Abstract
Estimates of seropositivity to a new infectious agent in a community are useful to public health. For severe acute respiratory syndrome (SARS), the figures are conflicting. Herein, we screened 12,000 people in a community stricken by SARS 10 months previously and found 53 individuals (0.44%) who had immunoglobulin G antibodies to the SARS coronavirus (SARS-CoV) nucleocapsid (N) produced in bacteria. However, only seven of these (group 1) had sera which also reacted with the native N antigen expressed in SARS-CoV-infected Vero cells, N-transfected 293T cells, and tissues of infected SARS patients. Of these, six individuals had had SARS previously. The remaining person, as well as the 46 other individuals (group 2), were healthy and had no history of SARS. Group 1 antibodies recognized epitopes located slightly differently in N from those of group 2 antibodies, and a mouse hybridoma antibody resembling the former type was generated. Unusually, group 2 antibodies appeared to recognize cross-reactive bacterial epitopes that presumably were posttranslationally modified in eukaryotes and hence were probably not induced by SARS-CoV or related coronaviruses but rather by bacteria. The N antigen is thus highly unique. The extremely low rate (0.008%) of asymptomatic SARS infection found attests to the high virulence of the SARS-CoV virus.
Severe acute respiratory syndrome (SARS) emerged suddenly in 2003 but disappeared soon afterwards. It has left behind a trail of questions, many of which have been promptly answered. Indeed, a voluminous amount of valuable information has been acquired regarding the etiological viral agent (SARS coronavirus [SARS-CoV]) (7, 18, 20, 22), the pathogenesis (10, 12, 21), the host immune response (15, 19, 26), and, more recently, determinants of host resistance (2).
However, one question has remained inadequately answered. This relates to the exposure of the general community to SARS-CoV in places which have experienced an outbreak of SARS. Determination of the exposure is usually carried out by serological methods which detect antibodies to the virus. Investigations conducted thus far are conflicting regarding the prevalence of individuals in a community who were seropositive but who did not have overt SARS previously (defined according to WHO guidelines). Based on this definition—which considers subclinical or nonpneumonic manifestations as asymptomatic—some studies found high exposure rates (0.48% to 4.60%) (9, 27, 31, 32, 34) while others found low (0.14% to 0.19%) (13, 16) to nil (1, 6, 9, 30) rates. Such differences may be due to the assay method used (for example, Vero cell immunofluorescence [IFA] versus enzyme-linked immunosorbent assay [ELISA]), the type of antigen used for detection (for example, crude viral lysate versus recombinant nucleocapsid [N] antigen), or the population size sampled (87 to 1,621 people). Accurate information is desirable because a high exposure rate suggests two possibilities: (i) the virus is not very virulent or (ii) the virus cross-reacts antigenically with other viruses or microorganisms that are found commonly in the community. A low exposure rate, on the other hand, suggests a highly virulent virus that is unique and novel to the community. This information is important not only for our general understanding of the pathogenic attributes and antigenic properties of the virus but also for public health measures and the development of appropriate immunodiagnostics.
Herein, we report the exposure rate of the Hong Kong community to SARS-CoV, determined after careful analysis of seropositives first identified by initial screening. Hong Kong had witnessed one of the first outbreaks of SARS in the world, which started in February 2003 and ended in May 2003. A total of 1,755 people were infected, of whom 298 died. The present study was carried out 9 to 10 months after the disease started. Serological screening was performed using an ELISA which detects immunoglobulin G (IgG) antibodies to recombinant SARS-CoV N antigens (15). We describe an unusual and important false-positivity problem associated with using antigens produced in bacteria for detection and report, after correcting for this shortcoming, the exceedingly low rate of exposure to SARS in the Hong Kong community. The findings are useful and applicable to other emerging infectious diseases such as bird flu.
MATERIALS AND METHODS
Study cohort.
Residents in the greater Hong Kong region were randomly selected by computer to participate in the study in September to October 2003. A total 50,000 households were invited, from which 12,000 people were recruited with due consent. Of these, 56.3% were females and 87.6% were aged 18 years and above.
Stored (−20°C) sera of SARS patients admitted to the Prince of Wales Hospital in Shatin, Hong Kong, obtained during the acute-convalescence phase (9 to 35 days after disease onset) of SARS were described previously (15).
SARS-CoV.
Native viral antigens were extracted from Vero cells infected with SARS-CoV (strain CUHK-W1; GenBank accession no. AY278554) by lysis in HEPES buffer (pH 7.0) containing 0.2% Nonidet P-40 and 1 mmol/liter 1,4-dithiothreitol (15). Recombinant viral antigens were produced as glutathione S-transferase (GST) fusion proteins in Escherichia coli BL21 using pGEX-2T (Amersham Bioscience) as a vector for the various N segments shown in Fig. 1 (15). The following forward (F) primers (containing a BamHI site, underlined) and reverse (R) primers (containing an EcoRI site, underlined) were used to synthesize the N segments by PCR from SARS-CoV cDNA: rNa (N-terminal N, 660 bp; F, 5′-CGTGGATCCATGTCTGATAATGGACCCCAA-3′; R, 5′-CGATGAATTCCGAGGGCAGTTTCACCACCTCC-3′), rNb (C-terminal N, 630 bp; F, 5′-CGTGGATCCGGAGGTGGTGAAACTGCCCTC-3′; R, 5′-CGATGAATTCCTGCCTGAGTTGAATCAGCAGA-3′), rNb1 (N-terminal rNb, 321 bp; F, 5′-CGTGGATCCGGAGGTGGTGAAACTGCCCTC-3′; R, 5′-CGATGAATTCCGCGTGACATTCCAAAGAATGC-3′), and rNb2 (C-terminal rNb, 330 bp; F, 5′-CGTGGATCCGCATTCTTTGGAATGTCACGC-3′; R, 5′-CGATGAATTCCTGCCTGAGTTGAATCAGCAGA-3′).
