Abstract
RNA can adopt different conformations in response to changes in the metabolic status of cells, which can regulate processes such as transcription, translation, and RNA cleavage. We previously proposed that an RNA conformational switch in an untranslated satellite RNA (satC) of Turnip crinkle virus (TCV) regulates initiation of minus-strand synthesis (G. Zhang, J. Zhang, A. T. George, T. Baumstark, and A. E. Simon, RNA 12:147-162, 2006). This model was based on the lack of phylogenetically inferred hairpins or a known pseudoknot in the “preactive” structure assumed by satC transcripts in vitro. We now provide evidence that a second pseudoknot (Ψ2), whose disruption reduces satC accumulation in vivo and enhances transcription by the TCV RNA-dependent RNA polymerase in vitro, stabilizes the preactive satC structure. Alteration of either Ψ2 partner caused nearly identical structural changes, including single-stranded-specific cleavages in the pseudoknot sequences and strong cleavages in a distal element previously proposed to mediate the conformational switch. These results indicate that the preactive structure identified in vitro has biological relevance in vivo and support a requirement for this alternative structure and a conformational switch in high-level accumulation of satC in vivo.
The inherent ability of RNA to switch conformations in response to different physiological conditions has fundamental implications for the regulation of many cellular processes, including transcription termination, protein translation, and RNA cleavage (1, 16). The need for RNA viruses to switch between mutually exclusive processes for genome amplification suggests that RNA switches may also control different steps in the virus life cycle. For example, plus-strand genomes must initially assume a conformation that is recognized by cellular ribosomes for translation of viral products such as the RNA-dependent RNA polymerase (RdRp). At some point, the RNA must switch to a form that is not available for translation but contains cis-acting elements recognized by the RdRp, leading to initiation of minus-strand synthesis (37). Following reiterative synthesis of plus strands from minus-strand templates, newly synthesized plus strands of some viruses may not be templates for further minus-strand synthesis (2, 3), suggesting that these strands need to adopt a structure that is incompatible with RdRp recognition.
RNA conformational switches control RNA dimerization in retroviruses (6, 8) and ribozyme activity in small virus-associated RNAs in vitro (10, 34). Changes in viral RNA conformation in 3′ regions of the genome that hide or expose the 3′ terminus (13, 18, 24) or permit the formation of important cis-acting structures (11) likely regulate initiation of minus-strand synthesis. Evidence for important alternative structures with no known function has also been reported (4, 5). Since RNA switches usually involve long-distance tertiary interactions that stabilize one of the RNA conformations (8, 11, 12), removal of cis-acting elements from their natural context for biochemical and biophysical analyses may lead to an oversimplification of how diverse elements are involved in many viral processes. However, efforts to identify conformational switches and equate RNA structure with biological function in intact viruses are complicated by large genome sizes. In addition, switches that may be replication specific are difficult to identify since both replication and translation occur on the same RNA. These problems can be overcome by analyzing the amplification of small, untranslated subviral RNAs (31). Subviral RNAs, such as satellite RNAs (satRNAs) and defective interfering RNAs, have limited genome sizes while containing all cis elements necessary to utilize the replication components provided by their helper viruses.
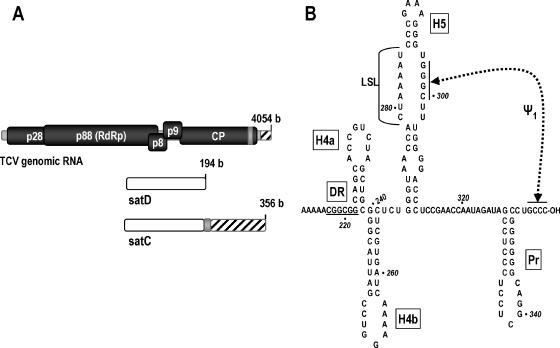
Viruses in the family Tombusviridae, genus Carmovirus, are among the smallest and simplest of the plus-strand RNA viruses. One member, Turnip crinkle virus (TCV), is associated with several small, untranslated subviral RNAs (194 to 356 nucleotides [nt]) (Fig. 1) (30). TCV contains five open reading frames within its 4,054-nt single RNA genome, beginning with p28 and the ribosomal readthrough product p88 (7) (Fig. 1A). While both p28 and p88 are required for viral replication in vivo, p88 produced as a fusion with maltose-binding protein in Escherichia coli can alone transcribe complementary strands of TCV-associated subviral RNAs in vitro (23, 40). TCV subviral RNA satC (356 nt), which originated from recombining a second satRNA, satD (194 nt), with two regions of TCV genomic RNA, contains the 3′-terminal 151 bases of TCV, making it an excellent model for studying 3′-proximal elements required by the TCV RdRp for robust and accurate initiation of minus-strand synthesis.
FIG. 1.
TCV system. (A) Schematic view of TCV genomic RNA and two of its associated satRNAs. p28 and ribosomal readthrough product p88 are the viral-encoded components of the replicase. p8 and p9 are movement proteins, and CP is the coat protein. satC is derived from satD and two regions of TCV. Similar sequences are shaded alike. (B) Structure of the 3′-terminal 145 nt of satC as determined by MPGAfold. This structure contains the four phylogenetically conserved hairpins and is thought to represent the active form of satC. H5 is required for accumulation in vivo. Pr is the satC core promoter. Ψ1 is conserved among carmoviruses and was identified by genetic and biochemical analyses (40). The DR (underlined) was identified as important for satC fitness by in vivo functional selection (35). H4a and H4b had no previously determined function.
By use of a combination of computer modeling and phylogenetic comparisons of carmoviral 3′-untranslated-region sequences, three plus-strand hairpins were determined to be structurally and spatially conserved among nearly all carmoviruses. These are (from 3′ to 5′) Pr, the core promoter for satC (33) and TCV (36); H5, a hairpin with a large internal symmetrical loop (LSL) of highly conserved sequence (42) that forms a pseudoknot with the 3′-terminal 4 nt (40); and H4b, a hairpin of unknown function. In addition, some carmoviruses contain a fourth hairpin of unknown function, H4a, located just upstream of H4b (Fig. 1B).
We recently proposed that a conformational switch affecting most of the 3′-terminal 140 nt of plus-strand satC may be an integral step leading to proper initiation of complementary-strand synthesis in vitro (41). This model was based on the finding that the initial (“preactive”) structure of satC transcripts prepared by in vitro transcription with T7 RNA polymerase does not contain H5, Pr, or the 3′ end/H5 interaction. Deletion of the 3′-terminal three cytidylates or 5′-terminal two guanylates, as well as specific mutations in the H5 region or addition of nontemplate bases to the 3′ end, caused a rearrangement of the Pr from its initial Pr-1 configuration to the alternative, phylogenetically inferred Pr-2 structure (41). This conformational rearrangement to Pr-2 correlated with a substantial (>20-fold) increase in the in vitro synthesis of complementary strands by purified, recombinant TCV RdRp. The Pr-2 form of the promoter was additionally associated with distinctive structural changes in a short element (CGGCGG) termed the derepressor (DR) and the adjacent H4a hairpin (Fig. 1B). Mutations in the DR reduced satC accumulation in vitro and in vivo (41). The proposal was made that a conformational switch requiring the DR region converts the preactive structure to the active structure prior to initiation of minus-strand synthesis in vitro (41). However, in the absence of in vivo confirmatory evidence, the biological relevance of this switch was unknown.
