protein motif involved in a number of protein-protein interactions that serve different biological functions, such as signal transduction events, regulation of transcription, and cytoskeleton mobility. Many of these interactions are dependent on the oligomerization properties of the coiled-coil motif (7, 28).
Using the MultiCoil prediction algorithm (44), coiled-coils spanning aa residues 1 to 34 and 38 to 80 have been predicted for the N protein of several hantaviruses: Hantaan virus, Seoul virus, Sin Nombre virus (SNV), Prospect Hill virus, and Tula virus (TULV) (2). In one study (3), synthetic peptides representing SNV N-protein residues 3 to 35, 43 to 75, and 3 to 75 were shown to oligomerize, and the longest peptide was shown to adopt an intramolecular helix-turn-helix conformation. In this paper, we study the role of the N-terminal coiled-coil structure in the oligomerization of TULV-N protein using a combination of computer modeling with in vitro techniques: mammalian two-hybrid (M2H) assay and immunofluorescence assay (IFA).
MATERIALS AND METHODS
Multiple-sequence alignment.
The N-protein sequences from the following hantavirus species were used: TULV, GenBank accession number Z69991; Topografov virus, AJ01164; Khabarovsk virus, U35255; Puumala virus (PUUV), X61035; Prospect Hill virus, Z49098; Blood Land Lake virus, U19303; Isla Vista virus, U19302; SNV, L25784; New York virus, U47135; Laguna Negra virus, AF005727; Rio Mamore virus, U52136; Andes virus (ANDV), U52136; Bayou virus, L36929; Black Creek Canal virus, L39949; El Moro Canyon hantavirus, U11427; Rio Segundo hantavirus, U18100; Hantaan virus (HTNV), M14626; Seoul virus (SEOV), AF288653; Dobrava virus, L41916; and Saaremaa virus, AJ009773.
Secondary structure prediction.
The following methods were used for predicting the N-protein secondary structure: (i) PsiPred (13, 29), which uses feed-forward neural networks and PSI-BLAST; (ii) Sam-T99 (14, 32), based on position-specific scoring matrices; (iii) Jufo (http://www.jens-meiler.de/jufo.html), which uses primary structure only; and (iv) PROFsec, an improved method of PHDsec (35). The program Coils (26) was used to predict coiled-coils.
3D structure prediction.
Due to the lack of significant sequence identity between the N-terminal domain of the TULV-N protein and proteins with a resolved structure, the three-dimensional (3D) structure prediction for this domain was performed using a meta server (http://www.bioinfo.pl/meta/) (12) that combines results of various threading methods. Threading methods are used to scan a representative set of the structures in the Protein Data Bank (4), taking into account both sequential and structural information of the query and the target proteins. This leads to the detection of hits that might not have sufficient sequence identity but nevertheless share important structural features with the query. The server then ranks the structure predictions generated with different methods for the given protein sequence and provides main-chain coordinates for the 20 best models. Phyre (http://www.sbg.bio.ic.ac.uk/∼phyre/), a successor to the server 3D-PSSM (20) that predicted the highest scoring model, was used in order to generate a model containing both main-chain and side-chain coordinates.
Plasmids.
The plasmids encoding the N protein of TULV, strain Moravia (39), with N-terminal truncations were created by PCR from the cDNA clone that contained the complete coding region of the S segment sequence (16). Point mutations were introduced into pM1-TULVN, pVP16-TULVN, and pcDNA3-TULVN plasmids (16) using a site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All the plasmids were characterized by restriction analysis and also by sequencing using an ABI PRISM Dye Terminator sequencing kit (Perkin-Elmer).
M2H assay.
The mammalian two-hybrid (M2H) assay was performed essentially as described previously (18). HeLa cells were cultivated in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin, and streptomycin on 24-well culture plates to approximately 70% confluence and transfected with 0.5 μg of pM1 and pVP16 DNA (expressing different N-protein constructs), 0.5 μg of reporter DNA pGluc (expressing firefly luciferase), and 0.01 μg of control DNA pRL-SV40 (expressing Renilla luciferase) (Promega). Transfections were performed in triplicate with 6 μl of FuGene6 reagent for each transfection according to the manufacturer's instructions (Roche Diagnostics Corporation), and after 24 h the luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega). The firefly luciferase values were normalized to the Renilla luciferase values, and the interaction was calculated as described earlier (16).