FIG. 1.
Serological identification of community subjects exposed to SARS-CoV. A) Map of SARS-CoV N protein showing the various fragments (Na, Nb, Nb1, and Nb2) produced as GST fusion proteins in bacteria used for serological detection. Numbers denote amino acid numbering of protein. B) Purity of the recombinant fusion proteins (rNa-GST and rNb-GST) isolated by affinity chromatography, separated on 14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, and stained by Coomassie blue. Starting material (crude lysate) and molecular weight markers are also shown. C) ELISA results of community subjects screened for IgG antibodies to a combination of rNa-GST and rNb-GST. Comparative data of SARS patients using sera obtained during acute-convalescence phase are shown (OD values >3.5 are plotted as 3.5). The shaded bar denotes the cutoff used to differentiate between elevated and normal levels of antibodies. ▵, CS individuals; ▿, CSN individual; •, CN individuals.
293T transient transfection.
The rNb segment was synthesized from SARS-CoV cDNA using the forward primer 5′-CCACCATGGGAGGAGGTGGTGAAACTGCCCTC-3′ (containing a Kozak sequence, underlined) and the reverse primer 5′-TGCCTGAGTTGAATCAGCAGA-3′ and ligated to the pcDNA3.1/V5-His-TOPO vector (Invitrogen). Clones with the right insert orientation were transiently transfected into 293T cells (ATCC) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty microliters of transfected cells was plated as droplet cultures on 12-well glass slides and incubated overnight at 37°C in a humidified incubator infused with 5% CO2. The cells were fixed with methanol-acetone (9:1) at −20°C for 10 min, air dried, and stored at −70°C until use.
Hybridoma production.
BALB/c mice were hyperimmunized with purified rNb-GST protein, and the spleen cells obtained from these animals were fused with Sp2/0 myeloma cells (29). Hybridomas obtained were screened for reactivity to rNb by ELISA (see below). Antibodies from selected clones were obtained from the spent-culture supernatant and purified partially by ammonium sulfate precipitation. In some cases, the antibodies were protein G selected and biotinylated.
Isolation of rNa- and rNb-specific antibodies from human sera.
Affinity adsorbents were made by coupling the rNa or rNb recombinant protein (100 μg) to cyanogen bromide-activated Sepharose 4B (Sigma) (0.5 ml, 50% vol/vol). The beads were blocked with 5% skim milk (2 h, room temperature [RT]), washed, and incubated with the test serum (0.24 ml) at 4°C for 16 h on a roller-mixer. After the beads were washed five times with phosphate-buffered saline (PBS) containing 0.1% Tween 20, the bound antibodies were eluted by incubating the beads with 0.1 M glycine-NaOH (pH 11.0) (0.5 ml) at RT for 5 min. The eluate was dialyzed against PBS at 4°C overnight, concentrated by Centricon (Millipore), and stabilized with 2.5% bovine serum albumin.
ELISA.
Serum IgG activity in response to SARS-CoV N antigen was detected by a direct ELISA which utilized a combination of bacterially produced rNa-GST and rNb-GST proteins. The protocol used was essentially as described previously (15). Basically, Immunon-2 plates (Dynex) were coated with equal proportions of rNa (1 μg/ml) and rNb (1 μg/ml) in bicarbonate buffer (pH 9.6) overnight at 4°C. The unknown serum (diluted 1:100) was incubated for 30 min at RT, while the developing peroxidase-labeled antibody (goat anti-human IgG; BD Biosciences) and substrate (3,3′,5,5′-tetramethylbenzidine) were each incubated for 15 min at RT. Results were read at 450 nm in a Dynex MRX II reader. Cutoff for seropositivity was calculated as mean ± 2 standard deviations (SD) for 50 healthy individuals. Other direct ELISAs were similarly performed using Immunon-2 plates coated with crude viral lysate (2 μg/ml), rNa-GST (1 μg/ml), or rNb-GST (1 μg/ml); the assay was developed with peroxidase-conjugated goat anti-human Ig (IgM, IgG, or IgA specific; BD Biosciences) or with goat anti-mouse Ig (all classes; BD Biosciences).
In the inhibition ELISA, serum antibody activity was detected by determining the ability of the serum to block the binding of the indicator antibody (mAb13) to the microplate-insolubilized antigen. The amount of mAb13 used was first determined by titration of the antibody. Thus, in the test, the unknown serum (diluted 1:100) was first incubated with the rNb-GST antigen for 16 h at 4°C. Following a washing, biotinylated mAb13 was added, the serum was incubated at 37°C for 2 h, and the assay was subsequently developed with peroxidase-conjugated streptavidin (BD Biosciences) for a further 1.5 h at 37°C. Inhibition was calculated as 1 − [(ODtest − ODmax)/(ODmin − ODmax)], where ODmin is the optical density (OD) without any inhibitor present (buffer control) and ODmax is the OD value at highest inhibition using unlabeled mAb13 as the inhibitor.