We now provide evidence for a pseudoknot between the loop sequence of H4b and sequence flanking the 3′ side of H5 that stabilizes the preactive structure. Mutations in either of two pseudoknot base pair partners resulted in a reduced accumulation of satC in vivo that was partially restored by compensatory mutations and caused distinctive, nearly identical structural alterations in the preactive structure in vitro that included specific changes in the DR region. These results indicate that the preactive structure identified in vitro has biological relevance in vivo and support a requirement for this alternative structure and a conformational switch in high-level accumulation of satC.
MATERIALS AND METHODS
Construction of satC mutants.
To generate plasmid CΔH4a, oligonucleotides T7C5′ and CΔH4a were used as primers for PCR with template pT7C(+), a plasmid containing wild-type (wt) satC cDNA downstream from a T7 RNA polymerase promoter (all oligonucleotide sequences are presented in Table S1 in the supplemental material). For all constructs, unless noted, digestion with appropriate restriction enzymes produced fragments for insertion into the analogous location in pT7C(+), which was treated with the same restriction enzymes. For H4aR, oligonucleotide CΔH4a was replaced by oligonucleotide H4aR. CΔH4b and H4bR were generated with primers CΔH4b and H4bR, respectively.
To construct CH5CCFVCA (names of these plasmids denote which hairpins were replaced in satC by the cardamine chlorotic fleck virus [CCFV] equivalent hairpins), PCR primers were 5′CCFV+C and 3′CCFV+A, and plasmid CH5CCFV (43) was the template. Following digestion, the fragment was inserted into the analogous location in satCE (43). satCE is similar to pT7C(+), except for the addition of an EcoRV site just downstream from H5 to aid in replacing the satC hairpins. Plasmids CH5CCFVCC, CH5CCFVAC, and CH5CCFVAA were generated in a similar fashion using PCR primers 5′CCFV+C and 3′CCFV+C, 5′CCFV+A and 3′CCFV+C, or 5′CCFV+A and 3′CCFV+A, respectively. For construction of CH5CCFV5′C, PCR was performed with primers 5′CCFV+C and oligo7 and template CH5CCFV. CH5CCFV3′A and H4a were generated using primers T7C5′ and either 3′CCFV+A or CCH4a, respectively, and template satCE. H4a/H5CA was generated in a similar fashion except that the fragment was inserted into the analogous location in H5CA. For H4b, PCR primers were T7C5′ and C5CC4b and the restriction fragment was cloned into satCE. H4a/H4b was generated similarly except that primer C5CC4b was replaced with CCFVH4ab and the template was H4a. To construct H5CA/H4b, PCR primers were T7C5′ and CCFVH4ab5 and the template was H4b, with the fragment inserted into the analogous location in H5CA. H4a/H4b/H5CA was constructed in a similar fashion except that the template was replaced by H4a/H4b. Pr and Pr/H5CA were generated using PCR primers T7C5′ and CCFV-Pr and template satCE or H5CA, respectively. PCR products were cloned into pUC19. Plasmids Pr/H4a, Pr/H4b, Pr/H4a/H4b, Pr/H4a/H5CA, Pr/H4b/H5CA, and Pr/H4a/H4b/H5CA were constructed by digestion of plasmids H4a, H4b, H4a/H4b, H4a/H5CA, H4b/H5CA, and H4a/H4b/H5CA, respectively, with EcoRV and NcoI and insertion of the fragments into the analogous location in Pr/H5CA.
To generate G253C and C313G, PCR primers T7C5′ and G253C or C313G were used with template pT7C(+) and restriction fragments inserted into pT7C(+). For G253C/C313G, satC fragments (SmaI-SpeI and SpeI-SmaI) containing the individual mutations were combined. G252C and G252C/C279A were constructed with PCR primers T7C5′ and G252C and template pT7C(+) and restriction fragments inserted into pT7C(+) or C279A, respectively. C314G and C314G/C279A were constructed using PCR primers T7C5′ and C314G with template pT7C(+) or C279A, respectively. PCR products were cloned into the SmaI site of pUC19. Plasmids G252C/C314G and G252C/C314G/C279A were generated using PCR primers T7C5′ and C314G and template pT7C(+) or C279A, respectively, and fragments cloned into plasmid G252C.
In vivo functional selection.
In vivo functional selection (35, 39) was performed to produce full-length satC RNAs with random sequence replacing the 16-base linker region between H5 and Pr. PCR was carried out using pC(+) as the template and primers T7C5′ and C327S (all oligonucleotide sequences are presented in Table S1 in the supplemental material). Five micrograms of satC transcripts containing randomized sequences was directly synthesized using T7 RNA polymerase and inoculated onto each of 29 turnip seedlings along with 2 μg of TCV genomic RNA transcripts. Total RNA was extracted from uninoculated leaves at 21 days postinoculation. Full-length satC was amplified by reverse transcription-PCR using primers C5′ and oligo7 and then cloned into the SmaI site of pUC19 and sequenced.
Inoculation of protoplasts and analysis of viral RNAs.
TCV genomic RNA and satC transcripts were synthesized from plasmids containing T7 RNA polymerase promoters and linearized with SmaI or directly from PCR products by use of T7 RNA polymerase. Protoplasts (5 × 106) prepared from callus cultures of Arabidopsis thaliana ecotype Col-0 were inoculated with 20 μg of TCV genomic RNA transcripts with or without 2 μg of satC RNA transcripts by using polyethylene glycol-CaCl2, as described previously (14). Total RNA isolated from protoplasts at 40 h postinoculation (hpi) was subjected to RNA gel blot analysis. The RNA was probed with a [γ-32P]ATP-labeled oligonucleotide complementary to positions 3950 to 3970 of TCV genomic RNA and positions 249 to 269 of satC. For analysis of minus-strand accumulation, the RNA was probed with an [α-32P]UTP-labeled riboprobe obtained from pT7C(+) by transcription with T7 RNA polymerase (17).
Preparation of RNA templates.
In vitro transcription assays were performed at 37°C for 2 h in a 60-μl reaction mixture containing 40 mM Tris-HCl (pH 8.0), 10 mM dithiothreitol, 8 mM MgCl2, 25 mM NaCl, 2 mM spermidine, 1 mM each of ATP, CTP, GTP, and UTP, 8 μg of DNA template, and 40 U of T7 RNA polymerase (Promega). RNA used for solution structure probing was treated with 8 U of RNase-free DNase (Promega) for an additional 20 min. Transcripts were phenol-chloroform extracted, ammonium acetate (NH4OAc)-isopropanol precipitated, and then purified through a 6% polyacrylamide gel electrophoresis (PAGE)-8 M urea sequencing-length gel. Full-length RNA was excised from the gel and the RNA removed from the gel slice by shaking overnight in an equal volume of 0.6 M NH4OAc, pH 5.3, 0.1% sodium dodecyl sulfate, and phenol-chloroform (1:1, vol/vol), followed by phenol-chloroform extraction and NH4OAc-ethanol precipitation. RNA was resuspended in H2O. RNA transcripts were quantified with a UV spectrophotometer.
In vitro transcription using recombinant TCV RdRp.