Immunofluorescence assay (IFA) was performed essentially as described previously (18). Briefly, COS7 cells were transfected with 0.5 μg of different pcDNA3-N constructs using Fugene reagent (Roche Diagnostics) according to the manufacturer's instructions. After 24 h, cells were fixed with 3.2% paraformaldehyde and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS). Cells were stained with monoclonal N-protein antibodies (1/50 in PBS, 1 h at room temperature) and then with fluorescein isothiocyanate-conjugated rabbit anti-mouse secondary antibodies (1/30 in PBS, 1 h at room temperature). The samples were examined using a Zeiss Axioplan microscope with a ×63 oil immersion lens.
RESULTS
Sequence analysis and structure predictions.
A multiple alignment of the 77 N-terminal aa residues of the N protein of 20 distinct hantavirus types/species is shown in Fig. 1A. The sequence is highly conserved between all hantaviruses (identity of 43 to 99%) and shows a pattern typical of coiled-coils: a heptad repeat with a hydrophobic residue in the first position and another hydrophobic residue in the fourth position. In our alignment, the most hydrophobic residue (the “a” residue in the abcdefg heptad) corresponds to positions 4, 11, 18, 25, and 32 in the first half of the sequence and to positions 44, 51, 58, and 65 in the second half. All these residues are highly conserved. Residues I11, L/I18, L/V25, L44, and L/I58 are shared by all hantaviruses. L4 is shared by all except Hantaan and Seoul viruses, V32 and V51 are shared by all except Murinae-carried viruses (group 3), and L65 is shared by all except Puumala virus. The “d” residue, which corresponds to positions 7, 14, 21, and 28 in the first half of the sequence and to positions 47, 54, 61, and 68 in the second half, is less conserved and not exclusively hydrophobic. At position 14 most of the sequences have histidines, and at position 47 all sequences have arginines.
FIG. 1.
A. Multiple alignment of 20 partial sequences of hantaviral N proteins. Symbols used in the Clustal program: *, positions that have a single fully conserved residue; :, “strong” groups that are fully conserved; ., “weaker” groups that are fully conserved. Hydrophobic residues comprising a heptad repeat have been shaded. GenBank accession numbers of hantaviral species used in the alignment are the following: Tula virus (TULV), Z69991; Topografov virus (TOPV), AJ01164; Khabarovsk virus (KHAV), U35255; Puumala virus (PUUV), X61035; Prospect Hill virus (PHV), Z49098; Blood Land Lake virus (BLLV), U19303; Isla Vista virus (ISLAV), U19302; Sin Nombre virus (SNV), L25784; New York virus (NYV), U47135; Laguna Negra virus (LANV), AF005727; Rio Mamore virus (RIOMV), U52136; Andes virus (ANDV), U52136; Bayou virus (BAYV), L36929; Black Creek Canal virus (BCCV), L39949; El Moro Canyon hantavirus (ELMCV), U11427; Rio Segundo hantavirus (RIOSV), U18100; Hantaan virus (HTNV), M14626; Seoul virus (SEOV), AF288653; Dobrava virus (DOBV), L41916 and Saaremaa virus (SAAV), AJ009773. B. Prediction of N-protein coiled-coil domains for TULV sequence with various algorithms. (The prediction was also performed for the following virus sequences, marked in boldface: PUUV, SNV, ANDV, HTNV, and SEOV). The two top lines show a prediction of coiled-coils and disordered regions. In the lower lines, “H” corresponds to helix and “E” to beta sheets.
The results of secondary structure predictions for the TULV-N protein sequence are summarized below the sequence alignment (Fig. 1B). The consensus of different prediction methods used was that the N-terminal part of the protein folds into two alpha-helices; the first helix includes residues 4 to 31(24), and the second one includes residues 38 to 72. Coiled-coils were predicted using all three window sizes, i.e., 14, 21, and 28 residues, for the regions spanning residues 4 to 31(24) and 44 to 71 with a probability of 1.0. This, too, indicated that the N-terminal part of the protein is alpha-helical. Almost identical results regarding the occurrence of coiled-coils were obtained for other hantaviral species: Puumala virus which, like TULV, belongs to the first group, SNV and Andes virus from the second group, and also Hantaan and Seoul viruses from the third group. It should be mentioned, however, that for Hantaan and Seoul viruses the probability of the second helix coiled-coil conformation was somewhat lower when bigger window sizes (21 and 28) were used: 0.2/0.4 and 0.65/0.85, respectively.