IFA.
SARS-CoV-infected Vero cells (Euroimmun), postmortem lung and intestinal tissues from SARS patients, and rNb-transfected 293T cells were used as substrates for the detection of antibodies to SARS-CoV by IFA. With the autopsy materials, sections (4 μm thick) were prepared from 10% formalin-fixed, paraffin-embedded blocks (25). Briefly, the cells or tissue sections were incubated with the test serum or mouse monoclonal antibody (MAb) for 45 min, followed by incubation (30 min) with fluorescein isothiocyanate-conjugated goat anti-human Ig (all classes) (Euroimmun) or goat anti-mouse Ig (all classes) (Sigma) (16, 25). With the tissue sections, images were captured individually in grayscale by Spot Insight 4 (Nikon Eclipse 80) and then pseudocolored.
Western blot (WB) analysis.
The detecting antigen (crude viral lysate, N fragments) was heat denatured, separated on 10 to 14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and transferred to polyvinylidene fluoride membrane (Bio-Rad) as described previously (15). The membrane was blocked and incubated (1 h at RT) with the test serum or mouse MAb and developed (1 h at RT) with horseradish peroxidase-conjugated goat anti-human IgG (BD Biosciences) or goat anti-mouse Ig (all classes) (Sigma). Thrombin proteolysis of GST fusion proteins was performed by incubating 5 μg of protein with 2 units of thrombin (Amersham) overnight at RT.
Statistics.
Comparison of groups was performed using the Mann-Whitney test (PRISM; GraphPad Software), and a P < 0.05 was considered significant.
RESULTS
Extremely low frequency of SARS seropositives found in community, most of whose sera were unreactive with native viral antigens.
A total of 12,000 people were recruited from the community to have their blood screened for the presence of antibodies to the SARS virus. Detection was done by a direct ELISA which detects IgG antibodies to viral N antigens. These antigens were produced as two separate halves (rNa and rNb) of the full-length protein in bacteria, each fused with the bacterial protein GST (Fig. 1A). Purity analysis of the affinity-isolated proteins used in the ELISA revealed no visible bacterial contaminants present (Fig. 1B).
Fifty-three individuals (0.44%) were positive in the test using a combination of rNa and rNb (Fig. 1C). The sera of these people were examined further for antiviral activity by Vero cell IFA. Only seven (0.058%) were positive (Table 1). Of these, six (designated CS) had SARS infection previously and were hospital patients. The remaining individual (designated CSN), a 27 year-old female, as well as the 46 other individuals (designated CN) who were IFA negative, had no history of SARS or had any known contact with SARS patients previously (Table 1).
TABLE 1.
Particulars of community seropositives
| Group | Designation | No. of individuals | Vero cell IFA titer(s) | Clinical history |
|---|---|---|---|---|
| 1 | CS | 6 | 1:80 (CS1), 1:320 (CS2, CS3), 1:640 (CS5, CS6), 1:1,280 (CS4) | All had SARS previously; age range: 21-47 yr (38.0 ± 9.5 yr, mean ± SD); 66.7% female |
| CSN | 1 | 1:640 | Healthy, no contact with SARS; age: 27 yr; female | |
| 2 | CN | 46 | <1:40 (all) | All healthy; age range: 9-78 yr (40.7 ± 15.3 yr, mean ± SD); 56.5% female |
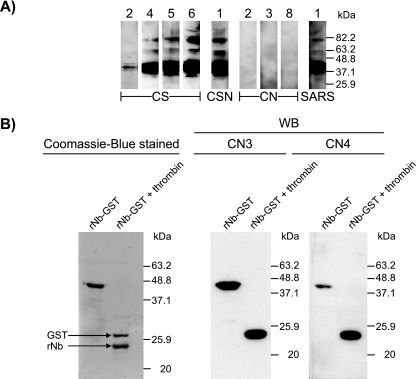
Sera from all 6 CS subjects and the single CSN individual, as well as all 46 individuals from the CN group, were also examined by WB analysis using native viral antigens (extracted from virally infected Vero cells) as the detecting antigen. While all the CS sera and the CSN serum showed appropriate reactivity, none of the CN sera were reactive (Fig. 2A). Taken together with the IFA results, the findings indicate that CN subjects did not really have antibodies directed to “native” SARS viral antigens, unlike the CS individuals or CSN individual.
FIG. 2.
Western blot results showing specificity of serum antibody activities of representative individuals from community groups (CS, CSN, and CN) against SARS-CoV crude, native antigens or a recombinant GST-fused antigen (rNb). Numbers refer to identification of individuals used throughout the text. A) Results for crude, native antigens separated on 10% gel. The reactivity of a SARS patient's convalescence serum is included for comparison. B) Specific reactivity of two individual CN sera to the viral protein moiety rather than the GST carrier shown in gel where the two components of the fusion protein (rNb-GST) were separated on 14% gel following thrombin digestion. The leftmost panel shows locations of the dye-stained components.
Community CN seropositives truly have antibodies reactive with the bacterial recombinant N antigens, but the target epitopes are different from those of antibodies derived from SARS patients.