In vitro RdRp assays were carried out using recombinant TCV p88. The p88-expressing plasmid, a kind gift from P. D. Nagy (University of Kentucky), was transformed into E. coli Rosetta pLacI-competent cells (DE3; Novagen). Expression and purification of the recombinant protein were carried out as described previously (23). In vitro RdRp assays were performed in the presence of 10 mM Mg2+, the concentration used for RNA structure probing (32). Briefly, 1 μg of purified RNA template was added to a 25-μl reaction mixture containing 50 mM Tris-HCl (pH 8.2), 100 mM potassium glutamate, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM each of ATP, CTP, and GTP, 0.01 mM UTP, 10 μCi of [α-32P]UTP (Amersham), and 2 μg of recombinant p88. After a 90-min incubation at 20°C, 1 μg of tRNA was added and the mixture subjected to phenol-chloroform extraction and ammonium acetate-isopropanol precipitation. Radiolabeled products were analyzed by denaturing 5% PAGE-8 M urea followed by autoradiography.
RNA solution structure probing.
RNA structure probing of satC and its mutants was performed using protocols and reagents obtained from Ambion. Briefly, gel-purified transcripts were 3′ end labeled in a final volume of 20 μl containing 6 μg of transcripts, 50 μCi of [32P]pCp (Amersham), 20 U of T4 RNA ligase, 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM dithiothreitol, and 1 mM ATP. After overnight incubation at 4°C, reactions were terminated by phenol-chloroform extraction and ethanol precipitation. Labeled transcripts were analyzed by 6% PAGE-8 M urea. Full-length RNAs were excised from the gel and eluted by soaking the slice overnight with constant shaking in a buffer containing 25 mM Tris-HCl (pH 7.5), 400 mM NaCl, and 0.1% sodium dodecyl sulfate. Following phenol extraction and ethanol precipitation, labeled transcripts were resuspended in H2O and added to a mixture containing RNA structure buffer (10 mM Tris-HCl, pH 7.0, 100 mM KCl, 10 mM Mg2+), yeast tRNA, and either H2O or RNase T1 (0.01 or 0.001 U/μl), RNase A (0.01 or 0.001 μg/ml), or RNase V1 (0.001 or 0.0001 U/μl) and incubated at 22°C for 15 min. Samples were precipitated, resuspended in 10 μl of loading buffer, and analyzed by sequencing-length 10% or 20% PAGE-8 M urea. Alkaline hydrolysis ladders were obtained by treatment of 3′-end-labeled transcripts with alkaline hydrolysis buffer (Ambion) at 95°C for 5 min. To obtain RNase T1 ladders, 3′-end-labeled transcripts were heated at 95°C for 5 min in buffer supplied by the manufacturer (Ambion) and then treated with RNase T1 (0.1 or 0.01 U/μl) at 22°C for 15 min.
Computational RNA structure determination.
MPGAfold (massively parallel genetic algorithm for RNA folding) (25, 26, 28, 29) was used to generate the secondary structure predictions for satC and satD. A detailed description of the algorithm is provided in the supplemental material. MPGAfold uses a set of population elements, each representing a stochastically evolving RNA secondary structure that is subject to the standard genetic algorithm operators of mutation, recombination, and selection over multiple generations in a run. To analyze the results from MPGAfold, STRUCTURELAB, a multifeatured RNA/DNA structure analysis workbench (27), and one of its components, Stem Trace (9), were used to depict the results of 20 MPGAfold runs of satC and satD at a population of 16,000.
RESULTS
H4a and H4b comprise a single functional unit that is important for satC accumulation in vivo.
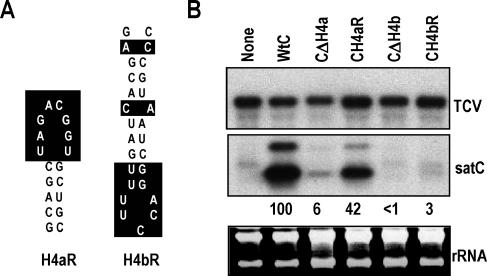
To better understand the structure and function of cis elements necessary for satC accumulation, we examined the phylogenetically conserved hairpins H4a and H4b for roles in satC replication. satC constructs containing deletion of each hairpin or replacement of each hairpin by their reverse complement were generated (Fig. 2A). Deletion of either H4a (CΔH4a) or H4b (CΔH4b) reduced satC levels to 6% and <1% of wt satC levels, respectively (Fig. 2B). satC with the reverse complement of H4a (CH4aR), which did not disrupt the stem but altered each position in the loop (Fig. 2A), accumulated to 42% of wt satC levels. In contrast, satC with the reverse complement of H4b (CH4bR), which contained both sequence and structural alterations, accumulated to only 3% of wt levels. These results indicate that both hairpins are important for satC accumulation and that both stem and loop sequences contribute to the function of H4a.
FIG. 2.
H4a and H4b are important for satC accumulation in protoplasts. (A) Sequences and predicted structures of the reverse complement of H4a (H4aR) and that of H4b (H4bR). Sequences that differ from wt H4a and H4b are boxed. (B) satC either containing precise deletions of H4a (CΔH4a) or H4b (CΔH4b) or containing H4a or H4b in their reverse complement orientations (CH4aR and CH4bR, respectively) was inoculated onto protoplasts with TCV genomic RNA. Total extracted RNAs were examined by RNA gel blot analysis at 40 hpi. The blot was probed with an oligonucleotide specific for both TCV genomic RNA and satC. rRNAs were used as a loading control. Values below the blots represent average satC percentages from three independent experiments. None, no satRNA added to the inoculum; wtC, wild-type satC.
To determine if H4a and H4b interact with each other or with H5 or Pr, satC constructs in which one or more hairpins were replaced with the equivalent hairpins from the carmovirus CCFV were generated. The parental satRNA for these constructs, satCE, contains a 2-base alternation in the linker sequence between H5 and Pr to aid in cloning. A previous replacement of satC H5 with that of CCFV (construct CH5CCFV) resulted in undetectable accumulation in protoplasts, although the 3′ end/H5 LSL interaction remained theoretically possible (43) (Fig. 3B). Before making the single and multiple exchanges of the remaining 3′-proximal hairpins for the current study, we needed to enhance the accumulation of CH5CCFV to detectable levels so that the effect of additional replacement hairpins could be more quantitatively evaluated.
FIG. 3.
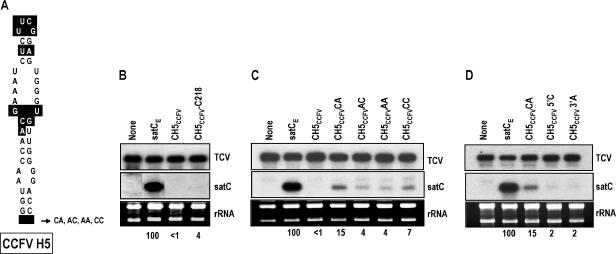
Base insertions and second site alterations improve accumulation of CH5CCFV in protoplasts. (A) Base differences between satC H5 and CCFV H5 are boxed. Location of residues inserted at the base of CCFV H5 in CH5CCFV is indicated. (B) Effect of a second site mutation on accumulation of CH5CCFV. Protoplasts were inoculated with satCE (satC with 2 bases altered downstream of H5 required for inserting CCFV hairpins into satC), CH5CCFV, and CH5CCFVG218C, and levels of accumulating satRNA were determined at 40 hpi. (C) Accumulation of CH5CCFV with inserted cytidylates and adenylates flanking the base of H5 as shown in panel A. (D) Accumulation of CH5CCFV with single inserts upstream or downstream of H5. rRNAs were used as a loading control. Values below the lanes denote the average satC percentages of at least two replicates. None, no satRNA added to the inoculum.
