Abstract
Recombinant adeno-associated viruses (AAVs) are promising vectors in the field of gene therapy. Different AAV serotypes display distinct tissue tropism, believed to be related to the distribution of their receptors on target cells. Of the 11 well-characterized AAV serotypes, heparan sulfate proteoglycan and sialic acid have been suggested to be the attachment receptors for AAV type 2 and types 4 and 5, respectively. In this report, we identify the receptor for the two closely related serotypes, AAV1 and AAV6. First, we demonstrate using coinfection experiments and luciferase reporter analysis that AAV1 and AAV6 compete for similar receptors. Unlike heparin sulfate, enzymatic or genetic removal of sialic acid markedly reduced AAV1 and AAV6 binding and transduction. Further analysis using lectin staining and lectin competition assays identified that AAV1 and AAV6 use either α2,3-linked or α2,6-linked sialic acid when transducing numerous cell types (HepG2, Pro-5, and Cos-7). Treatment of cells with proteinase K but not glycolipid inhibitor reduced AAV1 and AAV6 infection, supporting the hypothesis that the sialic acid that facilitates infection is associated with glycoproteins rather than glycolipids. In addition, we determined by inhibitor (N-benzyl GalNAc)- and cell line-specific (Lec-1) studies that AAV1 and AAV6 require N-linked and not O-linked sialic acid. Furthermore, a resialylation experiment on a deficient Lec-2 cell line confirmed a 2,3 and 2,6 N-linked sialic acid requirement, while studies of mucin with O-linked sialic acid showed no inhibition effect for AAV1 and AAV6 transduction on Cos-7 cells. Finally, using a glycan array binding assay we determined that AAV1 efficiently binds to NeuAcα2-3GalNAcβ1-4GlcNAc, as well as two glycoproteins with α2,3 and α2,6 N-linked sialic acids. Taken together, competition, genetic, inhibitor, enzymatic reconstitution, and glycan array experiments support α2,3 and α2,6 sialic acids that are present on N-linked glycoproteins as primary receptors for efficient AAV1 and AAV6 viral infection.
Adeno-associated viruses (AAVs), dependoviruses of the parvovirus family, rely on a helper virus, such as adenovirus or herpesvirus, to complete their life cycle. In recent years, AAVs have become promising vectors for gene therapy, due to their nonpathogenicity and their ability to mediate long-term transgene expression. Among the 11 AAV serotypes (AAV1 to AAV11) and over 100 variants (12, 24), AAV2 is the best characterized and the predominant serotype used in clinical trials. However, more and more studies suggest that AAV2-based vectors are rate limiting in certain tissues (5, 32). The availability of other AAV serotypes with tissue preference for transduction, such as AAV1 and AAV6 for muscle (2, 5) and AAV8 for liver (12, 14, 25), has overcome this restriction. The enhanced tropism of AAV serotypes in different organs is dictated by their capsid amino acid sequences and is believed in part to be related to the cumulative effects of viral binding to multiple cell surface receptors, cellular uptake, intracellular processing, nuclear delivery of vector genomes, uncoating, and second-strand DNA conversion (for reviews, see references 7 and 9).
The first step of AAV infection often requires the attachment of the capsid to carbohydrate moieties on the cell surface. Heparan sulfate proteoglycan (HSPG) has been suggested as the primary receptor of AAV2 (42). The HSPG interaction domain on AAV2 capsids has been identified as a clustering of basic residues, particularly R585 and R588, contributed by the icosahedral threefold-symmetry-related VP3 monomers, in the valley between the three protrusions surrounding the icosahedral threefold axis (21, 28). The attachment factor for AAV4 and AAV5 was determined to be sialic acid with different linkage forms (2,3 O linked versus 2,3 N linked) (19, 44), although the sialic acid capsid interaction domains have not been identified (29, 43). Besides these serotypes, identification of primary receptors for other serotypes has been lacking. In addition, AAV, like most viruses, requires the presence of coreceptors for productive infection. Integrin αVβ5, human fibroblast growth factor receptor 1, and hepatocyte growth factor receptor have all been suggested as coreceptors for AAV2 (20, 30, 41). The platelet-derived growth factor receptor has been shown to be a coreceptor for AAV5 (10). While receptor and coreceptors for other AAV serotypes remain to be identified, numerous studies have progressed using these reagents in vivo (for reviews, see references 13 and 15).
AAV1 and AAV6 are two closely related AAV serotypes. While AAV6 seems to be a naturally evolved recombinant between AAV1 and AAV2 (34, 46), differing in only six amino acids in the capsid region from AAV1, both AAV1 and AAV6 vectors transduced muscle very efficiently (2, 5). However, when these vectors were tested on other tissues such as liver, AAV6 showed much higher transduction efficiency than AAV1 (16). Whether use by these viruses of identical primary receptors and different secondary receptors explains the tissue preference described above remains unknown. Our recent work showed that use of a broad-spectrum neuraminidase decreased AAV1 transduction to vascular endothelial cells, suggesting that AAV1 may require sialic acid for some step in efficient cell entry (6). More recently, using both broad-spectrum and linkage-specific neuraminidases, Schmidt et al. showed that AAV6 and newly identified type 6-like variants also required a form of sialic acid for productive infection (35). However, Seiler et al. suggested that AAV6 vector transduction was sialic acid dependent or independent, depending on the target cells tested (37).