We investigated whether the ELISA reactivity seen with the CN sera when using the bacterial recombinant N antigens was real and not an artifact of the assay used. Eight CN individuals with the highest reactivity and serum availability >0.1 ml were selected from the group for further studies. From the WB analysis performed using thrombin-digested rNb in which the N moiety was separated from GST, the results clearly indicate that the serum reactivity was directed to N and not to GST (or to epitopes created artificially at the bridge between these fusion proteins) (Fig. 2B).
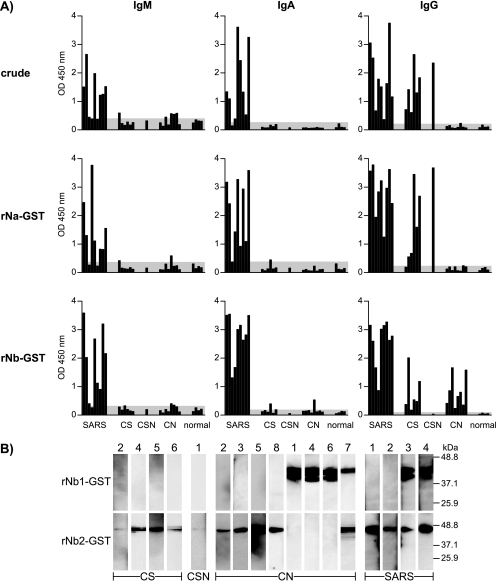
Further ELISA analysis demonstrated that the CN serum reactivity was directed at rNb and not rNa (Fig. 3A). In addition, the reactivity was due to IgG antibodies and not IgM or IgA.
FIG. 3.
More-detailed characterization of antibody activities in representative community group subjects, based on reactivity to various rNb subunits. A) ELISA results for individual subjects examined for type (IgM, IgA, or IgG) and specificity (crude native antigens or recombinant rNa or rNb) of antibodies present. Shown are nine sera obtained from SARS patients during acute-convalescence phase, 6 CS sera, 1 CSN serum, 8 CN sera, and 5 sera from healthy subjects. The shaded area denotes negative reactivity, based on a value of mean plus 2 SD for healthy (non-SARS) individuals. B) WB results for selected individuals showing reactivity of sera for rNb1-GST or rNb2-GST in 14% gel.
Further subdivision of the Nb region into two halves (Nb1 and Nb2) (Fig. 1A) as individual GST fusion proteins revealed that the WB reactivity of the CN sera was directed at rNb1 in 38% (3/8) of the subjects, at rNb2 in 50% (4/8), and at both segments in 12% (1/8) of the cases (Fig. 3B).
In contrast, the anti-N antibodies found in the CS and CSN subjects recognized different N epitopes. First, ELISA revealed the reactivity of these antibodies with both rNa and rNb, although reactivity with the latter was generally weaker than with the former and was absent for rNb in the case of CSN (Fig. 3A). The reactivity was due to IgG antibodies. For comparison, the reactive antibodies found in SARS patients within a month of infection belonged to other classes besides IgG, notably, IgM and IgA (Fig. 3A). IgA antibodies seemed to be particularly abundant for rNb.
Second, in WB analysis, the antibodies from CS subjects reacted only with rNb2, not rNb1 (5/6 cases), or with neither of these (Fig. 3B). A slightly different specificity was seen with the acute-convalescence sera of SARS patients, where reactivity not only to rNb2 was seen but also, in some cases (2/4), to rNb1 (Fig. 3B).
Nb-purified antibodies from CS individuals differ in reactivity from the Nb-specific antibodies of CN individuals.
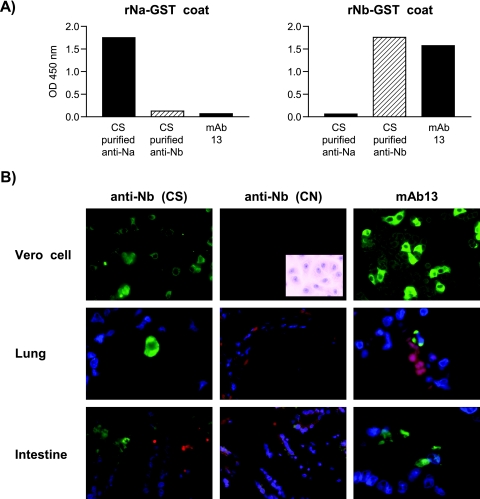
Since the anti-N antibodies found in CS individuals recognized both rNa and rNb, it was possible that the native antigen-binding property of CS sera might belong solely to antibodies specific for rNa and that the anti-rNb antibodies might not be reactive, just like those of CN individuals. We investigated this possibility by isolating the Nb-specific antibodies from CS sera. Firstly, these antibodies displayed exquisite specificity for the rNb fragment (Fig. 4A), whereas the rNa-specific antibodies similarly purified bound only to rNa (Fig. 4A).
FIG. 4.
Specificity of rNa- or rNb-purified antibodies from CS or CN individuals and related mouse MAbs. A) ELISA results showing absence of cross-reactivity between rNa and rNb used as the detecting antigen (coat) probed with rNa- or rNb-purified antibodies or with mAb13. B) IFA results showing reactivity of CS-purified rNb-specific antibodies and mAb13 with SARS CoV-infected Vero cells (×400) and the lung and intestinal tissues (×600) of SARS patients, and the lack of such reactivity by CN-purified rNb-specific antibodies. Inset shows Vero cells stained with hematoxylin. For the human tissues, green denotes reagent antibody fluorescence, blue (or purple) denotes DNA (nuclear) staining, and red (or orange) denotes nonspecific staining of red blood cells or the cytoplasm of other cells.