As an initial attempt to determine why satC with H5 of CCFV accumulated so poorly in protoplasts, CH5CCFV was inoculated onto six turnip plants along with TCV genomic RNA, and progeny at 3 weeks postinoculation were cloned and examined for any second site alterations that might have improved fitness. Seventy-two percent of recovered progeny (23 of 32 clones) had an identical transversion at position 218 (G218C), which is located within the DR sequence previously proposed to be important for the conformational switch between preactive and active structures in vitro (41). CH5CCFV containing G218C accumulated to 4% of wt levels (compared with undetectable accumulation of CH5CCFV), indicating that the second site alteration provided at least modest improvement (Fig. 3B). While G218C enhanced CH5CCFV accumulation to detectable levels, we did not want to proceed with hairpin exchanges using constructs containing an alteration in the DR region because of its apparent role in conformational changes in vitro (41).
Since poor accumulation of CH5CCFV might be caused in part by topological constraints due to the CCFV H5 lower stem missing 2 bases (1 bp) compared with satC H5, the 2 bases flanking natural CCFV H5 (C-A) were added, producing construct CH5CCFVCA. As shown in Fig. 3C, CH5CCFVCA accumulated to 15% of satCE levels, indicating that the additional nonpaired residues were beneficial for satC utilization of the CCFV H5. To determine if the added bases were sequence specific, CH5CCFV was also constructed to contain A-C, A-A, and C-C pairs flanking the CCFV H5 (constructs CH5CCFVAC, CH5CCFVAA, and CH5CCFVCC, respectively). These constructs accumulated to detectable levels that were two- to threefold lower than that of CH5CCFVCA (Fig. 3C), suggesting a preference for the natural flanking sequence. Constructs with a single insert of either the 5′ cytidylate or the 3′ adenylate (CH5CCFV5′C and CH5CCFV3′A, respectively) accumulated to levels 7.5-fold lower than that of CH5CCFVCA, demonstrating that inserted bases flanking both sides of the hairpin were important for enhanced accumulation. Based on these results, additional hairpin exchanges that included CCFV H5 also contained the CCFV H5 C-A flanking sequences (H5CA).
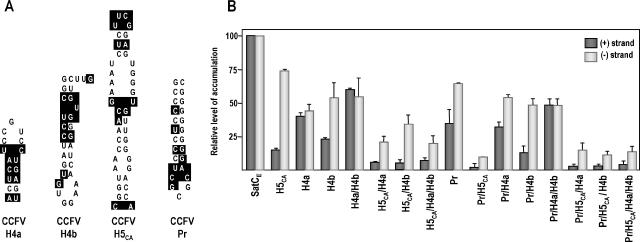
We incorporated CCFV H4a, H4b, H5CA, and Pr in all pairwise combinations into their respective positions in satC and determined the effect of the heterologous hairpins on satC plus- and minus-strand accumulation in protoplasts (Fig. 4). CCFV H4a and H4b reduced satCE accumulation to 40% and 24% of wt satC levels, respectively (Fig. 4B). Replacement of both H4a and H4b with those of CCFV restored accumulation to 61% of satCE levels. Enhanced accumulation when both CCFV hairpins are present suggests that H4a and H4b comprise a single unit and that function of the unit favors both hairpins originating from the same virus. When satC with CCFV H4a, H4b, or H4a/H4b replacements also contained CCFV H5CA or H5CA and Pr, all mutants accumulated to similar low levels (2 to 7% of satCE levels) (Fig. 4B). This indicated that poor satC accumulation due to a heterologous H5 was not improved by the presence of additional CCFV 3′ hairpins.
FIG. 4.
Replacements of satC hairpins with the equivalent hairpins of CCFV. (A) CCFV H4a, H4b, H5CA, and Pr. Sequences that differ from the satC equivalent hairpins are boxed. Hairpins are oriented to be compared more easily with the satC hairpins shown in Fig. 1B. H5CA is CCFV H5 with flanking cytidylate and adenylate residues (see the text). (B) RNA gel blots of total RNA accumulating at 40 hpi were probed with oligonucleotides specific for either the plus (+) or minus (−) strands. Data from two independent experiments were normalized to parental satCE levels, arbitrarily assigned a value of 100. Error bars denote standard deviations. satC plus strands accumulated to levels approximately 100-fold higher than those of minus strands.
satC with Pr of CCFV accumulated to 35% of satCE levels (Fig. 4B), and this level was reduced an additional 17.5-fold when CCFV H5CA was also replaced. However, when both Pr and H4a were replaced, satC accumulated to 33% of satCE levels, indicating that the negative effects of CCFV Pr and H4a on satC accumulation were not additive. In contrast, CCFV H4b reduced the accumulation of satC with the CCFV Pr to 12% of satCE levels. The poor accumulation of satC with CCFV Pr/H4b was enhanced fourfold to 48% of satCE levels when H4a also originated from CCFV.
We previously determined that mutations in H5 reduced accumulation of satC plus strands more than that of minus strands (compared with wt levels of plus and minus strands), despite H5 being a plus-strand element (43). This was suggested to reflect a secondary function of H5 as a scaffold for proper assembly of the RdRp in vivo (15). An assembly role was also suggested for the equivalent hairpin (SL3) in viruses from the genus Tombusvirus (21). Enhanced reduction in plus-strand synthesis due to TCV H5 mutations was attributed to an incorrectly assembled RdRp, which might exhibit differential impairment in transcription of plus and minus strands (43). When minus- and plus-strand levels of the hairpin replacement mutants were compared with levels for satCE, the ratio of minus strands to plus strands was higher than that for satCE for nearly all constructs (Fig. 4B). The ratio was substantially higher for replacements of H5CA (4.9-fold), H5CA/H4b (8.5-fold), Pr/H5CA (4.5-fold), and Pr/H5CA/H4b (5.5-fold). In contrast, replacement of H4a, H4a/H4b, and Pr/H4a/H4b had near-satCE ratios of minus and plus strands. These results suggest that CCFV H4b, in the absence of CCFV H4a, is responsible for an impairment to satC accumulation similar to that previously found for H5.
H4b sequence forms a pseudoknot with sequence flanking the 3′ side of H5.
