Abstract
The assembly of the alphavirus nucleocapsid core has been investigated using an in vitro assembly system. The C-terminal two-thirds of capsid protein (CP), residues 81 to 264 in Sindbis virus (SINV), have been previously shown to have all the RNA-CP and CP-CP contacts required for core assembly in vitro. Helix I, which is located in the N-terminal dispensable region of the CP, has been proposed to stabilize the core by forming a coiled coil in the CP dimer formed by the interaction of residues 81 to 264. We examined the ability of heterologous alphavirus CPs to dimerize and form phenotypically mixed core-like particles (CLPs) using an in vitro assembly system. The CPs of SINV and Ross River virus (RRV) do not form phenotypically mixed CLPs, but SINV and Western equine encephalitis virus CPs do form mixed cores. In addition, CP dimers do not form between SINV and RRV in these assembly reactions. In contrast, an N-terminal truncated SINV CP (residues 81 to 264) forms phenotypically mixed CLPs when it is assembled with full-length heterologous CPs, suggesting that the region that controls the mixing is present in the N-terminal 80 residues. Furthermore, this result suggests that the dimeric interaction, which was absent between SINV and RRV CPs, can be restored by the removal of the N-terminal 80 residues of the SINV CP. We mapped the determinant that is responsible for phenotypic mixing onto helix I by using domain swapping experiments. Thus, discrimination of the CP partner in alphavirus core assembly appears to be dependent on helix I sequence compatibility. These results suggest that helix I provides one of the important interactions during nucleocapsid core formation and may play a regulatory role during the early steps of the assembly process.
Alphavirus virions possess an icosahedral nucleocapsid core with T=4 symmetry. These cores are enveloped in a host-derived membrane embedded with viral glycoproteins (E1 and E2), which are also arranged in a T=4 icosahedral lattice (1, 5, 9, 14, 17, 31). The capsid protein (CP) is produced by autocatalytic cleavage from a structural polyprotein (CP-E3-E2-6K-E1). Viral genomic RNA is encapsidated by the CP, resulting in the rapid assembly of nucleocapsid cores in the cytoplasm. In parallel, the E1 and E2 glycoproteins are processed through the endoplasmic reticulum and Golgi and are transported to the plasma membrane. The cytoplasmic nucleocapsid cores interact with the E1-E2 glycoproteins at the plasma membrane, and they bud from the cells as mature virions (7, 10, 18, 20, 24, 30).
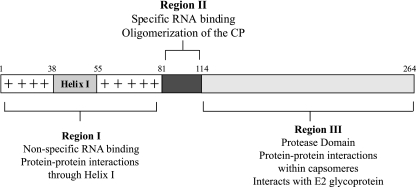
The nucleocapsid core in a mature alphavirus has a highly ordered structure (1, 12, 31). It consists of an RNA genome of ∼12 kb surrounded by 240 copies of a single species of a 30-kDa CP (21). The CP of Sindbis virus (SINV), the alphavirus prototype, is composed of 264 amino acids that can be divided into three regions based on structure and function: regions I, II, and III (Fig. 1). Region III (C-terminal amino acids 114 to 264) adopts a chymotrypsin-like protease fold (2) and forms the capsomeres visible on the outer shell of the nucleocapsid core (1, 31). Region II (amino acids 81 to 113) has been shown to be involved in specific binding to the encapsidation signal on viral genomic RNA (8, 13, 27). Region I (amino acids 1 to 80) contains many basic amino acids that have been implicated in nonspecific binding and charge neutralization with the viral genomic RNA. The highly basic nature of region I is interrupted at residue 38, with a stretch of 18 residues (helix I) that are conserved among alphaviruses and that have leucine heptad repeats (15).
FIG. 1.
Schematic representation of the SINV CP. Residues 1 to 80 (region I) has a high degree of positive charge implicated in nonspecific RNA binding. Helix I is embedded within this positive charge between residues 38 to 55. Residues 81 to 113 (region II) have the minimum sequence required for specific recognition of the 49S viral genomic RNA. The C-terminal protease domain (residues 114 to 264; region III) forms the capsomeres in the nucleocapsid core.
In the initial step of nucleocapsid assembly, viral genomic RNA is preferentially packaged into nucleocapsid cores, and the regions essential for this specific packaging have been mapped both on the genomic RNA and on the CP. In SINV, residues 81 to 113 of CP are involved in this specific recognition (8, 13, 27). The encapsidation signal of the RNA genome has been previously localized to a 132-nucleotide (nt) region spanning nt 945 to 1076 within the SINV nsP1 gene (26, 27). It has been suggested that the tertiary structure of the RNA triggers the binding of two CPs to the encapsidation signal sequence, forming the initiation complex, and that region II of the CP is involved in this binding (8). In addition to this putative CP-RNA interaction, it is important to identify CP-CP interactions that are involved in assembling the RNA and the CP into a nucleocapsid core structure.