Strikingly, the Nb-purified antibodies of CS individuals reacted very well with the native viral antigens found in Vero cells (Fig. 4B). In addition, these antibodies also stained the viral antigens present in the lung and intestinal autopsy materials obtained from SARS-infected patients (Fig. 4B). Thus, detached pneumocytes from the lung and surface enterocytes of the intestine were occasionally stained. As expected, Nb-purified antibodies similarly obtained from CN individuals which showed ELISA activity against rNb-GST (data not shown) did not react with either the Vero cells or the human tissues (Fig. 4B).
Mouse monoclonal antibody generated is representative of Nb-specific antibodies found in CS individuals.
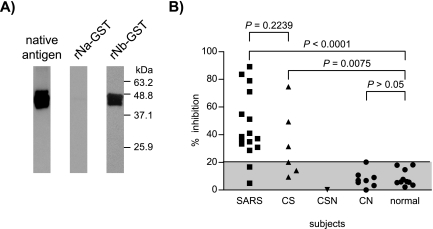
To understand more about the Nb-specific antibodies, monoclonal antibodies were generated from mice immunized with rNb. One hybridoma antibody (mAb13, IgG1) was obtained; this antibody resembled the Nb-specific antibodies of CS individuals (or SARS patients) in the IFA staining reactions on Vero cells and human biopsy samples (Fig. 4B), as well as in ELISA (Fig. 4A) and WB analysis (Fig. 5A) using either the Vero cell-derived native antigens or rNb as the substrate. To find out whether SARS patients would make antibodies to such an epitope, acute-convalescence sera from 16 patients were examined for the ability of these sera to block the binding of mAb13 (biotin labeled) to rNb in an ELISA. Fourteen of these sera (87.5%) were indeed found to be inhibitory (Fig. 5B). In contrast, none of the 8 CN and 10 normal sera were reactive. The CSN serum was also unreactive, but one-half (3/6) of the CS sera were inhibitory. Considering the SARS patients as a group, the reactivity of their serum was similar (P = 0.224) to that of the CS individuals, and both were significantly different from that of healthy people (P > 0.0001 and P = 0.0075, respectively).
FIG. 5.
Specificity of mAb13 and the prevalence of mAb13-like antibodies in human sera. A) WB results showing reactivity of MAb for rNb and the native N antigen but not rNa. B) Inhibition ELISA determining the presence of mAb13-like antibodies in serum of various groups of individuals. Antigen used, rNb-GST; indicator MAb used, biotinylated mAb13. Results indicate % inhibition of buffer control. Shaded area denotes negative reactivity, based on mean plus 2 SD for healthy subjects.
CN sera also fail to react with recombinant N antigens expressed in eukaryotic cells.
We reasoned that the CN sera most probably recognized epitopes that were unique to bacterial proteins and absent from eukaryotic proteins. To investigate this possibility, we transfected 293T human kidney epitheloid cells with Nb cDNA and examined transient transfectants with the CN sera. As expected, there was no reactivity with these sera, whereas cells stained in parallel with the CS sera or with mAb13 showed good cytoplasmic staining (Fig. 6).
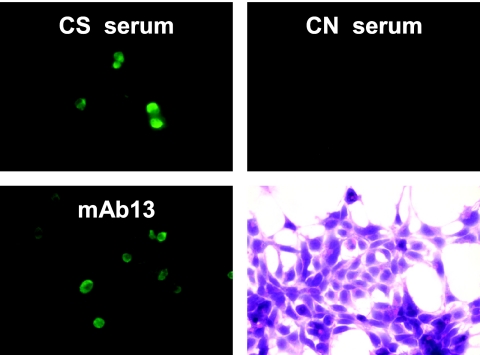
FIG. 6.
Examination of CN sera for reactivity with 293T cells transiently transfected with rNb. IFA results (×600) show reactivity of a CS serum and mAb13 but not the CN serum. Also shown are transfected cells stained with hematoxylin.
DISCUSSION
Based on the presence of IgG antibodies to the bacterially produced SARS-CoV N antigen, 0.44% of the community in Hong Kong were found seropositive for the SARS virus. Two groups of seropositives could be identified based on the fine specificity of the antibodies present: group 1, which resembles SARS patients, and group 2, which does not. In the first group, six of the seven individuals had had SARS previously. The seventh person had no apparent SARS and hence may be regarded as truly asymptomatic. We do not know how this young female got infected. Since it is unlikely that the antibodies found in the 46 group 2 seropositives were elicited by the SARS virus (see below), we estimate the asymptomatic exposure rate of the community to the virus as 1 in 12,000 or 0.008%. This is extremely low. This suggests that SARS-CoV is highly virulent, causing disease in virtually all people it infects and inducing an antibody response in them. This is not surprising in view of the high morbidity and mortality seen with SARS (http://www.who.int/csr/sars) and is also consistent with the behavior of a new emergent infectious agent. A similar conclusion was reached previously when health care workers who attended to SARS patients or their family members were examined for exposure to the virus, although the asymptomatic seropositive rates observed here, except for one study (19), were considerably higher (0.14% to 4.60%) than ours (6, 9, 13, 16, 31). The high rates observed in these studies may be due to the use of whole viral antigens or, as discussed below, the use of bacterially produced N fragments (31). They may also be due to the early sampling times used (within a few months of disease outbreak), although IgG anti-N antibodies have been reported to persist for as long as 16 months (17), longer than our sampling period.