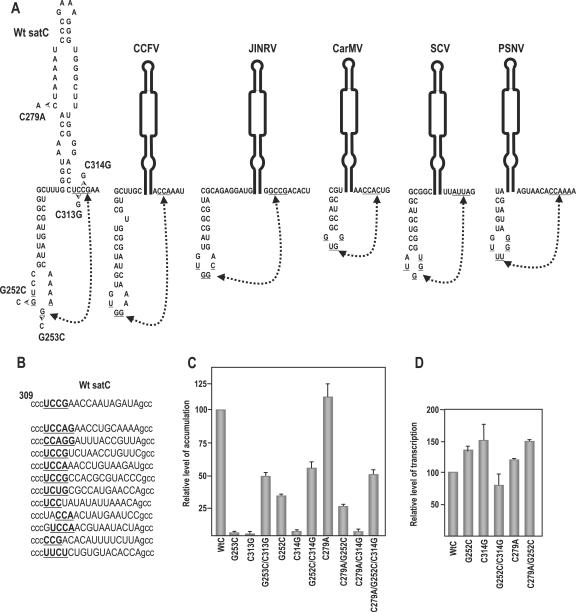
Since alterations to H4b were more detrimental to satC accumulation than similar alterations to H4a, it seemed likely that the H4b portion of the H4a/H4b unit contained an element important for robust RNA replication. Previous results indicated that the conformation of the 3′ region of satC contains substantial tertiary structure based on most guanylates in the region losing susceptibility to the single-stranded-specific RNase T1 in the presence of Mg2+, which stabilizes RNA tertiary structure (38). Six guanylates in the H4a/H4b region had conformations that were altered by Mg2+, including G252 and G253 in the H4b loop (41). Examination of the 3′ region of satC for sequences that might interact with the H4b loop revealed a possible candidate at positions 312 to 315 (UCCG), which flanks the 3′ side of H5 and could possibly pair with H4b positions 251 to 254 (UGGA) (Fig. 5A). Randomization of positions 312 to 327 and selection for satC fitness in six plants (in vivo functional selection) revealed that all satC recovered in the initially inoculated plants contained sequence between positions 312 to 316 that could maintain pairing with at least the UGG in the H4b loop and up to two additional flanking bases (Fig. 5B). Comparative analysis of other carmoviruses also revealed nearly universal conservation of UGG in their H4b loops and the existence of possible pairing partners flanking the 3′ side of their respective H5 (Fig. 5A).
FIG. 5.
Interaction between positions 251 and 254 and positions 312 and 315 is important for satC accumulation in vivo. (A) Phylogenetically inferred structure of the H4b/H5 region. Potential base pairing between underlined residues is denoted by connected arrowheads. Mutations generated in putative base-paired partners are shown. Location of alteration C279A, which stabilizes H5 by pairing the lower positions in the LSL, is shown. Also presented are putative pairings between H4b loop sequences and H5 flanking sequence that can occur in other carmoviruses. JINRV, Japanese iris necrosis virus; CarMV, Carnation mottle virus; SCV, Saguaro cactus virus; PSNV, Pea stem necrosis virus. While the interaction is shown in the active form of the 3′ region of satC for convenience, this does not imply that the interaction exists in this structural configuration. (B) Sequences found in satC accumulating in plants after in vivo functional selection. satC containing randomized sequence in positions 312 to 327 (uppercase) were inoculated onto turnip plants with TCV genomic RNA, and functional satC from six plants was recovered 3 weeks later. The top sequence is wt satC. Underlined, boldface bases maintain possible pairing with H4b loop sequences. (C) Accumulation of satC with various mutations in protoplasts at 40 hpi. Mutations are described above for panel A. Data from at least three independent experiments were normalized to wt satC levels, arbitrarily assigned a value of 100. (D) In vitro transcription of wt satC and mutant transcripts by use of purified recombinant TCV p88. Data are from three independent experiments. Error bars denote standard deviations.
To test for a possible pseudoknot interaction between the H4b UGGA and the UCCG flanking H5, two of the four presumptive paired bases (G252:C314 and G253:C313) were examined by construction of single compensatory mutations (Fig. 5A, left). satC with the guanylate at position 253 converted to a cytidylate (satC-G253C) or the cytidylate at position 313 converted to a guanylate (satC-C313G) accumulated to less than 2% of wt levels in protoplasts. satC with the compensatory exchange (satC-G253C/C313G) accumulated to 50% of wt levels, strongly suggesting that re-established base pairing between the two elements is responsible for partially restoring satC accumulation. The second compensatory exchange also supported the formation of the pseudoknot, with satC-G252C accumulating to 35% of wt levels, satC-C314G acuumulating to 2% of wt satC levels, and satC-G252C/C314G accumulating to 56% of the wt level. The differing levels of satC accumulation caused by mutations in the H4 loop (35% for satC G252C and 2% for G253C) could have reflected the possible formation of a three base-pair interaction between the mutant H4b sequence in satC-G252C (251UCG) and a one-base shifted sequence flanking H5 (314CGA). The pseudoknot between H4b UGGA and the UCCG flnking H5 has been termed Ψ2.
Alterations of individual interacting partners in Ψ2 generate very similar structural changes in the preactive form of satC.
While genetic and phylogenetic evidence supports the existence of Ψ2, this interaction could be topologically incompatible with the long H4b stem and H5 structure. However, previous solution structure probing suggested that the 3′ 140 nt of wt satC adopts an initial “preactive” conformation in vitro that does not appear to contain the phylogenetically conserved hairpin structures (40, 41). If Ψ2 is a feature of the preactive structure, then solution structure analysis should reveal similar or identical structural differences in satC-G252C and satC-C314G, which contain disruptions in one Ψ2 interacting sequence or the other.
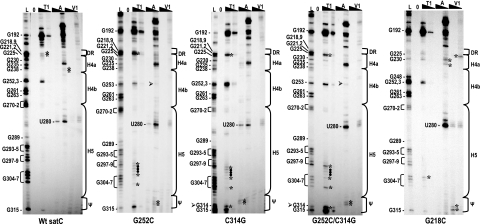
satC-G252G, satC-C314G, and satC-G252C/C314G were radioactively end labeled and subjected to partial digestion with RNase T1, RNase A (specific for single-stranded pyrimidines), or RNase V1 (specific for double-stranded or “stacked” nucleotides). Autoradiographs revealed that satC-G252C and satC-C314G exhibited strikingly similar structural changes from wt satC in three regions that were consistent in multiple independent experiments. These were (i) RNase A cleavages at C313 and C314 in the UCCG element in satC-G252C and RNase A cleavages at U312 and C313 and a new RNase T1 cleavage at the engineered 314 guanylate in satC-C314G (Fig. 6), (ii) three light cleavages in the wt DR region that were replaced by two strong RNase T1 cleavages at G221 and G222 and a weaker cleavage at G218 in satC-G252C and satC-C314G, and (iii) six identical RNase T1 cleavages at positions 299 to 304 and 307 in satC-G252C and satC-C314G. Four of these cleavages were unusual in that the residues are pyrimidines (C300, U301, U302, and U303), which are not normally susceptible to digestion with RNase T1. While it is unclear what these unusual cleavages imply structurally, nearly identical new cleavages in three regions of satC-G252C and satC-C314G transcripts, including new single-stranded-specific cleavages in the UCCG element, suggest that they result from disruption of Ψ2 in the satC preactive structure. In support of this conclusion, G252/G253 were also consistently cleaved more strongly by RNase T1 in satC-C314G than in wt satC, suggesting that these residues have also adopted a more single-stranded conformation (Fig. 6). Curiously, there were no obvious differences in H4b loop cleavages in satC-G252C beyond a new weak RNase A cleavage at the engineered 252 cytidylate. One possible explanation is if the alteration in the H4b loop sequence caused new pairing elsewhere, then the structure might not reflect the loss of pairing with the UCCG element. satC-G252C/C314G transcripts maintained all of the structural differences associated with the individual alterations (Fig. 6), indicating no evidence for re-formation of Ψ2 in the compensatory mutant under the conditions used for the in vitro assay.