The tertiary structure prediction for the N-terminal putative coiled-coil-forming domain (residues 1 to 79) gave the following results. Not less than 18 out of the 20 best models provided by the meta server were based on coiled-coil domains in different proteins. After manual inspection of the highest scoring models, the one generated with the 3D-PSSM method and ranked second was chosen as our working hypothesis. The model (Fig. 2A) is based on the coiled-coil forming a dispensable insert domain of human DNA topoisomerase I. To obtain the side chain coordinates for our model, we resubmitted the sequence of Tula virus N protein (aa residues 1 to 74) to the renewed 3D-PSSM server (Phyre). The part of TULV-N protein that was modeled included residues 3 to 72. This region was aligned to residues 643 to 712 of the human DNA topoisomerase I structure (Protein Data Bank entry 1k4tA). The alignment showed an identity of 25.7%. The e-value of the hit was 0.0072. Our further analysis of the hantaviral N protein relies on the model based on the DNA topoisomerase I structure. Our working model suggested that the N-terminal region of TULV-N protein forms a coiled-coil in which two antiparallel alpha-helices interact via their hydrophobic seams. The crucial aa residues predicted to interact are the hydrophobic residues L/I/V at the position “a” of the heptad repeats, namely, L4, I11, L18, L25, and V32 in the first helix and L44, V51, L58, and L65 in the second helix. This model agrees with the predictions published earlier (2) and with experimental data obtained using synthetic peptides (3). To verify this model we used two in vitro assays: M2H assay and IFA.
FIG. 2.
Model built according to the alignment between TULV sequence (aa residues 3 to 73) and DNA topoisomerase (DNA-TOP) sequence (aa residues 641 to 712). (A) The model shown is based on the 3D structure of human DNA topoisomerase I. Hydrophobic “a” residues, which are main forces in the coiled-coil formation, are depicted as space filled and are labeled. They were targets for point mutations. The arrows show N-protein truncations that were tested in this study. (B) The alignment. Shaded residues in the DNA-TOP sequence form the hydrophobic interface of the coiled-coil. TULV hydrophobic residues comprising a heptad repeat are in boldface. Numbers above the TULV-N sequence indicate “a” residues in heptad repeats. The symbols are described in the legend to Fig. 1. Identity between the aligned regions is 23.3%, and the bottom line shows the predicted secondary coiled-coil structure in the region. Shaded residues in the DNA-TOP sequence form the hydrophobic interface of the coiled-coil. In many cases, residues in the DNA-TOP conform to the residues in the TULV sequence..
Analysis of the N-terminal oligomerization domain of TULV-N protein.
In the first set of experiments, deletions of different lengths were introduced in the coiled-coil-forming part of TULV-N protein (Fig. 2B). Using immunoblotting, we ensured that all mutants are produced in comparable quantities (data not shown). Interacting capacities of the truncated molecules (fused either with the GAL4 DNA-binding domain [DBD] or the VP16 activation domain [AD]) were tested in the M2H system (Fig. 3). Altogether, four mutants lacking the first 50, 57, 64, and 77 aa residues, respectively, have been constructed. The molecule with the shortest deletion, N44-429, which was created earlier (16), retained approximately 80% of the interacting capacity of the full-length N protein. We therefore decided to introduce only deletions that are longer than 44 aa. The molecule with the longest deletion that included the whole coiled-coil-forming region plus several extra residues, N78-429, retained approximately 40% of the initial interacting capacity. This result was repeatedly observed when the deletion was introduced into the N protein fused with the DBD, AD, or both. Two other mutants, N51-429 and N58-429, showed a reduced interacting capacity as well. These results were in agreement with the earlier observations that the N-terminal domain is important for the homotypic interaction seen in the M2H assay (16). Surprisingly, in the mutant N65-429 the interacting capacity measured in the M2H assay remained unaffected. The reason(s) for this remains unknown.
FIG. 3.
Interaction of N- and C-terminal truncation mutants in the M2H assay. Deleted regions are shown in black. Numbers (center-left column) are averages of normalized luciferase activity values (%) ±standard deviations. Numbers are calculated from a majority of experiments (center-right column). Each mutant was tested in triplicate. Numbers marked with an asterisk are from Kaukinen et al. (16).
All five truncated N-protein molecules (fused with AD) were next tested together with the mutant N1-398 (fused with DBD). This mutant lacks the last 31 aa residues that form one of the two interacting alpha-helices in the C-terminal part of the molecule and therefore can react only with the partners carrying an intact C-terminal sequence. When used with wild-type N protein, the mutant N1-398 showed complete interacting capacity; when used with another N1-398 molecule, it showed no interaction at all (Fig. 3). We used this mutant to ensure that two N-protein mutants will react in our experiments exclusively “head-to-head” and “tail-to-tail.” Data presented in Fig. 3 show that mutants which carried N-terminal truncations reacted with the mutant N1-398 as efficiently as with the full-length N protein, resulting in reporter activity comparable to that observed in the first set of experiments. Truncated N-protein molecules with gradually removed coiled-coil-forming heptads showed a reduced interacting capacity. The mutant with the longest truncation, N78-429, showed the lowest interacting capacity. Notably, an interacting capacity of the mutant N65-429 again remained unaffected, and the mutant N58-429 showed a reduced interacting capacity (72%) in only one of three experiments performed.