In the present study, we focused on determining the linkage specificity of sialic acid binding for AAV1 and AAV6 transduction. We used cell-based assays to show that α2,3 and α2,6 sialic acids that are present on N-linked glycoproteins facilitate cellular transduction by both AAV1 and AAV6 vectors. The data from these assays were complemented by a glycan array analysis which determined that AAV1 efficiently binds to NeuAcα2-3GalNAcβ1-4GlcNAc as well as glycoproteins containing NeuAcα2-3/α2-6Galβ1-4GlcNAc N-linked glycans. Together, these results support the requirements for α2,3 and α2,6 sialic acids which are present on N-linked glycoproteins as primary receptors for efficient AAV1 and AAV6 viral infection.
MATERIALS AND METHODS
Cells and plasmids.
The six cell lines utilized in the present study were obtained from the American Type Culture Collection (Manassas, VA) and maintained at 37°C with 5% CO2 in their respective media, supplemented with 10% fetal bovine serum and penicillin-streptomycin. HEK 293 and Cos-7 cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco). HepG2 cells were cultured in Eagle's minimal essential medium (Gibco) supplemented with 1% nonessential amino acids. The parental CHO cell line Pro-5 and Pro-5 mutants Lec-1 and Lec-2 were maintained in α-minimum essential medium (α-MEM; Gibco) supplemented with ribonucleosides and deoxyribonucleosides.
The AAV serotype helper plasmids pXR1, pXR2, pXR4, and pXR5, as well as the plasmid pXX6-80 containing the adenovirus helper genes for AAV replication, have been described elsewhere (32, 47). AAV6 helper plasmid pXR6 was kindly provided by Joseph Rabinowitz (Thomas Jefferson University). The AAV vector plasmid pAAV-CBA-luc, which expresses firefly luciferase, was constructed in our laboratory. The plasmid pTR-lambda was generated as follows. The pSSV9 plasmid, which contains the wild-type AAV2 genome, was digested with XbaI and then incubated with the large Klenow fragment and deoxynucleoside triphosphate to make blunt ends. The ∼4.0-kb DNA fragment containing the two AAV2 inverted terminal repeats was recovered and ligated with an ∼2-kb HindIII fragment of λ phage DNA sequence to obtain the plasmid pTR-lambda.
Production of rAAV.
Recombinant AAV (rAAV) was produced using a previously described protocol (31) with some modifications. Briefly, a 15-cm dish of HEK 293 cells (ca. 2 × 107) was transfected using the Superfect reagent (QIAGEN, Valencia, CA) according to manufacturer's instructions with 6 μg of pAAV-CBA-luc, 12 μg of pXX6-80, and 10 μg of the AAV serotype helper plasmid. The cells were harvested at 48 h posttransfection by scraping, and all subsequent manipulations were performed on ice unless otherwise noted. A cell pellet was obtained by low-speed centrifugation and resuspended in a solution of 3 μg of leupeptin/ml, followed by sonication (Branson Sonifier 250; VWR Scientific) at 25 bursts under 50% duty and an output control setting of 2. Benzonase (Sigma, St. Louis, Mo.) was added to the lysate mixture at a final concentration of 1.2 U/μl, followed by incubation at 37°C for 30 to 60 min. Cesium chloride was added at 0.58 g/ml of lysate, and the mixtures were centrifuged for 4 h at 100,000 rpm in a TLN 100 rotor in an Optima TLX ultracentrifuge (Beckman, Palo Alto, Calif.). Peak gradient fractions (containing rAAV) were identified by dot blot hybridization as previously described using a 32P-labeled firefly luciferase DNA probe (33).
Competition assay for AAV1 and AAV6 vectors.
HepG2 or Pro-5 cells were seeded at a density of 2 × 105 cells/well in 24-well plates 24 h prior to transduction. The cells were coincubated with 2 × 108 AAV1-luc or AAV6-luc vectors in the presence of a 200-fold-excess amount of competing AAV1-, AAV6-, or AAV2-encapsidated λ phage DNA-containing vectors at 37°C for 1 h. The viruses were removed by washing the cells three times with medium, and luciferase expression was monitored at 24 h posttransduction on a VICTOR2 luminometer (Perkin-Elmer, Wellesley, MA) using a firefly luciferase assay kit (Promega).
Binding assays.
Cells from 15-cm-diameter plates were scraped and divided into aliquots of 1 × 105 cells per microcentrifuge tube. The cells were treated with 50 mU/ml neuraminidase type III from Vibrio cholerae (Sigma, St. Louis, Mo.) at 37°C for 2 h or were untreated and then chilled at 4°C for 15 min. Next, rAAV was added at 2 × 109 genome-containing particles per aliquot and incubated with cells at 4°C for 90 min to allow binding. Cells were then rinsed three times with medium, and cell-associated AAV DNA was extracted using a DNeasy kit from QIAGEN and detected by hybridization with a 32P-labeled luciferase DNA probe as described previously (33). Detection and quantification were performed using a PhosphorImager (Molecular Dynamics).