FIG. 6.
Disrupting either pseudoknot partner results in nearly identical structural changes in the preactive structure of satC in vitro. Names of the transcripts used for solution structure analysis are given below each autoradiograph. Transcripts were subjected to limited cleavage with two concentrations each of RNase T1 (T1), RNase A (A), or RNase V1 (V1). L, RNase T1 ladder; 0, no added enzymes. High and low concentrations of each enzyme are indicated by the filled triangles above the lanes. Positions of guanylates determined from the T1 ladder reactions are given at left, and the position of U280 is also shown. Locations of sequences within the phylogenetically inferred hairpins and flanking regions are denoted by brackets at right. Ψ denotes positions 312 to 315 (UCCG) involved in Ψ2. Arrowheads denote new guanylates or cytidylates in constructs containing C314G and G252C, respectively. Asterisks denote enzyme-specific cleavages that differ between mutants and wt satC. Filled circles denote nonspecific pyrimidine cleavages by RNase T1 using mutant transcripts.
Ψ2 stabilizes the preactive satC structure.
To determine if Ψ2 stabilizes the satC preactive structure, satC-G252C, satC-C314G, and satC-G252C/C314G were used as templates for in vitro transcription using purified recombinant TCV RdRp (p88). Our reasoning was that if satC-G252C or satC-C314G was a better template for in vitro transcription than wt satC, this would suggest that disrupting Ψ2 reduces the stability of the preactive structure, thus lowering the activation energy between conversion of the two structures and shifting the equilibrium towards the active structure. A more stable active structure, which might be detrimental for cyclic satC replication in vivo, should enhance transcription in vitro, as this assay reports only on initiation of minus-strand synthesis (products are double stranded and not templates for further transcription). As shown in Fig. 5D, transcription of satC-G252C and satC-C314G by p88 generated 37% and 51% more complementary products, respectively, than were produced using wt satC, which is consistent with a conformational shift that favors the active structure.
satC-G252C/C314G had reduced transcriptional activity, with an average of 84% of wt satC levels, which suggested re-formation of Ψ2 had occurred in the compensatory mutant. This is in contrast with the solution structure study, which indicated that satC-G252C/C314G did not detectably reform the wt satC preactive structure. However, if a portion of the transcripts had assumed the wt structure, this would not have been detected. In addition, the conditions used for in vitro transcription differ from the conditions used for structure probing. For example, in vitro transcription includes a lengthy incubation with the RdRp, which may promote reestablishment of Ψ2.
Altogether, these results indicate that disrupting Ψ2 is detrimental to satC accumulation in vivo (Fig. 5C) while increasing transcription of satC in vitro (Fig. 5D). This suggests that a preactive structure stabilized by Ψ2 may be a necessary feature for robust satC accumulation in vivo, which must produce asymmetric levels of plus and minus strands in the proper cellular location. If this hypothesis is correct, then alterations that stabilize the active structure of satC should further reduce mutant accumulation in vivo while enhancing complementary-strand synthesis in vitro. satC with C279A, which closes the lowest position in the H5 LSL and is predicted to slightly stabilize the H5 stem (Fig. 5A), accumulated 10% better than wt satC in vivo, suggesting that the mutation may modestly shift the equilibrium towards the active structure in a nondetrimental fashion (Fig. 5C) (a similar enhancement in accumulation [15%] was also found when satC contains the more stable H5 of TCV [43]). When combined with C279A, satC-G252C, satC-C314G, and satC-G252C/C314G accumulation decreased between 20 and 30%. Transcription of satC-C279A by p88 in vitro generated 20% more complementary strands than wt satC, supporting a stabilized active structure for this mutant (Fig. 5D). When satC-C279A was combined with G252C (satC-C279A/G252C), transcription in vitro was enhanced an additional 30%. These results support a requirement for a preactive structure that is stabilized by Ψ2 for robust accumulation of satC in vivo.
Possible interaction of the DR with Ψ2.
The strong cleavages in the DR region that accompany G252C or C314C alterations (Fig. 6) suggest that the DR may be structurally associated at or near the UGGA/UCCG interacting region in the preactive structure. The DR was first identified as important for satC accumulation by use of in vivo functional selection, which indicated that satC fitness was associated with the recovery of the sequence CGGCGG (35). An association between the DR and H5 was also indicated by the finding that the majority of progeny accumulating in plants inoculated with CH5CCFV had a second site alteration at position 218 (Fig. 3B). While this DR mutation enhanced accumulation of CH5CCFV, G218C reduced accumulation of wt satC by 72% in protoplasts and transcription in vitro to undetectable levels (41), leading to the suggestion that mutations in the DR inhibited the conformational switch.
We therefore wanted to determine if the G218C mutation affected the conformation of the satC preactive structure. Solution structure analysis of satC-G218C indicated a new RNase T1 site at position G304 within the H5 region and several other differences in the DR/H4a region (Fig. 6, right panel). These included a new RNase V1 cleavage at position 222, new RNase A cleavages at positions 228 and 232, and a new RNase T1 site at position 230. In addition, the RNase T1 ladder in the DR/H4a/H4b region was different than that of wt satC. Cleavages at positions G219, G221, G222, G238, and G235 in wt satC transcripts were absent, and there was a much stronger cleavage at G248. Since the ladder is generated using heat-denatured transcripts in the absence of Mg2+, lack of cleavage sites at known guanylates is generally taken as indicating rapid re-formation of local secondary structure, despite fast cooling of the transcripts prior to enzyme digestion. Interestingly, satC-G218C transcripts also contained two new RNase V1 cleavages at C314 and G315 in the Ψ2 UCCG sequence, suggesting that mutating the DR region affects the structure of Ψ2. The G218C mutation had no effect on the structure downstream of position G315, including the Pr region (41).
satD contains elements similar to H5 and H4b.
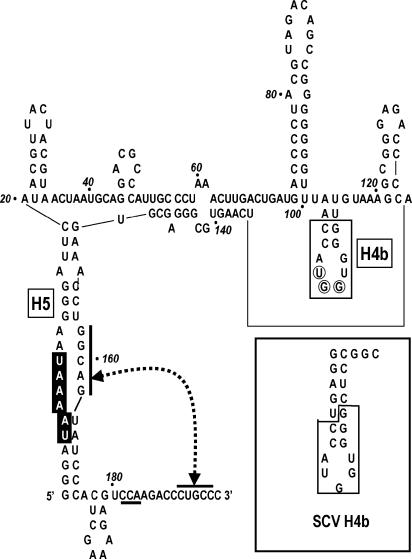
satD (Fig. 1A) shares no contiguous sequence similarity with TCV beyond seven 3′-terminal bases yet accumulates to levels similar to satC levels (30). The advanced RNA secondary structure folding program MPGAfold was used to determine if satD contains elements structurally equivalent to H5 and H4b. MPGAfold uses the concepts of mutation, recombination, and survival of the fittest to evolve a population of thousands of possible RNA structures (25, 26, 28, 29). Two major conformations of satD were present at the end of 20 MPGAfold runs (see Fig. S1 in the supplemental material). One of these structures, depicted in Fig. 7 and Fig. S2 in the supplemental material, contains an H5-like element composed of sequences from the 5′-end and 3′ regions that includes a nearly symmetrical interior loop with sequence similar to that of the satC H5 LSL, including the ability to form a pseudoknot with the 3′ end of satD. Both satD structural solutions had an H4b-like hairpin at positions 102 to 115 with 10 consecutive residues identical to H4b of saguaro cactus carmovirus, including the nearly invariant UGG (Fig. 7 and Fig. S2 in the supplemental material). Formation of Ψ2 is also possible with the CCA at positions 181 to 183 near the 3′ side of H5. The presence of H5- and H4b-like structures in satD is likely an example of convergent evolution that occurred following the initial generation of satD. H4b and H5 therefore appear to be essential for robust replication by the TCV RdRp in vivo.