It is somewhat surprising that, using 400 to 938 stored sera of healthy adults obtained 2 to 3 years prior to the SARS outbreak in Hong Kong, Woo et al. (27) and Zheng et al. (34) found considerable cross-reactivity of these sera with SARS-CoV. Thus, 4% and 0.75% seropositivities were observed in the ELISA and WB analysis used, respectively, both utilizing a bacterial full-length N protein (27). Questions have however been raised over these estimates (31). We suspect the bacterial antigen used (see below) may account for some degree of false positivity. The importance of using an accurate detection system was highlighted in a similar study by Yu et al. (32), who found 11 seropositives (0.68%) from 1,621 healthy military recruits in a pre-SARS period using an ELISA which utilized whole SARS-CoV antigens, but none of these people were positive in the viral neutralization test.
The low exposure rate found by us also suggests the lack of antigenic cross-reactivity between SARS-CoV and other viruses found commonly in the Hong Kong population, namely, HCoV-OC43 and HCoV-OC229E. A comparison of the N sequence data among these viruses, which indicates low amino acid homology between SARS-CoV and HCoV-OC43 (32.7%) or HCoV-229E (21.3%), supports this contention. In accordance, in a county in China adjacent to Hong Kong, Che et al. (3) found that, whereas 97% to 99% of the healthy adults there had IgG antibodies to the bacterially derived HCoV-229E and HCoV-OC43 N antigens, only 2% were reactive with the SARS-CoV counterpart. In another study using 13 or 14 pairs of serum samples from American individuals infected with HCoV-OC43 or HCoV-229E, which showed diagnostic rises for these viruses, Ksiazek et al. (11) found no reactivity of these sera with the SARS-CoV whole antigens when examined by Vero cell IFA or ELISA. Contradictory results, however, were observed by Woo et al. (28) in a repeat study using another group of American sera (7 to 21 pairs), where 14.3% cross-reactivity was found with both HCoV-OC43 and HCoV-229E. In other studies which used ELISAs to detect SARS-CoV N antigens, Che et al. (4) and Lau et al. (14) found no cross-detection of the HCoV-229E or HCoV-OC43 N antigen. Direct proof of the uniqueness of the SARS-CoV N antigen was provided by cross-WB analysis using sera of rabbits immunized with the various viral N antigens (3). Cross-reactivity, however, was observed in earlier studies when animal sera made to whole HCoV-229E virus were examined by SARS-CoV-infected Vero cell IFA (11) or when sera raised against group I coronaviruses, such as TGEV, CCoV, and FIPV, were used in WB analysis against the SARS-CoV bacterial N antigen (23).
In the present study, the antibody activities of group 2 (CN) individuals were found to be different from those of group 1 in that these were absent when assayed against the native N antigens expressed in Vero cells (Fig. 4B and 5A), in 293T cells (Fig. 6), or in human tissues (Fig. 4B). The ELISA activities of the group 2 sera were real, however, since the sera clearly reacted with the bacterially produced N antigens in WB analysis (Fig. 3), particularly with the cleaved N antigen moiety (Fig. 2B). These antibodies reacted specifically with rNb, but not rNa (Fig. 3), and with both rNb1 and rNb2 (Fig. 3B). In addition, Nb-specific antibodies could be purified from CN sera which retained ELISA activity against rNb.
In contrast, the antibody activities of group 1 individuals were directed at both rNa and rNb, particularly against rNb2 in the latter. An rNb-specific MAb (mAb13) was found that resembled these antibodies in IFA activity (Fig. 4B), as well as in ELISA and WB analyses (Fig. 4A and 5A). Importantly, such mAb13-like antibodies appeared to be dominantly and frequently produced by SARS patients (Fig. 5B). Altogether, our results suggest that SARS patients normally produce a great heterogeneity of anti-N antibodies.
It thus appears that the group 1 antibodies recognize different epitopes from those of group 2. Based on these differences, we contend that the group 2 (CN) antibodies were not elicited by SARS-CoV or by other coronaviruses. Rather, we argue that these antibodies were induced by some bacterial proteins that fortuitously share antigenic epitopes with the bacterially derived N antigens. Proof of this, and what these bacterial proteins are, will require more in-depth study. Since the antibodies are of the IgG class only (Fig. 3A), they are presumably the result of a distant infection.
Why the group 2 (CN) antibodies failed to react with the native N antigen reflects the subtle differences between bacterial and eukaryotic proteins. One possibility is the loss of fragile epitopes from the eukaryotic protein as a result of host enzymatic activity or during extraction. Although we have previously observed cleavage of the N protein in Vero cells (15), loss of epitopes is not likely since the epitopes involved are not localized at the C terminus of the protein or at the cleavage points. It is also not likely that the group 2 antibodies recognized cryptic epitopes that are absent in the eukaryotic proteins since Western blotting was also used in the investigation. It is more probable, however, that posttranslational modification of the eukaryotic protein—which is absent in the bacterial system—somehow masks the relevant epitope(s). Of the several types of modification known, phosphorylation is a distinct possibility. Thus, the N protein is known to be normally (33) and extensively phosphorylated at multiple residues (24), which involves many kinases that target the serine residue (24). The native antigen is, however, not glycosylated.