An in vitro system using recombinant CP has been established to examine possible steps in the assembly process (22, 23). The structure of an in vitro-assembled core revealed a T=4 icosahedral organization, suggesting that these particles are a reliable representation of the cores produced in the cytoplasm of infected cells (11). The recombinant SINV CP purified from Escherichia coli can form nucleocapsid core-like particles (CLPs) in the presence of single-stranded nucleic acid oligomers, and the requirement for single-stranded nucleic acid is necessary for CLP assembly. To date, no intermediates were found using the in vitro assembly system. However, a chemical cross-linker was used to capture a CP dimer in the in vitro assembly reaction (23). These purified cross-linked dimers of the SINV CP at residues 81 to 264 [CP(81-264)], together with a single-stranded DNA, could be used to reconstitute CLPs (25). Therefore, a nucleic acid-dependent CP dimer has been suggested as a possible building block of core assembly (23, 25).
A recent cryo-electron microscopic (cryo-EM) reconstruction of SINV at 9-Å resolution, together with the fitting of the atomic structure of the CP into this EM density, showed that there are minimal protein-protein contacts between region III (residues 114 to 264) CPs in the core (12). This result suggested that much of the driving force for core assembly results from CP-RNA interactions and CP-CP interactions involving regions I and II of the CP. Perera et al. (15) have shown that CP-CP contacts mediated through region I in the nucleocapsid core are also important during core assembly. By using molecular genetic approaches, the N-terminal helix I region (residues 38 to 55 in SINV) was shown to be important for the accumulation of stable cores in the cytoplasm. Furthermore, it was demonstrated that the helix I region can be functionally replaced by an unrelated sequence from the yeast transcription factor GCN4, which was previously shown to form a dimeric coiled coil (16).
To further examine the process of core assembly using the in vitro core assembly assay, we examined the ability of heterologous alphavirus CPs to dimerize and form CLPs. The CLPs that contain two different CPs are referred to as phenotypically mixed CLPs, as they have a distinct mobility in agarose gels. SINV CP and Western equine encephalitis virus (WEEV) CP can associate in vitro to form phenotypically mixed CLPs, whereas the CPs of SINV and Ross River virus (RRV) do not form phenotypically mixed CLPs but form homotypic CLPs exclusively. We have mapped the N-terminal helix I region as a determinant that controls the formation of phenotypically mixed CLPs by using deletion and domain-swapped SINV CP mutants. The interaction of helix I occurs as an early step in core assembly, and the compatibility of two interacting helix I regions is shown to be an important determinant for promoting the formation of the CP dimer in the assembly process. Based on these results, a role for the helix I region as a checkpoint for CP dimer formation and core assembly is suggested.
MATERIALS AND METHODS
Construction of domain-swapped mutants of the SINV CP and RRV CP. (i) Chimeric CP1.
The nucleotide sequence encoding N-terminal amino acid residues 1 to 89 of RRV CP (nt 7566 to 7832 of pRR64, a full-length cDNA clone of the wild-type T48 strain of RRV [6]) was amplified by PCR using primer pairs that generate an NdeI site 5′ of the start codon for the CP. The nucleotide sequence of amino acid residues 81 to 264 of SINV CP (nt 7887 to 8438 of pToto71, a full-length wild type SINV cDNA clone [15]) was amplified by PCR using primer pairs that generate an HindIII site 3′ of the stop codon for the CP. These two PCR products were used as templates for overlapping PCRs, and the CP coding region of the overlapping PCR product was inserted into the NdeI and HindIII sites of a pET30a vector (Novagen) for expression in E. coli. The cDNA clone was sequenced across the entire CP region.
(ii) Chimeric CP2.
The sequence of the RRV helix I (encoding amino acids 44 to 61), including some flanking sequences (residues 40 to 63, corresponding to nt 7683 to 7754 in pRR64), was amplified by PCR using the primer pairs that give NgoMI and MluI restriction sites at each 5′ and 3′ end, without changing any amino acid sequences. The resulting PCR product was digested with restriction enzymes NgoMI and MluI and inserted into pToto71 by using a three-fragment ligation as previously described (16). The CP coding region was subcloned into NdeI and HindIII sites of a pET30a vector for expression in E. coli. The cDNA clone was sequenced across the entire CP region.
(iii) Chimeric CP3.
Two PCR fragments were amplified: a 100-bp PCR fragment (designated fragment a) included nt 7662 to 7685 of the full-length cDNA of RRV, pRR64, and nt 7749 to 7815 of the full-length cDNA of SINV, pToto71; a 110-bp PCR fragment (fragment b) included nt 7566 to 7685 of pRR64 and nt 7749 to 7763 of pToto71. Both of these PCR fragments were used in overlap PCRs to amplify a 210-bp fragment. Another two PCR fragments were amplified independently: a 100-bp PCR fragment (fragment d) included nt 7755 to 7776 of pRR64 and nt 7758 to 7811 of pToto71; a 600-bp PCR fragment (fragment e) included nt 7755 to 8375 of pRR64 and nt 7803 to 7815 of pToto71. Fragments d and e were used in overlap PCRs to obtain PCR fragment f (700 bp). Fragments f and c were used in overlap PCRs to obtain a 910-bp product that was cloned into the bacterial pET30a expression vector using NdeI and HindIII enzymes.
Expression and purification of the CPs.
Recombinant wild-type SINV CP(19-264) in a pET11a vector was expressed in E. coli strain BL21 DE3-RP and purified as previously described (22). The same conditions were used for the expression and purification of the chimeric CP1, CP2, and CP3. RRV CP and WEEV CP were expressed and purified as previously described (11).
In vitro CLP assembly.