Lectin staining and competition.
Lectin staining of HepG2 cells, Pro-5 cells, and Cos-7 cells was performed by incubation with fluorescein isothiocyanate (FITC)-labeled wheat germ agglutinin (WGA), Maackia amurensis lectin (MAA), or Sambucus nigra lectin (SNA) (Vector Laboratories Inc.), as described in reference 44. Briefly, cells growing in 24-well plates (approximately 2 × 105 cells/well) were chilled to 4°C, and then lectin (10 μg/ml) was added to the respective media. Cultures were then incubated at 4°C for 20 min, rinsed three times with medium, and then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). Binding of lectin to cells was detected using fluorescence microscopy. Lectin competition experiments were done by preincubating cells with 100 μg/ml of either WGA, MAA, or SNA (Vector Laboratories Inc.) in medium at 4°C for 10 min. The preincubation solution was removed, and medium containing 100 μg/ml lectin and 2 × 108 rAAV particles was added. Cultures were maintained at 4°C for 1 h and then rinsed three times with medium. The cells were grown at 37°C for an additional 24 h in serum-containing media and then assayed for luciferase expression.
Cell surface modifications.
Sialic acid was biochemically removed from the surfaces of Pro-5 cells, HepG2 cells, and Cos-7 cells by neuraminidase treatment. Briefly, cells were rinsed with medium and then incubated with nonserum medium containing 50 mU/ml neuraminidase type III from Vibrio cholerae for 2 h at 37°C. The cells were then washed three times with medium prior to binding or transduction experiments.
Sialic acid was restored to the surfaces of Lec-2 cells in defined linkages using purified sialyltransferases as described previously (19). Briefly, Lec-2 cells were incubated with either α2,3(O)-sialyltransferase, α2,3(N)-sialyltransferase, or α2,6(N)-sialyltransferase (Calbiochem) in the presence of 1 mM CMP-sialic acid (Calbiochem). Resialylation was carried out with 50 mU/ml sialyltransferase in α-MEM for 2 h at 37°C. In the control groups, Lec-2 cells were untreated or incubated with either sialyltransferase or CMP-sialic acid. The cells were rinsed three times with α-MEM prior to transduction.
Proteinase K treatment.
Proteinase K treatment of cells was carried out as previously described (26) with some modifications. Cos-7 and Pro-5 cells were cultured in 15-cm-diameter culture dishes until cells were 80% confluent. Cells were washed twice with PBS, scraped, resuspended in 10 ml PBS, and divided into two aliquots (∼4 × 106 cells). Aliquots were treated with 200 μg/ml proteinase K or mock treated for 1 h at 37°C. An equal volume of PBS containing 6% fetal bovine serum, 2 mM phenylmethylsulfonyl fluoride, and 2× concentrated protease inhibitor cocktail (Sigma) was added to inactivate proteinase K. After a 10-min incubation at room temperature, the cells were washed twice with medium and seeded at 5 × 104 cells/well in a 48-well plate. After incubation for 1 h at 37°C, cells were transduced at a multiplicity of infection (MOI) of 2 × 103 for 1 h with virus, and luciferase expression was analyzed 24 h after transduction.
Inhibition of glycolipid synthesis.
Inhibition of glycolipid synthesis was carried out as described previously (36) with some modifications. Briefly, Cos-7 cells were plated at a density of 2 × 104 cells/well in a 48-well plate. Eight hours after seeding, cells were incubated for 40 h with the glucosylceramide synthase inhibitor dl-threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP) (Sigma, St. Louis, MO). Cells were then washed with medium and transduced with 1 × 108 particles of rAAV for 1 h. Luciferase expression was analyzed 24 h after transduction.
Glycosylation inhibitor studies.
Glycosylation inhibitor studies were performed using a previously described method (19) with some modifications. Briefly, Cos-7 cells were plated at 20% confluence in 48-well plates 24 h before the addition of inhibitors tunicamycin (Oxford) or N-benzyl-GalNAc (Calbiochem). The cells were then cultured with the aforementioned inhibitors for 24 h and transduced with rAAV. After a 1-h infection, the medium was removed and the cells were washed and incubated for 24 h before a luciferase assay.
Mucin competition.
Mucin competition was carried out by pretreating rAAV with bovine submandibular gland mucin (Sigma) for 30 min at 20°C. AAV vectors, in the presence or absence of mucin, were added to Cos-7 cells in equal volumes of DMEM for 1 h at 37°C. Cells were rinsed twice with DMEM and incubated at 37°C. Two thousand particles/cell were pretreated with mucin (concentrations up to 0.5 mg/ml), and cells were assayed for luciferase expression 24 h posttransduction.
Screening for the glycan binding specificity of AAV1 capsids on a glycan array.