These results confirmed that both N-terminal coiled-coils and C-terminal alpha-helices contribute to the N-N interaction. In a control experiment, when the N-terminal oligomerization domain was removed from one partner, AD-N78-429, and the C-terminal oligomerization domain was removed from the other partner, DBD-N1-392, an interacting capacity of the pair was almost completely abolished.
Intracellular localization and immunofluorescence pattern of transiently expressed truncated N-protein molecules.
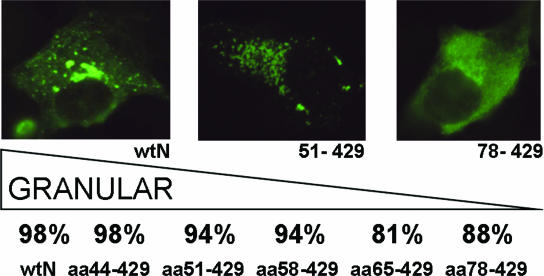
Our previous study (18) showed good correlation between an interacting capacity of the mutated N protein in the M2H assay and a perinuclear localization, as well as a granular fluorescence pattern of the transiently expressed polypeptide. While intact N protein localized in the perinuclear region and showed a bright granular pattern, similar to what was seen in the virus-infected cells, the N-protein mutants (both point mutants and truncations) showed a more diffuse localization. To study the intracellular localization of the N-protein mutants with the N-terminal truncations, we expressed them transiently in COS7 cells and then visualized them with TULV-specific monoclonal antibody (MAb) 3D3, which recognizes the central part of the protein spanning aa residues 226 to 293 (25). The results are summarized in Fig. 4. There was a steady increase in the number of transfected cells, showing a diffuse immunofluorescence pattern with a stretch of aa removed from the N terminus of the molecule. An increase as high as sixfold (from 2% to 12%) was observed. Interestingly, a substantial portion of cells transfected with the mutant N65-429 showed diffuse immunofluorescence patterns, suggesting that the oligomerization capacity of this mutant was affected.
FIG. 4.
Percentages obtained in three independent counting sessions of localization patterns of transiently expressed N-protein mutants in COS7 cells. Immunofluorescence was used to visualize N protein in cells transfected with a construct expressing wild-type N protein (wtN), pictured on the left (a granular pattern was observed in 98% of the cells). Truncation mutant N51-429 is shown in the center (granular pattern in 94% of cells), and N78-429 is on the right (diffuse pattern in 12% of cells).
To exclude the possibility that the appearance of a diffuse immunofluorescence in this experiment was artificial (e.g., caused by unusually high background fluorescence), we checked the transfected cells with another MAb, 1C12, which recognizes a conservative, genus-specific epitope located within the first 79 aa residues (24). All preparations, except the ones with a positive control, showed no detectable fluorescence (data not shown), thus confirming specificity of the observations made with the MAb 3D3.
Analysis of point mutations at position “a” in the coiled-coil heptads.
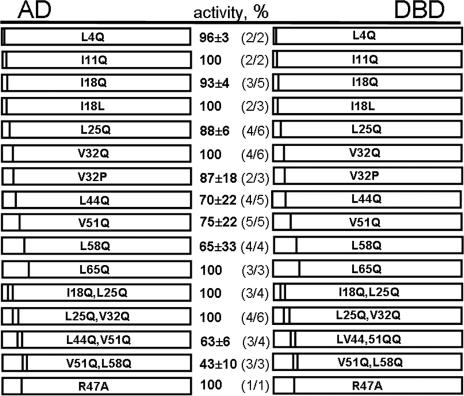
Our data suggested that the removal of the whole coiled-coil-forming region or its parts affected the oligomerization capacity of the N-protein molecule. In the next set of experiments, we mutated L/I/V residues located at positions 4, 11, 18, 25, 32, 44, 51, 58, and 65 in order to evaluate their contribution in keeping together the 3D structure of the N-terminal coiled-coil (supposedly crucial for the homotypic interaction). We constructed a set of the N-protein mutants in which these residues were replaced with glutamine (Q), a polar amino acid with an aliphatic chain similar to that of leucine. In theory, mutation of leucine, isoleucine, or valine to glutamine should have a negative effect on the coiled-coil formation. In addition to single point mutants, four double mutants, I18Q-L25Q, L25Q-V32Q, L44Q-V51Q, and V51Q-L58Q, were produced.