In vitro assembly reactions of different E. coil-expressed CPs and 48-mer oligonucleotide were performed as previously described (22). Briefly, 1:1 molar ratios of purified CPs and 48-mer oligonucleotide were mixed in a final volume of 100 μl in assembly buffer (25 mM HEPES [pH 7.4], 100 mM potassium acetate, 1.7 mM magnesium acetate). Typical reaction mixtures contained 50 μl of 400 μg/ml CP (740 pmol) and 50 μl of 240 μg/ml 48-mer oligonucleotide (740 pmol), and reaction mixtures were incubated for 20 min at room temperature. The products of in vitro assembly reactions were assayed by mobility in a 0.8% agarose gel in Tris-acetate-EDTA buffer. The presence of CLPs was demonstrated by differences in mobility between CLPs and the free 48-mer band after ethidium bromide staining. The presence of CPs in the shifted bands was confirmed by Coomassie staining of the gel.
In vitro assembly of phenotypically mixed CLPs.
The standard in vitro CLP assembly assay was modified to include two different CPs. The two different CPs, at 1:1 molar ratios, were mixed and incubated for 5 min at room temperature, and 48-mer oligonucleotide was added to the reactions, maintaining 1:1 molar ratios of total protein and 48-mer oligonucleotide. Reactions were incubated for 20 min at room temperature. The presence of phenotypically mixed CLPs was determined by the presence of intermediate migrating species between two homotypic CLPs in the agarose gel assay.
Sucrose gradient sedimentation and fractionation of CLPs.
Sucrose gradient sedimentations were performed as described previously (22). Briefly, the in vitro assembly reaction was layered onto 12 ml of a 25% freeze-thaw sucrose gradient prepared with assembly buffer. The samples were centrifuged at 4°C in an SW41 rotor (Beckman) for 100 min at 38,000 rpm. Following sedimentation, gradients were fractionated into 1-ml aliquots for Western blot analysis using polyclonal anti-SINV CP antibody. The polyclonal anti-SINV CP antibody is cross-reactive to RRV CP and WEEV CP.
Cross-linking of CPs on the phenotypically mixed CLPs.
Following the in vitro core assembly reaction, dimethyl suberimidate (DMS; Pierce) was added to a final concentration of 0.75 mM, and the mixture was incubated for 1 h at room temperature. Reactions were terminated by the addition of 200 mM glycine and then a 15-min incubation. The products of cross-linking were assayed by sucrose gradient sedimentation fractionation followed by Western blot analysis.
Negative-stain EM of CLPs.
Following assembly, 3.5 μl of sample was placed on a prewashed 400-mesh copper grid coated with Formvar and carbon. After 2 min of sample absorption and extensive washing with water, 7 μl of a 2% (wt/vol) uranyl acetate stain was applied. After 4 min of staining, grids were wick dried with Whatman no. 1 filter paper and allowed to air dry for 20 min. Samples were then viewed with a Philips EM300 electron microscope at a magnification of ×45,000, and images were captured on Kodak SO-163 EM film.
RESULTS
SINV CP(19-264) and RRV CP cannot form phenotypically mixed CLPs.
Previous experiments of in vitro CLP assembly were performed using full-length and truncated forms of recombinant SINV CP (22, 23, 25). The SINV CP expressed in E. coli is cleaved at its N terminus by an endogenous protease, and the purified protein starts at residue 19 and is designated SINV CP(19-264). It appears that the first 18 amino acids are not necessary for its function; therefore, SINV CP(19-264) has been used as the full-length SINV CP. This well-established in vitro system has been extended to other CPs from different alphaviruses, such as RRV and WEEV (11). Full-length RRV and WEEV CPs with no observed N-terminal cleavage were expressed and purified from E. coli. These CPs can form CLPs under the same conditions as those used for SINV CLP assembly. EM of in vitro-assembled CLPs of RRV and WEEV demonstrated that the overall size and morphology of these CLPs were similar to those of SINV CLPs (11).
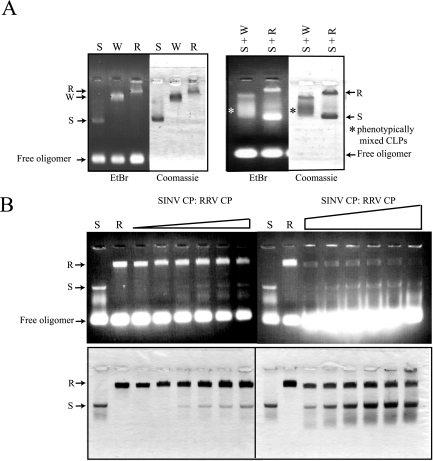
In order to examine the specific nature of interactions involved in CP contacts during the assembly process, 1:1 molar ratios of two CPs from different viruses were mixed and added to the standard assembly oligonucleotide to determine whether assembly intermediates or phenotypically mixed particles could be formed in vitro. In vitro-assembled CLPs of SINV, RRV, and WEEV CPs migrated differentially in an agarose gel assay (Fig. 2A), thereby allowing easy discrimination of CLP assembly during mixed CP experiments. An intermediate species of migration would represent heterogeneous particles containing both species of CP. Consistent with previous observations (22), mixed protein assembly reactions between SINV and RRV CPs failed to generate any detectable intermediate species on an agarose gel. In contrast, when SINV and WEEV CPs were mixed in an assembly reaction, intermediate species indicative of mixed particles were observed (Fig. 2A). CLPs containing a mixture of two CPs were indistinguishable from homotypic CLPs by negative-stain EM (data not shown). The failure to generate phenotypically mixed cores in the reaction containing SINV and RRV CPs was observed at various molar ratios of two proteins (Fig. 2B). Molar ratios of RRV CP and SINV CP of 1:0.1 to 1:10 in the presence of standard oligomer formed only homotypic CLPs. In addition, immunoblotting of the agarose gel with an anti-SINV CP antibody, which is cross-reactive to all three CPs, failed to detect any intermediate species in the mixed reaction of SINV and RRV. Heterotypic CLP formation in the presence of SINV and WEEV CPs occurred across the range of molar ratios assayed (data not shown). We reasoned that there are some important interactions for core assembly that are missing between SINV CP(19-264) and RRV CP but that are present between SINV CP(19-264) and the WEEV CP. An experiment to measure mixing between RRV and WEEV CPs was not included, because the difference in migration between RRV and WEEV CLPs was not sufficient to determine the existence of phenotypically mixed CLPs.