Empty AAV1 capsids were produced using a baculovirus system expressing the VP proteins and purified using a sucrose cushion followed by a 5 to 40% (wt/vol) sucrose gradient as previously described for AAV5 (8). Using a high-throughput glycan array screening assay developed by cores D and H of the Consortium for Functional Glycomics (CFG; an NIH National Institute of General Medical Sciences initiative) for identifying specific carbohydrate binding partners for proteins, we screened the glycan binding capabilities of AAV1 capsids (3). The printed array (Printed Array [PA] V2, http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh5.shtml) contained 264 different natural and synthetic glycans (including sialylated sugars with different linkages and modifications, for example, sulfation, but not heparin sulfate). The method used for generating the printed array is detailed in a publication by Blixt et al. (3). Briefly, the array is created using a robotic printing technology that uses amine coupling to covalently link amine-functionalized glycans or glycanconjugates to amine-reactive N-hydroxysuccinimide-activated glass slides. The slides contain six addresses per glycan or glycoconjugate. A printed slide was incubated with AAV1 capsids (at 200 μg/ml), and then a capsid monoclonal antibody (generated in collaboration with Colin Parrish) was overlaid on the bound capsids followed by a FITC-labeled secondary antibody (at 5 μg/ml). The fluorescence intensity was detected using a ScanArray 5000 (Perkin-Elmer Inc.) confocal scanner. The image was analyzed using the IMAGENE image analysis software (BioDiscovery, El Segundo, CA). The data were plotted using the Microsoft EXCEL software.
RESULTS
AAV1 and AAV6 compete for transduction.
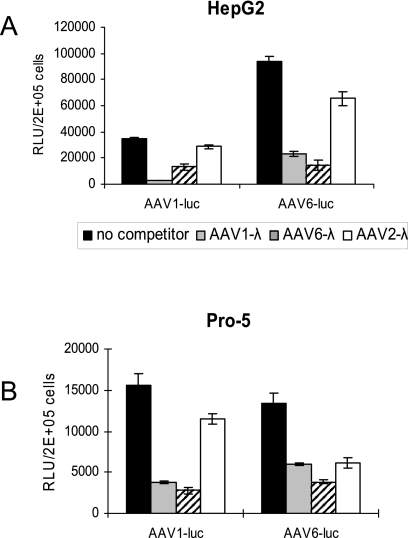
AAV1 and AAV6 are two closely related viruses, with only six amino acid differences in their capsid regions. Previous studies have shown that AAV1 and AAV6 are almost identical serologically (14, 16). To test if these two viruses use the same receptor(s) for transduction, we carried out a competition assay on HepG2 and Pro-5 cells. While the HepG2 cell line is commonly used in many laboratories as a representative cell of liver origin, Pro-5 cell line is a derivative of the original CHO cell line and is the parental line for several glycosylation mutants. The cells were transduced with a constant amount of either AAV1 or AAV6 vector expressing luciferase in the presence of a 200-fold excess of AAV1, AAV6, or AAV2 as competitor. To avoid compounding effects such as the competition for transcription factors between vector and competitor, a lambda phage DNA sequence instead of an expression cassette was packaged into AAV1, -2, and -6 capsids when used as competitors. As shown in Fig. 1, in both HepG2 and Pro-5 cells, rAAV1 vector transduction was inhibited not only by the AAV1 competitor but also by the AAV6 competitor (Fig. 1). Similarly, rAAV6 vector transduction was inhibited by both AAV6 and AAV1 competitors. These results suggest that AAV1 and AAV6 may share a common receptor for transduction. As a negative control, we show that the AAV2 competitor, which uses heparan sulfate proteoglycan as a receptor, does not markedly inhibit AAV1 and AAV6 transduction on HepG2 cells and AAV1 transduction on Pro-5 cells. However, the AAV2 competitor was able to inhibit 50% of Pro-5 cell transduction by rAAV6 (Fig. 1B). Although the exact reason for this partial inhibition of AAV6 on Pro-5 cells with the AAV2 competitor is unknown, it is interesting that AAV6 binds heparin (17), and it is possible that the binding of the AAV2 competitor with heparan sulfate proteoglycan interferes with the binding of the AAV6 vector with its receptor on Pro-5 cells. It is also possible that AAV2 and AAV6 might share some common receptors or coreceptors on Pro-5 cells.
FIG. 1.
Transduction of cells by luciferase-expressing AAV1 or AAV6 vectors in the presence of competing λ phage DNA-containing AAV1, AAV6, or AAV2 vectors. HepG2 (A) and Pro-5 (B) cells were transduced with a constant amount of AAV1-luc or AAV6-luc vectors in the presence of a 200-fold-excess amount of competing AAV1, AAV6, or AAV2 encapsidated λ phage DNA-containing vector at 37°C for 1 h. The viruses were removed, and the cells were washed three times with medium. The cells were then grown in serum-containing medium. Luciferase expression was tested 24 h posttransduction. RLU, relative light units.