Data presented in Fig. 5 show the interacting capacity of the wild-type N protein, as seen in the majority of the experiments. Three out of 10 single mutants, I11Q, V32Q, and L65Q, retained the interacting capacity of the wild-type N protein; in 7 others this capacity was reduced. Notably, for the mutations residing in the first alpha-helix, L4Q, I18Q, and L25Q, the reduction was minimal (Fig. 5). The mutant I18L served as a control for this set. We anticipated that the “quasi-neutral” mutation of isoleucine at position 18 to leucine should not have an inhibitory effect, which was indeed the case. The mutant V32P was included to test whether this aa residue is located within the helix (as it is shown in Fig. 2) or in the loop/turn which connects two alpha-helices of the coiled-coil. If V32 is located within the helix, the change to proline (known as a “disruptor” of alpha-helices) should have a greater effect on the N-protein-interacting capacity. The results suggested that V32 is located within the helix.
FIG. 5.
Interaction of N-terminal point mutants in the M2H assay. Point mutations are indicated by bars. Numbers (center-left column) are averages of normalized luciferase activity values (%) ±standard deviations. Numbers are calculated from a majority of experiments (center-right column). Each mutant was tested in triplicate.
Of the four double mutants, two (I18Q-L25Q and L25Q-V32Q) showed unaffected interacting capacity, while this capacity in the other two mutants, L44Q-V51Q and V51Q-L58Q, was reduced to approximately half. Interestingly, this reduction was significantly more pronounced than that in either of the single mutants (70%, 75%, and 63%, as well as 75%, 65%, and 43%, respectively), i.e., a cumulative effect of the L/V and V/L replacements was observed.
Taken together, these results suggested that L44, V51, and L58 from the second alpha-helix of the coiled-coil contribute to the stability of the 3D structure. The residues in the first alpha-helix, especially L25 and V32, also made a contribution, albeit a modest one.
Unfortunately, IFA was not really useful in the evaluation of point mutations. For instance, the double mutant L44Q-V51Q that demonstrated reduced interacting capacity in the M2H system showed an immunofluorescent pattern indistinguishable from that of the wild-type N protein (data not shown). It seems that the remaining oligomerization capacity of the N-terminal domain is still sufficient to bring together interacting C-terminal alpha-helices and hence to secure both perinuclear localization of the mutant protein and the granular pattern of immunofluorescence. It is worth mentioning that MAbs 3D3 and 1C12 recognized the double mutant equally well.
Finally, we tested one of the charged “d” residues in the coiled-coil, namely R47. When this aa residue was replaced with neutral alanine, the interacting capacity of the N protein remained unaffected. This result suggested that the contribution of the positive R47 to the overall stability of the 3D structure of the coiled-coil is minimal, if any.
DISCUSSION
Data presented in this paper confirm an involvement of the N-terminal coiled-coils in the oligomerization of hantaviral N protein. Taken together with the previously published results by us (15, 16, 18) and others (2, 3, 42), they support the model of trimerization based on the “head-to-head, tail-to-tail” mode of interaction. According to this model (18), three monomers are brought together via interaction of their N-terminal domains. This initial contact is followed by the interaction between two C-terminal alpha-helices that form a shared hydrophobic space and thus consolidate the trimer formation. Details of the interaction between the N-terminal coiled-coils remain largely unknown. Some possibilities are discussed below.
Here we would like to stress that in vitro assays used in this study have had to perform on the “margins” of contribution of the N-terminal domain to the two-step interaction process, and those appeared to be rather small. Besides, in the M2H assay, our main experimental tool, the N terminus of the N protein was fused to the C terminus of either DBD or AD. This could complicate evaluation of some mutations within the coiled-coil, especially those that would have a modest impact on the overall folding. Fortunately, the efficiency of mutant evaluation using the M2H assay was reasonably high: four of five truncation mutants, 7 of 10 “a” residue point mutants, and two of four double mutants showed a reduced interacting capacity.