FIG. 2.
Agarose gel assay of in vitro-assembled CLPs and phenotypic mixing between different CPs. (A) In vitro assembly reactions were performed at 1:1 molar ratios of purified CPs and 48-mer single-stranded DNA. Products of the in vitro assembly reactions were run on a 0.8% agarose gel. The gel was stained with ethidium bromide to visualize nucleic acid, and the same gel was dried and stained with Coomassie blue to visualize proteins. S, SINV CP(19-264); W, WEEV CP; R, RRV CP; and EtBr, ethidium bromide. (B) Different molar ratios of SINV CP(19-264) and RRV CP were mixed in the in vitro assembly system. The molar ratio of SINV CP(19-264) to RRV CP ranged from 0.1:1 to 1:1 (left panel) and 1:1 to 1:10 (right panel). The 48-mer DNA was added to the mixed CPs for CLP formation, maintaining 1:1 molar ratios of total protein to 48-mer DNA. Reactions were run on a 0.8% agarose gel, and CLPs were visualized by ethidium bromide staining and Coomassie staining.
SINV CP(81-264) can form an intermediate migrating species with RRV CP in the agarose gel assay.
SINV CP(81-264), which has a deletion of the entire charged N-terminal portion of the CP and the N-terminal helix I region, was able to form CLPs under the same in vitro conditions as full-length CP (Fig. 3). Previously, SINV CP(81-264) CLPs were not detectable by negative-stain EM when a sucrose gradient purification step was incorporated after the assembly reaction (22). With the purification step, SINV CP(81-264) CLPs were able to assemble only when the CP(81-264) dimers were preformed through the use of a chemical cross-link (23). Without this purification step, the SINV CP(81-264) CLPs have a morphology similar to the SINV CLPs, as determined by negative-stain EM (data not shown). Since SINV CP(81-264) can form CLPs, region II-III of CP presumably retains all the molecular interactions required for CP-nucleic acid oligomerization during assembly. The dimerization domain of the CP has been suggested to reside within residues 81 to 264, based on the successful core reconstitution with purified chemically cross-linked dimers of SINV CP(81-264) (25). Therefore, we reasoned that we could map the determinants that prevent phenotypic mixing of cores onto residues found within region II-III of the CP. SINV CP(81-264) was used for a phenotypic mixing experiment with different CPs, and assembly reactions were analyzed by agarose gel assay (Fig. 3). Surprisingly, and in contrast to full-length CP, SINV CP(81-264) can be mixed with RRV CP as well as with SINV CP(19-264) and WEEV CP, generating an intermediate migrating CLP species on an agarose gel. This result suggested that the determinant of phenotypic mixing was not located in region II-III of the CP.
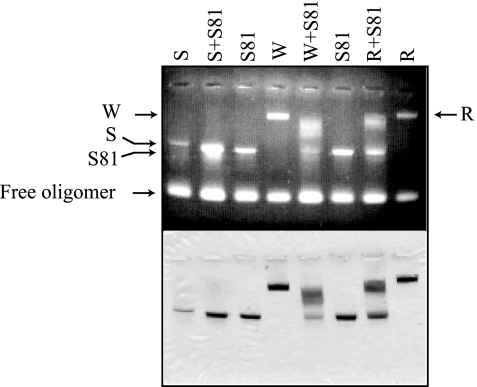
FIG. 3.
Agarose gel assay of phenotypically mixed CLPs between SINV CP(81-264) and other full-length CPs. SINV CP(81-264) was mixed with SINV CP(19-264), WEEV CP, and RRV CP, respectively, in a molar ratio of 1:1:2 (SINV CP(81-264):full-length CP:48-mer DNA). S, SINV CP(19-264); S81, SINV CP(81-264); W, WEEV CP; R, RRV CP. The reactions were run on an 0.8% agarose gel, and the gel was stained with ethidium bromide (top panel) followed by Coomassie blue (bottom panel).
SINV CP(81-264) and RRV CP can form phenotypically mixed CLPs.