FIG. 7.
Computer-generated structure for satD contains both H5- and H4b-like elements. Full-length satD was subjected to computational structural prediction using MPGAfold. Putative H5 and H4b hairpins are indicated. Bases boxed in black are identical to the 5′-side LSL of satC H5. Potential interaction between interior loop sequences and the 3′ terminus is indicated by connected arrowheads and overlined sequences. The boxed sequence in satD putative H4b is identical to the Saguaro cactus virus (SCV) H4b sequence (inset). The putative satD H4b loop contains the conserved UGG motif (circled), and pairing of this sequence is possible with sequence proximal to the 3′ side of putative H5 (CCA, underlined).
DISCUSSION
We previously proposed that initiation of satC minus-strand synthesis in vitro requires two mutually exclusive structures, an initial preactive structure that contains extensive tertiary interactions and an active structure that contains the phylogenetically inferred hairpins and a required pseudoknot (Ψ1) between the 3′ end and H5 (41). The existence of the preactive structure was based solely on in vitro solution structure probing, which indicated that satC templates correctly recognized by the TCV RdRp did not appear to contain any of the phylogenetically inferred hairpins. The structure assumed by the transcripts persisted despite a heat/cool treatment, which is routinely used to reduce or eliminate kinetically trapped intermediates (20).
Our current results provide important evidence that the preactive structure is biologically relevant, and thus a conformational switch is likely required to initiate satC minus-strand synthesis in vivo (Fig. 8). This evidence centers on a newly discovered pseudoknot, Ψ2, which forms by pairing positions 251UGGA254 in the loop of H4b with 312UCCG315, which flanks the 3′ side of H5. Evidence for the pseudoknot includes the following: (i) compensatory alterations between positions 252 and 314 and between positions 253 and 313 that reestablish pairing enhance accumulation of satC in vivo compared with satC containing the individual mutations (Fig. 5), (ii) in vivo functional selection of satC with randomized linker sequences produces progeny that all have between 3 and 5 residues at the base of H5 capable of pairing with H4b loop sequence including the phylogenetically conserved UGG (Fig. 5B), and (iii) satC constructs containing mutations at either position 252 or position 314 have identical structural changes in vitro (Fig. 6). In addition, some progeny accumulating in plants inoculated with satC-C314G contain new alterations in either the H5 flanking sequence or the H4b loop sequence, each of which reforms the pseudoknot (R. Guo and A. E. Simon, unpublished). Compensatory analysis of the comparable interaction in TCV also supports the formation of Ψ2 in the viral genomic RNA (J. C. McCormack and A. E. Simon, unpublished).
FIG. 8.
Model for a structural switch in the 3′ region of satC that activates the template for minus-strand synthesis. The complete preactive structure of satC is not yet known. The pairing interaction between UGGA and UCCG described in this report is tentatively described as a pseudoknot, which may change when the complete preactive structure is known. Results from hairpin replacements (Fig. 4) suggest that H4a and H4b may form a single functioning unit in the preactive structure. The DR is proposed to interact in the pseudoknot region, reducing the stability of Ψ2. It is likely that host or viral factors, such as the viral RdRp, are required to promote the conformational switch. See the text for more details.
Results from the in vitro RdRp transcription assays (Fig. 5D) and solution structure probing (Fig. 6) indicate that Ψ2 is present in and stabilizes the preactive structure. This interpretation is consistent with our previous finding that release of the 3′ end, which led to structural changes throughout the 3′ 140 bases in vitro (and is equated with formation of the active structure), resulted in new RNase A cleavage sites in the UCCG element (41). Since disruption of Ψ2 did not lead to the same structural alterations in the DR/H4a, H5, and Pr regions as release of the 3′ end, disruption of Ψ2 is likely necessary, but not sufficient, for conversion to the active structure.
The DR mutation G218C was previously proposed to inhibit the conformational switch to the active structure, based on very poor transcription of satC-G218C by the TCV RdRp in vitro (41). Our current results suggest that a structural relationship between the DR and Ψ2 may exist. Mutations in either Ψ2 partner sequence caused identical strong RNase T1 cleavages in the DR sequence. In addition, satC-G218C transcripts contained new RNase V1 sites in the Ψ2 UCCG sequence. While it is possible that the G218C mutation stabilizes Ψ2 and thus the preactive structure, it is also possible that the structural changes in the DR/H4a/H4b regions caused by the G218C mutation may be the primary basis of conformational switch inhibition, which could explain the reduced template activity of satC-G218C transcripts in in vitro RdRp transcription assays. Either explanation is consistent with a previous finding that satC-G218C together with H5 mutation C279A (which stabilizes H5 and thus the active structure) accumulated twofold better than satC-G218C in vivo (41).
The model shown in Fig. 8 also suggests that the preactive structure contains an interaction between H4a and H4b sequences. This is based on the finding that H4a and H4b are more functional when originating from the same virus source (Fig. 4). The structural similarity of CCFV and satC H4a and the presence of similar loop sequences suggest that the function of H4a is not merely to act as a scaffold for cohelical stacking of H4b. Although the CCFV H4b loop contains the UGGA necessary for formation of Ψ2, poor accumulation of satC with CCFV H4b suggests that maintaining the H4a/H4b interaction may be a necessary requirement for Ψ2 formation.
In addition to a connection with H4b, H4a may have a separate association with the conformational switch of the core promoter from Pr-1 to Pr-2. This is suggested by the finding that accumulation of satC with Pr of CCFV is not further depressed by coreplacement of H4a (Fig. 4B). With 10 consecutive base pairs, 8 of which are GC or CG pairings, the CCFV Pr may be limited to a single conformation, and thus H4a would not be required to support any Pr conformational switch. An association between H4a/H4b and Pr could explain why all satC mutants with the active Pr-2 structure also contain distinctive structural changes in the H4a region compared with wt satC (41).
If satC plus strands transcribed in vivo fold into the preactive structure, an important question is how satC converts to its active form for initiation of minus-strand synthesis in newly infected cells. Depending on the energy barrier between the two structures, the switch could be self-induced (16) or require a trans-acting element. In Alfalfa mosaic virus, coat protein binding to specific 3′-end sequences may compact the genomic RNA into its preactive structure, which would require a conformational switch to an active form consisting of a series of short hairpins (18, 22). Structural rearrangement of the human immunodeficiency virus type 1 leader RNA to a thermodynamically less stable, dimerization-competent form in vitro is mediated by the viral nucleocapsid protein that is thought to stabilize the branched, multihairpin structure (8). However, structure probing of the RNA in infected cells and virus particles did not confirm the dimerization-incompetent structure, suggesting the RNA already exists in dimeric form even while nuclear localized (19). If a trans-acting factor is required to mediate the satC structural switch, the most probable candidate is the viral RdRp, which could also promote an analogous switch in the genomic RNA between translation-competent and replication-competent forms.