Sialic acid is required for efficient AAV1 and -6 binding and transduction.
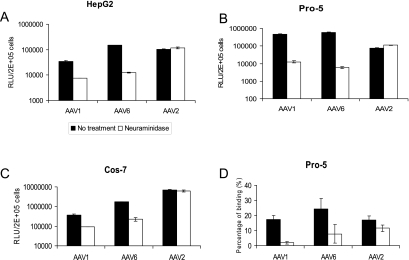
Heparan sulfate proteoglycan is a primary receptor for AAV2 infection (42). It is not a receptor for AAV1, and AAV6 transduction is not blocked by heparin sulfate, despite the fact that the latter capsid binds to it (17, 31). Sialic acid is another abundant carbohydrate moiety on the cell surface, which is required for both AAV4 and AAV5 binding and transduction (19, 44). We observed that treatment of HepG2, Pro-5, or Cos-7 with the neuraminidase isolated from V. cholerae, which has a broad specificity and cleaves all α2,3, α2,6, and α2,8 sialic acids, did not reduce AAV2 transduction (Fig. 2A to C). In contrast, neuraminidase treatment markedly decreased gene transduction by either AAV1 or AAV6 vectors on all the cells tested (Fig. 2A to C). These data suggest that sialic acid present on the cell surface is required for efficient AAV1 and AAV6 transduction.
FIG. 2.
Neuraminadase treatment of cells reduces AAV1 and AAV6 binding and transduction. (A, B, and C) Transduction assay. Cell surface sialic acid was removed from HepG2 (A), Pro-5 (B), and Cos-7 (C) cells by neuraminadase from Vibrio cholerae prior to transduction with AAV1-luc, AAV6-luc, and AAV2-luc. Twenty-four hours after transduction, cells were analyzed for luciferase expression with a luminometer. RLU, relative light units. (D) Cell binding assay. Pro-5 cells were treated with neuraminidase from Vibrio cholerae or mock treated prior to adding AAV1-luc, AAV6-luc, and AAV2-luc at 4°C. Virus binding was determined by quantitative dot blot hybridization. Values are means from three experiments. Error bars represent standard deviations.
To test if the decreased transduction of AAV1 and AAV6 on neuraminidase-treated cells is related to reduced virus binding, a binding assay based on dot blot hybridization was performed (Fig. 2D). AAV2 binding to Pro-5 cells was not significantly reduced after neuraminidase treatment. In contrast, 89% and 68% inhibition of AAV1 and AAV6 binding, respectively, was observed after neuraminidase treatment. AAV6 binding appears to be less affected by neuraminidase compared with AAV1 binding, suggesting that AAV6 may also bind to moieties other than sialic acid on the cell surface. However, binding to sialic acid seems to be the major determinant of AAV6 transduction, since about 98% inhibition of transduction was observed following neuraminidase treatment (Fig. 2B).
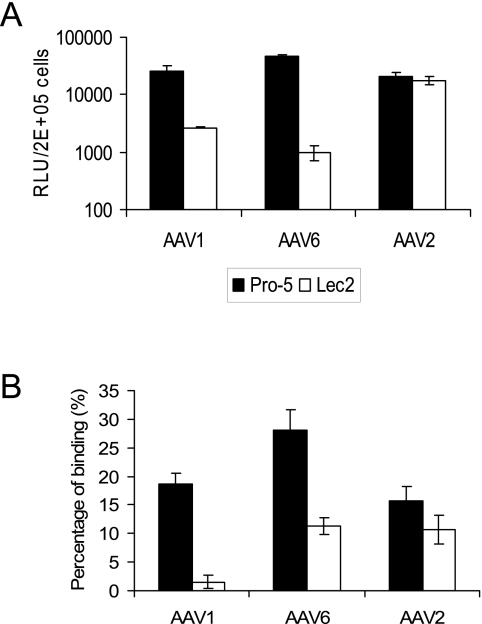
To further test if sialic acid is needed for efficient transduction by AAV1 and AAV6, we measured binding to and transduction of CHO cells deficient in cell surface sialic acid. Lec-2 cells are from a mutant clone derived from the parental CHO cell line Pro-5 (39). They are deficient in the transport of CMP-sialic acid into the Golgi compartment. As shown in Fig. 3, AAV2 bound and transduced both the parental cell line (Pro-5) and the sialic acid-deficient mutant (Lec-2) with similar efficiencies. In contrast, AAV1 and AAV6 bound and transduced Lec-2 cells much less efficiently than Pro-5 cells. Similar to the neuraminidase treatment experiment (Fig. 2D), a larger amount of AAV6 bound to the Lec-2 cells compared to AAV1, consistent with its possible utilization of additional carbohydrates in binding to these cells. Taken together, these results show that both enzymatic removal and genetic removal of sialic acid on the cell surface reduce AAV1 and AAV6 binding and transduction.
FIG. 3.