To determine whether the observed intermediate species in the mixed assembly reactions were phenotypically mixed CLPs and not CP aggregates, sucrose gradient sedimentation analysis of the mixed assembly reactions was performed. This assay was used to detect whether SINV CP(81-264) was incorporated into different CLPs by looking at the cosedimentation of two CPs in the gradient. In the control reactions, where CPs in the absence of nucleic acid were sedimented through the sucrose gradient, CPs were detected only in the top fraction of the gradient (data not shown). CLPs of SINV CP(81-264), although they are morphologically similar to full-length CP CLPs, showed mobility different from that of full-length CP CLPs (Fig. 4a) and were found predominantly in fractions 3 and 4. CLPs of SINV CP(19-264), RRV, and WEEV CPs sedimented mainly in fractions 6 and 7 (Fig. 4b to d), a result which is consistent with the mobility of cores purified from virus. When in vitro assembly reactions contained a mixture of SINV CP(81-264) and SINV CP(19-264), WEEV, or RRV CP, the CPs migrated together in the gradient predominantly in fractions 4 and 5, indicating that two CPs produced phenotypically mixed CLPs with an intermediate mobility in the gradient (Fig. 4e to g). Migration data for SINV CP(81-264) CLPs, SINV CP(19-264) CLPs, and phenotypically mixed CLPs through the sucrose gradient were different, suggesting that the molecular weights, densities, or shapes of these CLPs could be different. However, negative-stain EM results for each type of CLP and for phenotypically mixed CLPs were indistinguishable (data not shown).
FIG. 4.
Sucrose gradient sedimentation of the phenotypically mixed CLPs between SINV CP(81-264) and full-length CPs. In vitro-assembled CLPs [(a) SINV CP(81-264), (b) SINV CP(19-264), (c) WEEV CP, and (d) RRV CP] and the mixed assembly reactions involving two CPs [(e) SINV CP(81-264) plus SINV CP(19-264), (f) SINV CP(81-264) plus WEEV CP, and (g) SINV CP(81-264) plus RRV CP] were monitored by sucrose gradient sedimentation followed by fractionation and Western blot analysis using polyclonal anti-SINV CP antibody. S, SINV CP(19-264); S81, SINV CP(81-264); W, WEEV CP; R, RRV CP.
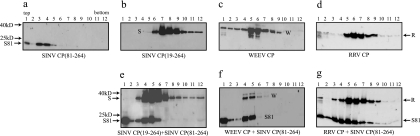
Chemical cross-linking can capture both homodimers and heterodimers from the phenotypically mixed CLPs of SINV CP(81-264) and RRV CP.
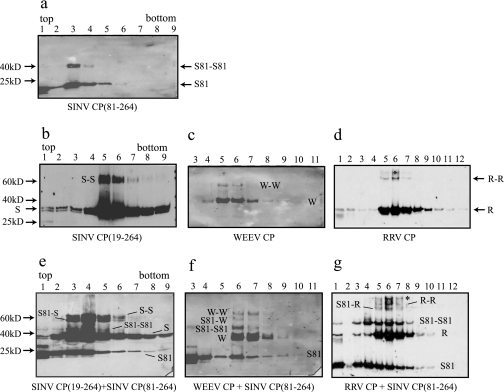
DMS, a lysine-specific chemical cross-linker, was previously used to capture the CP dimer intermediate in SINV in vitro core assembly. This cross-linking occurred between CPs and not between CP and the nucleic acid. The position of this cross-link was assigned to lysine 250 on each of the two SINV CPs (23). This lysine residue is conserved in most alphavirus CPs and, because of this conservation, an analogous dimer can be cross-linked from RRV and WEEV CLPs. In order to determine how SINV CP(81-264) and RRV CP are arranged in the phenotypically mixed CLPs, a chemical cross-linker was added to the phenotypically mixed CLPs after their formation (Fig. 5) in order to capture the CP dimers present in the phenotypically mixed CLPs. In the control reactions, where free CPs in the absence of nucleic acids were cross-linked with DMS and sedimented through sucrose gradients, CPs were detected only in the top fractions, and no cross-linked dimers were generated (data not shown). In the reactions where CLPs of SINV CP(81-264), SINV CP(19-264), WEEV, and RRV were cross-linked with the same concentration of DMS, cross-linked dimers of each CP were detected from the sucrose sedimentation fractions where CLPs were present (Fig. 5a to d). The efficiencies of cross-linking of RRV and WEEV CLPs were lower than those of SINV CLPs. In addition, the cross-linking of RRV CLPs generated higher-order cross-linked species, possibly because the cross-linking conditions were optimized for the detection of dimers from SINV CLPs. DMS cross-linking of phenotypically mixed cores generated three different CP dimers, and these dimers were detected from the sucrose sedimentation fractions where the phenotypically mixed CLPs were present (Fig. 5e to g). In the mixing reaction of SINV CP(19-264) and SINV CP(81-264), fractions 4 and 5, where the SINV CP(19-264) and CP(81-264) cosedimented, contained three different species of cross-linked dimer bands. The molecular weights of these three species corresponded approximately to a homodimer of SINV CP(19-264), a heterodimer of SINV CP(19-264) and SINV CP(81-264), and a homodimer of SINV CP(81-264) (Fig. 5e). Three similar DMS cross-linked dimer bands were captured from assembly reactions containing phenotypically mixed CLPs of SINV CP(81-264) plus WEEV CP (Fig. 5f) and from the phenotypically mixed CLPs of SINV CP(81-264) plus RRV CP (Fig. 5g). These data suggest that there are both homotypic and heterotypic dimeric contacts between CPs in the phenotypically mixed CLPs of SINV CP(81-264) plus RRV CP. The higher-order cross-linked species that was seen in the cross-linking of RRV CLPs appeared in the cross-linking of SINV CP(81-264) plus RRV CLPs (Fig. 5g).