In summary, our results suggest that a preactive structure is a necessary component for robust accumulation of a nontranslated viral RNA in vivo. Formation of a preactive structure by newly synthesized satC plus strands could allow satellite progeny to keep their 3′ ends hidden and promoters unavailable to the RdRp. Restricting access of RdRp to progeny plus strands would promote the continued transcription of initially infecting plus strands into minus strands and the continued synthesis of progeny plus strands from the minus-strand intermediates. This process would allow progeny to be “stamped” off of the original parental genome, reducing the number of potentially deleterious mutations (3). Recent evidence for alternative RNA structures in the 5′ and 3′ regions of plant, animal, and bacterial RNA viruses (5, 8, 13, 18, 24) suggests that conformational switches leading to initiation of minus-strand synthesis and possibly restricting RdRp access to de novo-synthesized plus strands may be a contributing factor to the replication of a number of plus-strand RNA viruses.
Supplementary Material
Acknowledgments
Funding was provided by grants from the U.S. Public Health Service (GM61515-01) and the National Science Foundation (MCB-0086952) to A.E.S. This research was also supported in part by a grant from the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, to B.A.S.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Brantl, S. 2004. Bacterial gene regulation: from transcription attenuation to riboswitches and ribozymes. Trends Microbiol. 12:473-475. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao, L., C. U. Rang, and L. E. Wong. 2002. Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage Φ6. J. Virol. 76:3276-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutkiewicz, M., and J. Ciesiolka. 2005. Structural characterization of the highly conserved 98-base sequence at the 3′ end of HCV RNA genome and the complementary sequence located at the 5′ end of the replicative viral strand. Nucleic Acids Res. 33:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greatorex, J. 2004. The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology 1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacker, D. L., I. T. D. Petty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasprzak, W., and B. A. Shapiro. 1999. Stem Trace: an interactive visual tool for comparative RNA structure analysis. Bioinformatics 15:16-31. [DOI] [PubMed] [Google Scholar]
- 10.Ke, A. L., K. H. Zhou, F. Ding, J. H. D. Cate, and J. A. Doudna. 2004. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature 429:201-204. [DOI] [PubMed] [Google Scholar]
- 11.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klovins, J., V. Berzins, and J. Van Duin. 1998. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 4:948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koev, G., S. J. Liu, R. Beckett, and W. A. Miller. 2002. The 3′-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3′ end. Virology 292:114-126. [DOI] [PubMed] [Google Scholar]
- 14.Kong, Q., J. Wang, and A. E. Simon. 1997. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell 9:2051-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack, J. C., and A. E. Simon. 2004. Biased hypermutagenesis associated with mutations in an untranslated hairpin of an RNA virus. J. Virol. 78:7813-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagel, J. H. A., and C. W. A. Pleij. 2002. Self-induced structural switches in RNA. Biochimie 84:913-923. [DOI] [PubMed] [Google Scholar]
- 17.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsthoorn, R. C. L., S. Mertens, F. T. Brederode, and J. F. Bol. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 18:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paillart, J.-C., M. Dettenhofer, X.-F. Yu, C. Ehresmann, B. Ehresmann, and R. Marquet. 2004. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 279:48397-48403. [DOI] [PubMed] [Google Scholar]
- 20.Pan, J., D. Thirumalai, and S. A. Woodson. 1997. Folding of RNA involves parallel pathways. J. Mol. Biol. 273:7-13. [DOI] [PubMed] [Google Scholar]
- 21.Panaviene, Z., T. Panavas, and P. D. Nagy. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrillo, J. E., G. Rocheleau, B. Kelley-Clarke, and L. Gehrke. 2005. Evaluation of the conformational switch model for alfalfa mosaic virus RNA replication. J. Virol. 79:5743-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuppli, D., J. Georgijevic, and H. Weber. 2000. Synergism of mutations in bacteriophage Qβ RNA affecting host factor dependence of Qβ replicase. J. Mol. Biol. 295:149-154. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro, B. A., and J. Navetta. 1994. A massively parallel genetic algorithm for RNA secondary structure prediction. J. Supercomput. 8:195-207. [Google Scholar]
- 26.Shapiro, B. A., and J.-C. Wu. 1996. An annealing mutation operator in the genetic algorithm for RNA folding. Comput. Appl. Biosci. 12:171-180. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro, B. A., and W. Kasprzak. 1996. STRUCTURELAB: a heterogeneous bioinformatics system for RNA structure analysis. J. Mol. Graph. 14:194-205. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro, B. A., J.-C. Wu, D. Bengali, and M. Potts. 2001. a. The massively parallel genetic algorithm for RNA folding: MIMD implementation and population variation. Bioinformatics 17:137-148. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro, B. A., D. Bengali, W. Kasprzak, and J.-C. Wu. 2001b. RNA folding pathway functional intermediates: their prediction and analysis. J. Mol. Biol. 312:27-44. [DOI] [PubMed] [Google Scholar]
- 30.Simon, A. E., and S. H. Howell. 1986. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′-end of the helper virus genome. EMBO J. 5:3423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon, A. E., M. J. Roossinck, and Z. Havelda. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42:415-447. [DOI] [PubMed] [Google Scholar]
- 32.Song, C., and A. E. Simon. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)- and (−)-strands of virus-associated RNAs. Proc. Natl. Acad. Sci. USA 91:8792-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 34.Song, S. I., and W. A. Miller. 2004. cis and trans requirements of rolling circle replication of a satellite RNA. J. Virol. 78:3072-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, X., G. Zhang, and A. E. Simon. 2005. Short internal sequences involved in RNA replication and virion accumulation in a subviral RNA of turnip crinkle virus. J. Virol. 79:512-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, X., and A. E. Simon. 2006. A cis-replication element functions in both orientations to enhance replication of turnip crinkle virus. Virology 352:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dijk, A. A., E. V. Makeyevt, and D. H. Bamford. 2004. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85:1077-1093. [DOI] [PubMed] [Google Scholar]
- 38.Woodson, S. A. 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 9:104-109. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, G., and A. E. Simon. 2003. A multifunctional turnip crinkle virus replication enhancer revealed by in vivo functional SELEX. J. Mol. Biol. 326:35-48. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, G., J. Zhang, and A. E. Simon. 2004. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 78:7619-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, G., J. Zhang, A. T. George, T. Baumstark, and A. E. Simon. 2006. Conformational changes involved in initiation of minus-strand synthesis of a virus-associated RNA. RNA 12:147-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, J., R. Stuntz, and A. E. Simon. 2004. Analysis of a viral replication repressor: sequence requirements for a large symmetrical internal loop. Virology 326:90-102. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, J., and A. E. Simon. 2005. Importance of sequence and structural elements within a viral replication repressor. Virology 333:301-315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.