Of course, the M2H assay is not an ideal one. For instance, truncation of the first 65 aa residues did not affect the interacting capacity of the N protein, while the removal of either 58 aa residues or 77 aa residues did. One possible explanation is that the truncations shorter than 65 aa residues destroy the structure of the coiled-coil and hence induce a misfolding of the remaining part of the N-terminal region. This might create a steric obstacle(s) for moving C-terminal domains of the interacting N-protein molecules into close proximity to each other and hence for the reporter gene expression. In the mutant N65-429, almost the entire coiled-coil-forming region is removed, and therefore these steric obstacles may not exist. In addition, the part of the molecule between positions 65 and 77 might contain aa residues crucial for the oligomerization. This hypothesis awaits further investigation. Most recently we produced three point mutants, D67A, K73A, and V69Q, in which conserved charge residues or a conserved hydrophobic “d” residue was replaced, but we have not seen major changes in the oligomerization capacity of the N protein.
IFA, in which the conclusions are drawn on the basis of a somewhat changed pattern of immunofluorescence (that, for the wild-type protein, is granular and mostly perinuclear), was less efficient. In our earlier work (18), several mutations introduced into C-terminal oligomerization domains caused drastic changes in the IFA pattern of transiently expressed N proteins. In the present study, no pronounced changes were seen with point mutants, and only an exhaustive search revealed some changes with the truncated mutants. These IFA results, however, cannot be taken as direct evidence for a reduced oligomerization capacity of the N-protein mutants. An intracellular transport of the N protein is supposedly a complex process which may include an interaction with a cellular component(s). Although it is quite possible that this interaction would occur only if the N protein exists in a proper form (e.g., trimer/oligomer), the results also suggest that mutations affect the ability of the N protein to interact with its cellular counterparts. Further studies are needed to clarify this issue.
Model for the trimerization of the N-terminal coiled-coils.
There are at least three different ways in which the N-terminal coiled-coils can form a trimer (Fig. 6). The first possibility is that the antiparallel coiled-coils of three monomers remain intact (Fig. 6A). In this case, the interacting forces are generated mainly by charged or polar residues, which are located on the opposite side of the helix relative to the hydrophobic “a” residues. Alternatively, the hydrophobic seams of the three coiled-coils are shared, thus, the main contribution is due to the hydrophobic forces. The second scenario includes two steps: (i) opening of three intramolecular coiled-coils and (ii) formation of three intermolecular coiled-coils (Fig. 6B). The first helix of one monomer interacts with the second helix of another monomer. Thus, both helices of each monomer are involved, and three separate antiparallel coiled-coils are formed. In this case, the interacting forces are supposedly mostly hydrophobic. The third option is that the first helices of three monomers form a parallel coiled-coil trimer (Fig. 6C). Here, interactions between the helices are expected to be predominantly hydrophobic as well (1, 43). In model A, antiparallel coiled-coils of the monomers apparently remain intact, while the other scenarios include conformational changes that involve opening of the intramolecular coiled-coils. Examining the properties of synthetic peptides representing N-terminal residues of Sin Nombre virus N protein, Alfadhli et al. (3) came up with a similar suggestion on the two alternative ways of forming a trimer: (i) bundling of intramolecular antiparallel coils or (ii) a conformational switch from intramolecular to intermolecular coiled-coils.
FIG. 6.
Models of N-terminal coiled-coil trimerization. (A) Coiled-coils are intramolecular, and the trimerization forces are due either to shared hydrophobic space (conserved leucines, isoleucines, and valines) or to other residues that form intermolecular hydrogen bonds. (B) Coiled-coils are intermolecular and interact to form a trimer. The first helix of one monomer interacts with the second helix of another monomer. (C) Coiled-coils form a trimer in a manner in which hydrophobicity is shared between three helices.
We favor model A for the following reasons. First, it is the simplest model that does not require an opening of the coiled-coils of three monomers. Second, this conclusion is not contradicted by results of in silico docking. Three N-protein monomers using the 3D model predicted by the Rosetta ab initio protocol (5, 6, 37, 38) were docked (as rigid bodies) into a trimer using the ClusPro server (9). One of the obtained solutions was a trimeric structure similar to model A (V. Halttunen, A. Alminaite, and A. Plyusnin, unpublished data). Third, some data of Alfadhli et al. (3) point in this direction as well. These authors observed that at low concentrations, the peptide representing aa residues 3 to 75 adopted an intramolecule helix-turn-helix conformation, i.e., antiparallel coiled-coil, and only at high concentrations did it assemble dimers and/or trimers. Furthermore, the idea that charged and/or polar aa residues of the coiled-coils might be important for the N-protein trimerization (as model A suggests) is in agreement with our early data that an increase in the ionic strength inhibits interaction(s) between N-protein molecules (16). It is also supported by the observation that charged and polar residues are abundant in the coiled-coil-forming region, and most of them are highly conserved (Fig. 1). Our preliminary data on mutants with the aspartic acid residues at positions 37 and 38 (presumably located in the loop/turn structure on the coiled-coil) replaced with alanines showed that at least one of these charges contributes to the interaction registered in the M2H system. Efforts are under way to pinpoint other interacting charged and polar aa residues and also to model interacting surfaces presumably formed by the N-terminal domains of three monomers.