FIG. 5.
Chemical cross-linking of the phenotypically mixed CLPs between SINV CP(81-264) and full-length CPs. In vitro-assembled CLPs [(a) SINV CP(81-264), (b) SINV CP(19-264), (c) WEEV CP, and (d) RRV CP] and the mixed assembly reactions involving two CPs [(e) SINV CP(19-264) plus SINV CP(81-264), (f) WEEV CP plus SINV CP(81-264), and (g) RRV CP plus SINV CP(81-264)] were chemically cross-linked by DMS. The cross-linking reactions were assayed by sucrose gradient sedimentation and Western blot analysis as described in Materials and Methods. The higher-order cross-linked species was designated with an asterisk (d and g). S81-S81, S-S, W-W, and R-R are homodimers of two CP of each; S81-S, S81-W, and S81-R are heterodimers. S, SINV CP(19-264); S81, SINV CP(81-264); W, WEEV CP; R, RRV CP.
Domain-swapped helix I mutants of SINV CP can form phenotypically mixed CLPs.
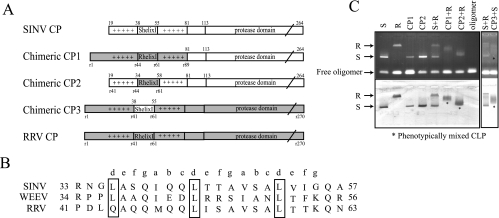
Since SINV CP(19-264) was not able to form phenotypically mixed CLPs with the RRV CP, but SINV CP(81-264) was capable of forming phenotypically mixed CLPs with RRV CP, the N-terminal 80 residues of SINV CP would be predicted to have distinct molecular interactions that permit or prevent heterotypic mixing in core assembly. In order to identify the residues that regulate phenotypic mixing, three chimeric CPs were constructed (Fig. 6A). Based on the amino acid alignment between the SINV and RRV CPs, the N-terminal 80 residues of SINV CP were replaced by residues 1 to 89 of RRV CP, resulting in chimeric CP1. Previous experiments have shown that the helix I had a prominent role in NC assembly and was proposed to stabilize cores through a coiled-coil dimeric protein interaction (15, 16). Therefore, it was of interest to see if the helix I region could influence the outcome of phenotypic mixing. Chimeric CP2 has the RRV helix I (residues 41 to 63, including helix I and two flanking residues at each end) in place of the SINV helix I (residues 35 to 57). The reciprocal construct containing SINV helix I residues 38 to 55 embedded in the RRV CP was also made (CP3). The E. coli-expressed and purified chimeric CPs were tested for their abilities to assemble CLPs using the standard established in vitro assembly conditions (Fig. 6C). The CLPs of CP1 and CP2 migrated in the agarose gel in a manner similar to that of SINV CP(19-264), and negative-stain EM of assembled particles indicated no observable morphological differences between SINV or RRV CLPs (data not shown). Likewise, CLPs generated using CP3 looked indistinguishable from RRV CLPs by EM and migrated with them in the agarose gel assay (data not shown). The phenotypic mixing competence of chimeric CPs was tested by an agarose gel assay (Fig. 6C). Interestingly, both chimeric CPs that contained RRV helix I (CP1 and CP2), when mixed with the RRV CP in the in vitro assembly system, gained the ability to generate intermediate species, as determined by migration, whereas the parental SINV CP was not able to generate a phenotypically mixed core with the RRV CP in the control reaction (Fig. 6C). Similarly, CP3 could form mixed CLPs with SINV CP(19-264) (Fig. 6C). CP3 mixing with the parental RRV CPs was not attempted, because the mobilities of RRV CPs in the agarose assay are identical and the predicted negative result would not be observable. Thus, the failure to form phenotypically mixed CLPs with the SINV and RRV CPs could be due to missing interactions promoted by region I, specifically in the helix I regions. Several single substitutions of residues within helix I of RRV CP were made, and the resulting proteins were assembled in the presence of SINV CP(19-264). In no cases did complete mixing occur (data not shown) and, thus, the requirement of helix I to permit mixed CLPs must require more than one amino acid substitution in the helix I region.
FIG. 6.
In vitro CLP assembly of chimeric SINV CPs and phenotypic mixing of chimeras with RRV CP. (A) Three chimeric SINV CPs were constructed. Chimeric CP1 contains the first 89 residues of the RRV CP replacing SINV CP residues 1 to 80. Chimeric CP2 has helix I of RRV CP replacing the corresponding SINV CP helix I. Chimeric CP3 has helix I of SINV CP replacing the corresponding RRV CP helix I. (B) Amino acid alignment of the helix I regions from three alphaviruses. Letters above the alignment identify amino acid positions in a coiled-coil helix. (C) The three chimeric CPs were assayed for their ability to generate phenotypically mixed CLPs by agarose gel analysis. CP1, chimeric CP1; CP2, chimeric CP2; CP3, chimeric CP3; S, SINV CP(19-264); R, RRV CP.