Transduction and binding on sialic acid-deficient cell lines. (A) Transduction of CHO cells (Pro-5) and sialic acid-deficient CHO cells (Lec-2) with AAV1, AAV6, and AAV2. RLU, relative light units. (B) Binding of AAV1, AAV6, and AAV2 to Pro-5 cells and Lec-2 cells. Values are means from three experiments. Error bars represent standard deviations.
Interestingly, AAV6 transduction appears to be more dependent on sialic acid than AAV1 (Fig. 2A and B and 3A), although AAV6 binding to the cell surface was less affected by the removal or absence of sialic acid (Fig. 2D and 3B). There was a 12- or 98-fold decrease in AAV6 transduction following neuraminidase treatment on HepG2 and Pro5 cells, respectively, in contrast to a 5- or 37-fold decrease in AAV1 transduction on these two cell lines. In addition, there was a 60-fold difference between Pro-5 cells and Lec-2 cells for AAV6 transduction while only an 8-fold difference for AAV1 transduction. These data most likely reflect the difference between AAV1 and AAV6 interactions with sialic acid during the initial binding and cell entry steps.
Both α2,3 and α2,6 sialic acids facilitate transduction by AAV1 and AAV6.
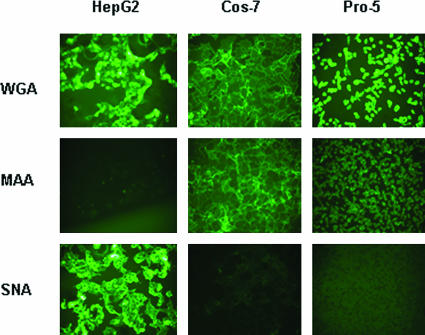
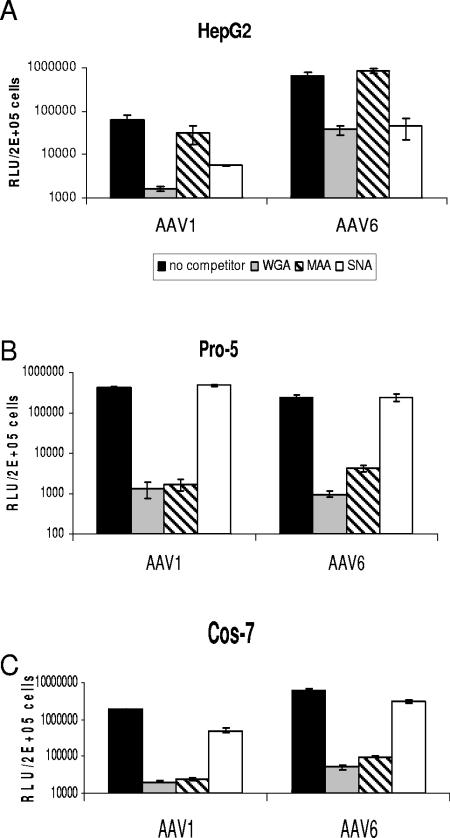
Previous studies have shown that both AAV4 and AAV5 use α2,3 sialic acid for efficient binding and transduction (19). We asked if AAV1 and AAV6 have a preference for a sialic acid linkage(s) for efficient transduction. The above results (Fig. 2) show that AAV1 and AAV6 transduce Pro-5, Cos-7, and HepG2 cells efficiently; therefore, we tested the sialic acid linkages on the surfaces of these cell lines. We stained cell lines with lectins that have been used to recognize the following three different epitopes: (i) WGA recognizes all sialic acids, (ii) MAA recognizes α2,3 sialic acid, and (iii) SNA recognizes α2,6 sialic acid. WGA bound to all three cell lines tested, while MAA bound both Pro-5 and Cos-7 cells but not HepG2 cells (Fig. 4). SNA did not bind to Pro-5 cells; however, it did bind well to HepG2 cells and, to a lesser extent, Cos-7 cells. Therefore, Pro-5 cells and HepG2 cells display α2,3 sialic acid or α2,6 sialic acid on their surfaces, respectively, while Cos-7 cells display α2,3 sialic acid and a relatively small amount of α2,6 sialic acid. Since AAV1 and AAV6 transduced all these cell lines efficiently, it appears that there is no obvious preference for a particular sialic acid linkage(s). To confirm if both α2,3 and α2,6 sialic acids facilitate AAV1 and AAV6 transduction, we carried out a lectin competition assay on these three cell lines (Fig. 5A to C). WGA, which binds to all linkage forms of sialic acid, blocked both AAV1 and AAV6 transduction on all the cells tested. MAA blocked AAV1 and AAV6 transduction on Pro-5 and Cos-7 cells, which display α2,3 sialic acid on their surfaces, but not on HepG2 cells, which display α2,6 sialic acid. Similarly, SNA blocked AAV1 and AAV6 transduction on HepG2 cells and, to a lesser extent, on Cos-7 cells but not on Pro-5 cells. As a control, AAV2 transduction was not affected by lectin competition on all cell lines (data not shown). These results again support the idea that sialic acid facilitates AAV1 and AAV6 transduction, in particular both α2,3 and α2,6 sialic acids.
FIG. 4.