Coiled-coils are among the most abundant domains involved in protein interactions. They play key roles in virtually every physiological system. It is a common motif that is often used to control oligomerization. Very long coiled-coils are the basis for some of the fibrous proteins, while shorter coiled-coils are used in several types of proteins, such as transcription factors and tRNA synthetases. Coiled-coils have one of the simplest dimerization interfaces, yet they can mediate highly selective protein associations (28). As a well-defined structure, coiled-coil regions have been chosen as targets for antiviral drugs (8, 33). Future studies are needed to explore this possibility to control hantaviral infections.
Acknowledgments
We thank Pasi Kaukinen and Kirsi Tulimäki for their help with the M2H system and Anna Katz for help in reading IFA slides and stimulating discussion.
This work was supported by The Academy of Finland (grants 2022012, 212313, and 105210), the EU (grant QLK2-CT-2002-01358), and the Sigrid Jusélius Foundation.
REFERENCES
- 1.Akey, D. L., V. N. Malashkevich, and P. S. Kim. 2001. Buried polar residues in coiled-coil interfaces. Biochemistry 40:6352-6360. [DOI] [PubMed] [Google Scholar]
- 2.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 4.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. Protein data bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonneau, R., C. E. Strauss, C. A. Rohl, D. Chivian, P. Bradley, L. Malmstrom, T. Robertson, and D. Baker. 2002. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322:65-78. [DOI] [PubMed] [Google Scholar]
- 6.Bonneau, R., J. Tsai, I. Ruczinski, D. Chivian, C. Rohl, C. E. Strauss, and D. Baker. 2001. Rosetta in CASP4: progress in ab initio protein structure prediction. Proteins Suppl. 5:119-126. [DOI] [PubMed] [Google Scholar]
- 7.Burkhard, P., J. Stetefeld, and S. V. Strelkov. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comeau, S. R., D. W. Gatchell, S. Vajda, and C. J. Camacho. 2004. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20:45-50. [DOI] [PubMed] [Google Scholar]
- 10.Crick, F. H. C. 1953. The packing of α-helices: simple coiled coils. Acta Crystallogr. 6:689-697. [Google Scholar]
- 11.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Petterson, A. Plyusnin, and C. Schmaljohn. 2000. Bunyaviridae, p. 599-621. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: the classification and nomenclature of viruses. The seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 12.Ginalski, K., A. Elofsson, D. Fischer, and L. Rychlewski. 2003. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19:1015-1018. [DOI] [PubMed] [Google Scholar]
- 13.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 14.Karplus, K., C. Barrett, and R. Hughey. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14:846-856. [DOI] [PubMed] [Google Scholar]
- 15.Kaukinen, P., V. Koistinen, O. Vapalahti, A. Vaheri, and A. Plyusnin. 2001. Interaction between molecules of hantavirus nucleocapsid protein. J. Gen. Virol. 82:1845-1853. [DOI] [PubMed] [Google Scholar]
- 16.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Mapping of the regions involved in homotypic interactions of Tula hantavirus N protein. J. Virol. 77:10910-10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 92:37-45. [DOI] [PubMed] [Google Scholar]
- 18.Kaukinen, P., V. Kumar, K. Tulimaki, P. Engelhardt, A. Vaheri, and A. Plyusnin. 2004. Oligomerization of hantavirus N protein: C-terminal alpha-helices interact to form a shared hydrophobic space. J. Virol. 78:13669-13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2005. Hantavirus nucleocapsid protein: a multifunctional molecule with both housekeeping and ambassadorial duties. Arch. Virol. 150:1693-1713. [DOI] [PubMed] [Google Scholar]
- 20.Kelley, L. A., C. M. McCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3DPSSM. J. Mol. Biol. 299:501-522. [DOI] [PubMed] [Google Scholar]
- 21.Kukkonen, S. K. J., A. Vaheri, and A. Plyusnin. 2004. Tula hantavirus L protein is a 250 kDa perinuclear membrane-associated protein. J. Gen. Virol. 85:1181-1189. [DOI] [PubMed] [Google Scholar]
- 22.Lee, B. H., K. Yoshimatsu, A. Maeda, K. Ochiai, M. Morimatsu, K. Araki, M. Ogino, S. Morikawa, and J. Arikawa. 2003. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 98:83-91. [DOI] [PubMed] [Google Scholar]