DISCUSSION
The assembly process to generate an icosahedral core structure from 240 copies of the alphavirus CP and the viral genomic RNA has been investigated in vivo by genetic analyses (3, 4, 15, 16, 18, 19) and in vitro by using an in vitro assembly system (22, 23, 25, 28, 29) The assembly of the nucleocapsid core requires numerous CP-CP and CP-RNA interactions to generate the highly organized icosahedral core structure. An in vitro core assembly system was developed in order to avoid the complexity of in vivo conditions and has identified several requirements but no clear intermediates for core assembly. An indirect approach using a chemical cross-linker has demonstrated that a nucleic acid-dependent CP dimer could function as a building block for core assembly. This dimerization occurs through CP-CP contacts between residues 81 to 264 and, therefore, a nucleic acid-dependent CP dimer-based assembly model has been proposed for alphavirus core assembly (15, 23, 25).
Although this CP dimer has been suggested as an intermediate in core assembly, not much is known about the CP sequences involved in protein dimerization or oligomerization. Purified recombinant SINV CP behaves as a monomer in solution. Since the addition of nucleic acid to the CP induces a rapid assembly of CLPs, the nucleic acid-driven CP dimer has not been observed in solution. However, the use of a chemical cross-linker can capture this dimer, and region II and region III of the CP have been shown to be sufficient for CP dimer formation (residues 81 to 264) in vitro. In a binding experiment using a peptide of region II (residues 81 to 113) and the RNA encapsidation signal sequence found within the viral genomic RNA, two molecules of region II peptide bound to one RNA molecule, suggesting that region II is involved in the initial CP dimer formation (8), although there is no evidence for an interaction between the peptides. A different dimer has been observed in SINV CP crystals. The X-ray crystal structure showed that peptide arm (amino acids 108 to 111) binds to a specific hydrophobic pocket (methinine 137, phenylalanine 166, and tryptophan 247) in an adjacent molecule (7). This CP interaction does not occur in the native virion (1), but mutagenesis studies have shown that amino acids 108 and 111 of the CP are important for some undefined step in core assembly (4, 7). The interaction of residues in the hydrophobic pocket may be involved in a later step of oligomerization in core assembly, since arm region-mutated Semliki Forest virus CP had a defect in core formation in the cytoplasm and formed a CP-RNA complex that sedimented slower than wild-type cores in the cytoplasm (19).
As the in vitro assembly conditions for SINV, RRV, and WEEV were identical in protein and nucleic acid requirements (12) and no assembly intermediate had been isolated, a logical next step was to mix two heterologous CPs in the assembly reaction in order to capture possible assembly intermediates, such as a CP dimer or a higher-order oligomer, or to generate phenotypically mixed CLPs. The rationale is that, although they are similar to one another, these CPs might not assemble complete CLPs because of subtle amino acid differences and, hence, might be blocked at specific steps in assembly. The agarose gel system developed to examine CLP assembly provided a convenient assay for these experiments because of the differences in the migration of cores from different viruses. The mixing of SINV and RRV CPs has previously shown a failure to form mixed CLPs, suggesting that the CP-CP interactions involved in the in vitro assembly of SINV and RRV CLPs are homotypic (22). In the present experiment, SINV and WEEV CPs generated phenotypically mixed CLPs, whereas SINV and RRV CPs did not form mixed CLPs, consistent with the previous report. These results suggest that there are distinct CP-CP interactions in the core that prevent heterotypic mixing between SINV and RRV CPs but allow heterotypic mixing between SINV and WEEV CPs. Since previous results have suggested that region II-III is involved in dimeric interactions, it was reasonable to postulate that the CP-CP dimeric and oligomeric interactions found in region II-III were missing between SINV and RRV CPs and, therefore, only homotypic CLPs could assemble. However, further mapping of the residues that are responsible for the phenotypic mixing showed that the N-terminal helix I, not the residues within region II-III, was responsible for the lack of phenotypic mixing.
In vitro assembly results showed that the N-terminal 80 residues of the CP, which include helix I and many basic amino acids, are not essential for CLP formation. However, in vivo studies have suggested the importance of region I and especially of helix I (3, 15, 16). Complete or partial deletion of helix I, or single-site substitutions at the conserved leucine residues, caused a significant decrease in virus replication and a defect in core formation in the cytoplasm. Replacing helix I with the leucine zipper domain of GCN4, which was previously shown to promote formation of a dimeric coiled coil, produced a virus with a wild-type phenotype, and this chimeric virus accumulated cores in the cytoplasm.
In this study, we have determined that helix I acts as a checkpoint for CP-CP interaction during the initial steps of core assembly. Chimeric CPs containing either RRV helix I or the first 89 residues of the RRV CP restored the ability of these CPs to form phenotypically mixed CLPs with SINV CPs. The reciprocal combination containing helix I of RRV CP replaced with the SINV helix I also permitted the chimeric protein to form phenotypically mixed CLPs with SINV CP. This result indicates that the incompatibility observed between wild-type SINV and RRV CPs during dimer formation and phenotypically mixed CLP assembly was corrected in the chimeras and was defined by the N-terminal sequences of the CP, specifically helix I. It seems that the dimeric interactions mediated through region II-III can be stabilized or destabilized by helix I, depending on its sequence compatibility with its partner CP, providing a checkpoint for CP dimerization. However, it is also possible that the interaction through helix I occurs first in the assembly process and drives or prevents the interaction through region II-III. It remains to be demonstrated which dimeric interaction occurs first during core assembly.