Lectin binding to HepG2, Cos-7, and Pro-5 cells. Cultures were stained with FITC-labeled lectins that bind to three different carbohydrates as follows: WGA binds sialic acid in any linkage, MAA binds 2,3-linked sialic acid, and SNA binds 2,6-linked sialic acid.
FIG. 5.
Lectin competition on HepG2 (A), Pro-5 (B), and Cos-7 (C) cells. Cells growing in 24-well plates (approximately 2 × 105 cells/well) were preincubated at 4°C with 100 μg/ml of each lectin for 10 min. The preincubation solution was removed, and medium containing 100 μg/ml lectin and 2 × 108 virus particles of AAV1-luc, AAV6-luc, or AAV2-luc was added. Cultures were maintained at 4°C for 1 h and then rinsed with medium three times. The cells were grown at 37°C for an additional 24 h in serum-containing media and then assayed for luciferase expression. RLU, relative light units.
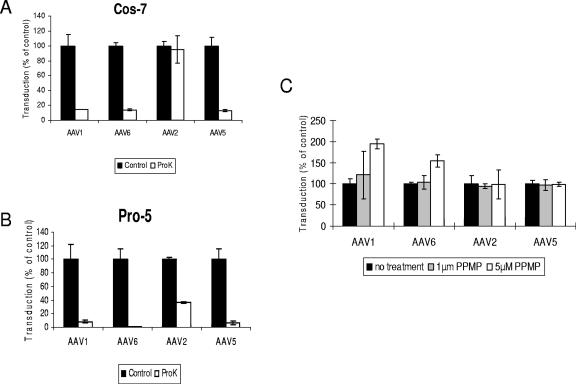
The sialic acid that facilitates AAV1 and AAV6 transduction localizes on glycoproteins rather than on glycolipids.
Sialic acid saccharides are expressed on both glycoproteins and glycosphingolipids. To analyze if the sialylated receptors of AAV1 and AAV6 are glycoproteins or glycolipids, we treated Cos-7 and Pro-5 cells with 200 μg/ml proteinase K. Cell viability was assessed immediately after proteinase K treatment, and no cytotoxic effect was observed. As shown in Fig. 6, for both cells lines, proteinase K treatment inhibited more than 80% of transduction by AAV1, AAV6, and AAV5, suggesting that the receptors of these viruses are glycoproteins. AAV2, which has been demonstrated to use integrin αVβ5, basic fibroblast growth factor receptor, or hepatocyte growth factor receptor as its coreceptor for cell entry (20, 30, 41), was much less sensitive to proteinase K treatment for transduction (Fig. 6). The lack of AAV2 sensitivity may be related to the fact that these coreceptors are more resistant than others to proteinase K treatment under the conditions we used, or it could be that, after virus binding to the heparin sulfate primary receptor, viral entry can proceed although not optimally.
FIG. 6.
The AAV1 and AAV6 receptor is a glycoprotein rather than a glycolipid. (A and B) Proteinase K treatment reduced AAV1 and AAV6 transduction. Cos-7 (A) and Pro-5 (B) cells were incubated in the presence or absence of 200 μg/ml proteinase K for 1 h at 37°C. The cells were transduced at an MOI of 1 × 103 for 1 h with AAV1-luc, AAV-6-luc, AAV2-luc, or AAV5-luc, respectively. Luciferase expression was analyzed 24 h after transduction. (C) Glycosphingolipid synthesis inhibitor does not block AAV1 and AAV6 transduction. Cos-7 cells were treated for 40 h with PPMP before transduction with AAV1-luc, AAV6-luc, AAV2-luc, or AAV5-luc. Luciferase expression was analyzed 24 h after transduction.
PPMP is a glucosylceramide synthase inhibitor, which acts to deplete glycosphingolipids from the cell membrane (22). To test if glycolipids on cell membrane are also required for AAV1 and AAV6 transduction, Cos-7 cells were incubated with PPMP for 40 h prior to transduction with AAV1, AAV6, AAV2, or AAV5. At the two concentrations of PPMP we tested (1 μM and 5 μM), no decrease was observed for transduction with either one of the four vectors (Fig. 6C). However, these concentrations of PPMP significantly inhibited bovine AAV infection, which requires gangliosides as receptors (36). Therefore, glycolipids seem not to be required for AAV1 and AAV6 transduction.
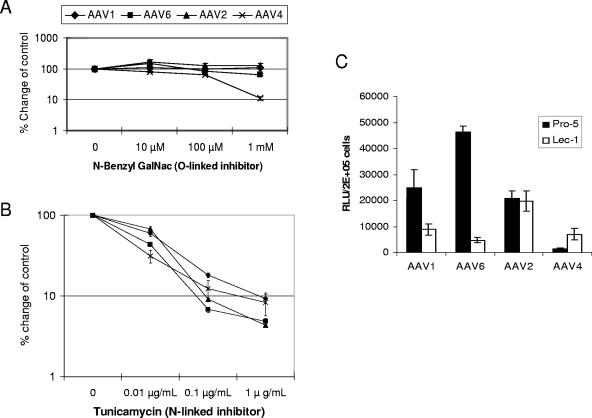
N-linked, not O-linked, sialic acid facilitates AAV1 and AAV6 transduction.