The findings reported herein are important for the development of appropriate immunodiagnostics for SARS. The employ of 12,000 subjects in the present study provides a robust evaluation of the specificity of the ELISA used. Even when bacterial N antigens were used as the detecting antigens, when 46 individuals were falsely positive, the specificity of the test was extremely high (99.62%). Specificity can conceivably be enhanced even further if rNb is omitted from the test. Alternatively, the N antigens can be produced in eukaryotic cells and used, but such production is more cumbersome and expensive than production in bacteria. Or, if the bacterial rNb is to be kept, assaying for IgM or IgA antibodies instead of IgG would avoid the cross-reactivity problem. Cases of the CSN type, however, would be missed, but these numbers would be small. An abundance of IgA antibodies appears to be produced by SARS patients early in the disease, particularly to rNb (Fig. 3A). This suggests strong viral stimulation of the mucosal lymphoid tissues during the infection, similar to that seen with poliovirus (8) or Epstein-Barr virus (5).
Acknowledgments
We thank Peggy Fung for secretarial assistance in preparing the manuscript and Po Ki Ma for technical assistance in the ELISA screening.
This work was supported by the Research Grants Council earmarked grant CUHK 4527/03 M.
REFERENCES
- 1.Chan, P. K. S., M. Ip, K. C. Ng, C. W. Rickjason, A. Wu, N. Lee, T. H. Rainer, G. M. Joynt, J. J. Sung, and J. S. Tam. 2003. Severe acute respiratory syndrome-associated coronavirus infection. Emerg. Infect. Dis. 9:1453-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, V. S., K. Y. Chan, Y. Chen, L. L. Poon, A. N. Cheung, B. Zheng, K. H. Chan, W. Mak, H. Y. Ngan, X. Xu, G. Screaton, P. K. Tam, J. M. Austyn, L. C. Chan, S. P. Yip, M. Peiris, U. S. Khoo, and C. L. Lin. 2006. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 38:38-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Che, X. Y., L. W. Qiu, Z. Y. Liao, Y. D. Wang, K. Wen, Y. X. Pan, W. Hao, Y. B. Mei, V. C. Cheng, and K. Y. Yuen. 2005. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 191:2033-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Che, X. Y., L. W. Qiu, Y. X. Pan, K. Wen, W. Hao, L. Y. Zhang, Y. D. Wang, Z. Y. Liao, X. Hua, V. C. Cheng, and K. Y. Yuen. 2004. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 42:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien, Y. C., J. Y. Chen, M. Y. Liu, H. I. Yang, M. M. Hsu, C. J. Chen, and C. S. Yang. 2001. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N. Engl. J. Med. 345:1877-1882. [DOI] [PubMed] [Google Scholar]
- 6.Chow, P. K., E. E. Ooi, H. K. Tan, K. W. Ong, B. K. Sil, M. Teo, T. Ng, and K. C. Soo. 2004. Healthcare worker seroconversion in SARS outbreak. Emerg. Infect. Dis. 10:249-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 8.Herremans, T. M., J. H. Reimerink, A. M. Buisman, T. G. Kimman, and M. P. Koopmans. 1999. Induction of mucosal immunity by inactivated poliovirus vaccine is dependent on previous mucosal contact with live virus. J. Immunol. 162:5011-5018. [PubMed] [Google Scholar]
- 9.Ip, M., P. K. S. Chan, N. Lee, A. Wu, T. K. C. Ng, L. Chan, A. Ng, H. M. Kwan, L. Tsang, I. Chu, J. L. K. Cheung, J. J. Y. Sung, and J. S. Tam. 2004. Seroprevalence of antibody to severe acute respiratory syndrome (SARS)-associated coronavirus among health care workers in SARS and non-SARS medical wards. Clin. Infect. Dis. 38:e116-e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffers, S. A., S. M. Tusell, L. Gillim-Ross, E. M. Hemmila, J. E. Achenbach, G. J. Babcock, W. D. J. Thomas, L. B. Thackray, M. D. Young, R. J. Mason, D. M. Ambrosino, D. E. Wentworth, J. C. Demartini, and K. V. Holmes. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 101:15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 12.Kuba, K., Y. Imai, S. Rao, H. Gao, F. Guo, B. Guan, Y. Huan, P. Yang, Y. Zhang, W. Deng, L. Bao, B. Zhang, G. Liu, Z. Wang, M. Chappell, Y. Liu, D. Zheng, A. Leibbrandt, T. Wada, A. S. Slutsky, D. Liu, C. Qin, C. Jiang, and J. M. Penninger. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, T. S. T., T. K. Ng, W. H. Seto, L. Yam, K. I. Law, and J. Chan. 2005. Low prevalence of subclinical severe acute respiratory syndrome-associated coronavirus infection among hospital healthcare workers in Hong Kong. Scand. J. Infect. Dis. 37:500-503. [DOI] [PubMed] [Google Scholar]