Despite the ability of SINV CP(81-264) to assemble CLPs that are morphologically similar to native cores, the phenotypic mixing experiments have suggested that the interactions mediated by helix I play an important regulatory role in core assembly. The recruitment of monomeric CP together with nucleic acid to form a dimer requires neutralization of the negative charge on the nucleic acid. The basic residues of the CP throughout regions I and II have been implicated in this neutralization process, which allows CP-CP dimerization to occur. At this step, helix I checks its ability to form a proper coiled-coil interaction with its partner CP and, if allowed, stabilizes the CP dimer through a coiled-coil interaction. If the sequences are not compatible, helix I acts to destabilize the CP interaction, and dimers do not form. Further determination of how dimers interact during dimer formation and subsequent oligomerization is necessary to more fully define the mechanism of core assembly.
Acknowledgments
We acknowledge Ranjit Warrier and Chanakha Navaratnarajah for their help with data analysis. We thank Anita Robinson for help with the preparation of the manuscript.
This research was supported by Public Health Service Grant GM56279 from the National Institutes of Health.
REFERENCES
- 1.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H.-K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi, H. K., L. Tong, W. Minor, P. Dumas, U. Boege, M. G. Rossmann, and G. Wengler. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354:37-43. [DOI] [PubMed] [Google Scholar]
- 3.Forsell, K., M. Suomalainen, and H. Garoff. 1995. Structure-function relation of the NH2-terminal domain of Semliki Forest virus capsid protein. J. Virol. 69:1556-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsell, K., G. Griffiths, and H. Garoff. 1996. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 15:6495-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuller, S. D., J. A. Berriman, S. J. Butcher, and B. E. Gowen. 1995. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell 81:715-725. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn, R. J., H. G. Niesters, Z. Hong, and J. H. Strauss. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430-441. [DOI] [PubMed] [Google Scholar]
- 7.Lee. S., K. E. Owen, H. K. Choi, H. Lee, G. Lu, G. Wengler, D. T. Brown, M. G. Rossmann, and R. J. Kuhn. 1996. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4:531-541. [DOI] [PubMed] [Google Scholar]
- 8.Linger, B. R., L. Kunovska, R. J. Kuhn, and B. L. Golden. 2004. Sindbis virus nucleocapsid assembly: RNA folding promotes capsid protein dimerization. RNA 10:128-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 10.Melancon, P., and H. Garoff. 1987. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol. 61:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay, S., P. R. Chipman, E. M. Hong, R. J. Kuhn, and M. G. Rossmann. 2002. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76:11128-11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhopadhyay, S., W. Zhang, S. Gabler, P. R. Chipman, E. G. Strauss, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owen, K. E., and R. J. Kuhn. 1996. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 70:2757-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paredes, A. M., D. T. Brown, R. Rothnagel, W. Chiu, R. J. Schoepp, R. E. Johnston, and B. V. Prasad. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA 90:9095-9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perera, R., K. E. Owen, T. L. Tellinguisen, A. E. Gorbalenya, and R. J. Kuhn. 2001. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 75:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perera, R., C. Navaratnarajah, and R. J. Kuhn. 2003. A heterologous coiled coil can substitute for helix I of Sindbis virus capsid protein. J. Virol. 77:8345-8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skoging, U., M. Vihinen, L. Nilsson, and P. Liljestrom. 1996. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4:519-529. [DOI] [PubMed] [Google Scholar]
- 19.Skoging-Nyberg, U., and P. Liljestrom. 2001. M-X-I motif of Semliki Forest virus capsid protein affects nucleocapsid assembly. J. Virol. 75:4625-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soderlund, H., and I. Ulmanen. 1977. Transient association of Semliki Forest virus capsid protein with ribosomes. J. Virol. 24:907-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. Genetic and biochemical studies on the assembly of an enveloped virus. Genet. Eng. 23:83-112. [DOI] [PubMed] [Google Scholar]
- 25.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 75:2810-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss, B., H. Nitschko, I. Ghattas, R. Wright, and S. Schlesinger. 1989. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J. Virol. 63:5310-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss, B., U. Geigenmuller-Gnirke, and S. Schlesinger. 1994. Interactions between Sindbis virus RNA and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 22:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wengler, G., U. Boege, G. Wengler, H. Bischoff, and K. Wahn. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 118:401-410. [DOI] [PubMed] [Google Scholar]
- 29.Wengler, G., G. Wengler, U. Boege, and K. Wahn. 1984. Establishment and analysis of a system which allows assembly and disassembly of alphavirus core-like particles under physiological conditions in vitro. Virology 132:401-412. [DOI] [PubMed] [Google Scholar]
- 30.Wirth, D. F., F. Katz, B. Small, and H. F. Lodish. 1977. How a single Sindbis virus mRNA directs the synthesis of one soluble protein and two integral membrane glycoproteins. Cell 10:253-263. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, W., S. Mukhopadhayay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]