- 23.Li, X. D., T. P. Makela, D. Guo, R. Soliymani, V. Koistinen, O. Vapalahti, A. Vaheri, and H. Lankinen. 2002. Hantavirus nucleocapsid protein interacts with the Fas-mediated apoptosis enhancer Daxx. J. Gen. Virol. 83:759-766. [DOI] [PubMed] [Google Scholar]
- 24.Lundkvist, Å., and B. Niklasson. 1992. Bank vole monoclonal antibodies against Puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch. Virol. 126:93-105. [DOI] [PubMed] [Google Scholar]
- 25.Lundkvist, Å., O. Vapalahti, A. Plyusnin, K. B. Sjolander, B. Niklasson, and A. Vaheri. 1996. Characterization of Tula virus antigenic determinants defined by monoclonal antibodies raised against baculovirus-expressed nucleocapsid protein. Virus Res. 45:29-44. [DOI] [PubMed] [Google Scholar]
- 26.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 27.Maeda, A., B. H. Lee, K. Yoshimatsu, M. Saijo, I. Kurane, J. Arikawa, and S. Morikawa. 2003. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). Virology 305:288-297. [DOI] [PubMed] [Google Scholar]
- 28.Mason, J. M., and K. M. Arndt. 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 6:170-176. [DOI] [PubMed] [Google Scholar]
- 29.McGuffin, L. J., K. Bryson, and D. T. Jones.2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 30.Mir, M. A., and A. T. Panganiban. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78:8281-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mir, M. A., and A. T. Panganiban. 2006. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA 12:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, J., K. Karplus, C. Barrett, R. Hughey, D. Haussler, T. Hubbard, and C. Chothia. 1998. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 284:1201-1210. [DOI] [PubMed] [Google Scholar]
- 33.Pinon, J. D., S. M. Kelly, N. C. Price, J. U. Flanagan, and D. W. Brighty. 2003. An Antiviral peptide targets a coiled-coil domain of the human T-cell leukemia virus envelope glycoprotein. J. Virol. 77:3281-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravkov, E. V., S. T. Nichol, C. J. Peters, and R. W. Compans. 1998. Role of actin microfilaments in Black Creek Canal virus morphogenesis. J. Virol. 72:2865-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 36.Severson, W. E., X. Xu, M. Kuhn, N. Senutovitch, M. Thokala, F. Ferron, S. Longhi, B. Canard, and C. B. Jonsson. 2005. Essential amino acids of the hantaan virus N protein in its interaction with RNA. J. Virol. 79:10032-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons, K. T., C. Kooperberg, E. Huang, and D. Baker. 1997. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268:209-225. [DOI] [PubMed] [Google Scholar]
- 38.Simons, K. T., I. Ruczinski, C. Kooperberg, B. A. Fox, C. Bystroff, and D. Baker. 1999. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins 34:82-95. [DOI] [PubMed] [Google Scholar]
- 39.Vapalahti, O., Å. Lundkvist, S. K. Kukkonen, Y. Cheng, M. Gilljam, M. Kanerva, T. Manni, M. Pejcoch, J. Niemimaa, A. Kaikusalo, H. Henttonen, A. Vaheri, and A. Plyusnin. 1996. Isolation and characterization of Tula virus, a distinct serotype in the genus Hantavirus, family Bunyaviridae. J. Gen. Virol. 77:3063-2067. [DOI] [PubMed] [Google Scholar]
- 40.Vapalahti, O., J. Mustonen, A. Lundkvist, H. Henttonen, A. Plyusnin, and A. Vaheri. 2003. Hantavirus infections in Europe. Lancet Infect. Dis. 3:653-661. [DOI] [PubMed] [Google Scholar]
- 41.Xu, X., W. Severson, N. Villegas, C. S. Schmaljohn, and C. B. Jonsson. 2002. The RNA binding domain of the Hantaan virus N protein maps to a central, conserved region. J. Virol. 76:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimatsu, K., B. H. Lee, K. Araki, M. Morimatsu, M. Ogino, H. Ebihara, and J. Arikawa. 2003. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J. Virol. 77:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiltscheck, R., R. A. Kammerer, S. A. Dames, T. Schulthess, M. J. Blommers, J. Engel, and A. T. Alexandrescu. 1997. Heteronuclear NMR assignments and secondary structure of the coiled coil trimerization domain from cartilage matrix protein in oxidized and reduced forms. Protein Sci. 6:1734-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf, E., P. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]