- 14.Lau, S. K., P. C. Woo, B. H. Wong, H. W. Tsoi, G. K. Woo, R. W. Poon, K. H. Chan, W. I. Wei, J. S. Peiris, and K. Y. Yuen. 2004. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in SARS patients by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 42:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung, D. T. M., F. C. H. Tam, C. H. Ma, P. K. S. Chan, J. L. K. Cheung, H. Niu, J. S. L. Tam, and P. L. Lim. 2004. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 190:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung, G. M., P. H. Chung, T. Tsang, W. Lim, S. K. K. Chan, P. Chau, C. A. Donnelly, A. C. Ghani, C. Fraser, S. Riley, N. M. Ferguson, R. M. Anderson, Y. L. Law, T. Mok, T. Ng, A. Fu, P. Y. Leung, J. S. M. Peiris, T. H. Lam, and A. J. Hedley. 2004. SARS-CoV antibody prevalence in all Hong Kong patient contacts. Emerg. Infect. Dis. 10:1653-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, W., A. Fontanet, P. H. Zhang, L. Zhan, Z. T. Xin, L. Baril, F. Tang, H. Lv, and W. C. Cao. 2006. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 193:792-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 19.Nie, Y., G. Wang, X. Shi, H. Zhang, Y. Qiu, Z. He, W. Wang, G. Lian, X. Yin, L. Du, L. Ren, J. Wang, X. He, T. Li, H. Deng, and M. Ding. 2004. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J. Infect. Dis. 190:1119-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, L. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perlman, S., and A. A. Dandekar. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rassmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 23.Sun, Z. F., and X. J. Meng. 2004. Antigenic cross-reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group I animal coronaviruses: implication for SARS diagnosis. J. Clin. Microbiol. 42:2351-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surjit, M., R. Kumar, R. N. Mishra, M. K. Reddy, V. T. Chow, and S. K. Lal. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 79:11476-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tse, G. M., K. F. To, P. K. Chan, A. W. Lo, K. C. Ng, A. Wu, N. Lee, H. C. Wong, S. M. Mak, K. F. Chan, D. S. Hui, J. J. Sung, and H. K. Ng. 2004. Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS). J. Clin. Pathol. 57:260-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Y. D., W. Y. Sin, G. B. Xu, H. H. Yang, T. Y. Wong, X. W. Pang, X. Y. He, H. G. Zhang, J. N. Ng, C. S. Cheng, J. Yu, L. Meng, R. F. Yang, S. T. Lai, Z. H. Guo, Y. Xie, W. F. Chen, and H. H. Yang. 2004. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J. Virol. 78:5612-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo, P. C., S. K. Lau, H. W. Tsoi, K. H. Chan, B. H. Wong, X. Y. Che, V. K. Tam, S. C. Tam, V. C. Cheng, I. F. Hung, S. S. Wong, B. J. Zheng, Y. Guan, and K. Y. Yuen. 2004. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 363:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo, P. C., S. K. Lau, B. H. Wong, K. H. Chan, W. T. Hui, G. S. Kwan, J. S. Peiris, R. B. Couch, and K. Y. Yuen. 2004. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid enzyme-linked immunosorbent assay due to HCoV-OC43 and HCoV-229E rectified by Western blotting with recombinant SARS-CoV spike polypeptide. J. Clin. Microbiol. 42:5885-5888. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wun, H. L., D. T. M. Leung, K. C. Wong, Y. L. Chui, and P. L. Lim. 2001. Molecular mimicry: anti-DNA antibodies may arise inadvertently as a response to antibodies generated to microorganisms. Int. Immunol. 13:1099-1107. [DOI] [PubMed] [Google Scholar]
- 30.Yip, C. W., C. C. Hon, F. Zeng, K. Y. Chow, and F. C. Leung. 2004. Prevalence of non-pneumonic infections with SARS-correlated virus. Lancet 363:1825-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, F., M. Q. Le, S. Inoue, H. T. Thai, F. Hasebe, M. Del Carmen Parquet, and K. Morita. 2005. Evaluation of inapparent nosocomial severe acute respiratory syndrome coronavirus infection in Vietnam by use of highly specific recombinant truncated nucleocapsid protein-based enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 12:848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, S., M. Qiu, Z. Chen, X. Ye, Y. Gao, A. Wei, X. Wang, L. Yang, J. Wang, J. Wen, Y. Song, D. Pei, E. Dai, Z. Guo, C. Cao, J. Wang, and R. Yang. 2005. Retrospective serological investigation of severe acute respiratory syndrome coronavirus antibodies in recruits from mainland China. Clin. Diagn. Lab. Immunol. 12:552-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakhartchouk, A. N., S. Viswanathan, J. B. Mahony, J. Gauldie, and L. A. Babiuk. 2005. Severe acute respiratory syndrome coronavirus nucleocapsid protein expressed by an adenovirus vector is phosphorylated and immunogenic in mice. J. Gen. Virol. 86:211-215. [DOI] [PubMed] [Google Scholar]
- 34.Zheng, B. J., K. H. Wong, J. Zhou, K. L. Wong, B. W. Young, L. W. Lu, and S. S. Lee. 2004. SARS-related virus predating SARS outbreak, Hong Kong. Emerg. Infect. Dis. 10:176-178. [DOI] [PMC free article] [PubMed] [Google Scholar]