Although both AAV4 and AAV5 bind to α2,3-linked sialic acid for transduction, AAV4 binds sialic acid present on O-linked oligosaccharides, whereas AAV5 binds sialic acid present on N-linked oligosaccharides (19). To determine whether O-linked or N-linked sialic acid is used by AAV1 and AAV6 for transduction, Cos-7 cells were cultured with inhibitors of O-linked (N-benzyl GalNac) or N-linked (tunicamycin) glycosylation. As shown in Fig. 7A, consistent with previous reports, the O-linked inhibitor decreased AAV4 transduction in a dose-dependent manner. At the concentration of 1 mM, N-benzyl GalNac inhibited AAV4 transduction by 10-fold. In contrast, only marginal or no inhibition was seen for AAV1, AAV6, or AAV2 transduction, indicating that AAV1 and AAV6 do not use O-linked sialic acid for transduction. The N-linked inhibitor tunicamycin inhibited both AAV1 and AAV6 transduction; however, it also inhibited AAV2 and AAV4 transduction (Fig. 7B). Similar to a previous study (19), the inhibited transduction with AAV1, -2, -4, and -6 by tunicamycin may be due to its broad effect on intracellular activity, ranging from protein folding and secretion to signal transduction and transcription activation.
FIG. 7.
N-linked, not O-linked, sialic acid facilitates AAV1 and AAV6 transduction. Cos-7 cells were treated with the indicated doses of N-benzyl GalNAc (A) or tunicamycin (B) for 24 h prior to transduction. Then the cells were transduced with AAV1-luc, AAV6-luc, AAV4-luc, or AAV2-luc. Luciferase expression was analyzed 24 h after transduction. The relative transduction efficiency was determined by comparison with control untreated cells and is presented in log scale. (C) Transduction of CHO cells (Pro-5) and N-linked-glycan-deficient CHO cells (Lec-1) with AAV1-luc, AAV6-luc, AAV2-luc, and AAV4-luc. RLU, relative light units.
To determine if N-linked sialic acid is necessary for efficient AAV1 and AAV6 transduction, we tested AAV1 and AAV6 transduction on another Pro-5-derived cell line, Lec-1 (40), that is deficient in N-linked glycans but is not deficient in O-linked glycans (Fig. 7C). AAV1 and AAV6 transduced Lec-1 cells 3- and 10-fold less efficiently than Pro-5 cells. In contrast, AAV4, which uses O-linked sialic acid for transduction, transduced Lec-1 cells fourfold more efficiently than Pro-5 cells, suggesting that removal of the N-linked glycan facilitates AAV4 interaction with O-linked glycan. There was no difference for AAV2 transduction between Pro-5 and Lec-1 cells. These data suggested that AAV1 and AAV6 use N-linked sialic acid for efficient transduction.
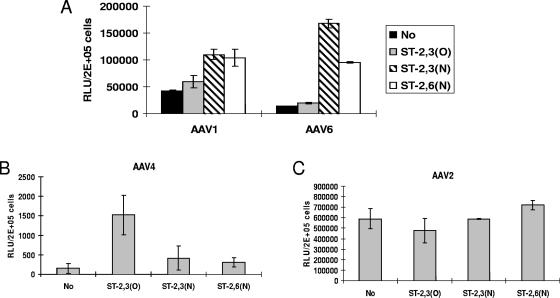
To further investigate the role of α2,3 and α2,6 sialic acids and their linkage specificities in AAV1 and AAV6 transduction, we carried out resialylation on sialic acid-deficient Lec-2 cells using sialyltransferase and CMP-sialic acid. Consistent with previous report, resialylation with α2,3(O)-sialyltransferase markedly increased AAV4 transduction, while no effect was seen by resialylation with α2,3(N)- or α2,6(N)-sialyltransferase (Fig. 8B). In contrast, resialylation with α2,3(O)-sialyltransferase did not affect AAV1 and AAV6 transduction, while resialylation with α2,3(N)- or α2,6(N)-sialyltransferase resulted in substantial increases in AAV1 and AAV6 transduction (Fig. 8A). Treatment of Lec-2 cells with either sialyltransferase or CMP-sialic acid alone did not result in successful resialylation and any increased transduction by the viruses (data not shown). Consistent with the results shown in Fig. 2 and 3, transduction by AAV6 appears to be more dependent on sialic acid than AAV1. Resialylation by α2,3(N)-sialyltransferase and α2,6(N)-sialyltransferase increased AAV6 transduction by 11-fold and 6-fold, respectively, but only 2.5-fold for AAV1. As a negative control, transduction by AAV2 did not obviously change after resialylation with each sialyltransferase (Fig. 8C). Resialylation experiments confirmed that AAV1 and AAV6 use α2,3 or α2,6 N-linked sialic acid for efficient transduction.
FIG. 8.
AAV transduction of resialylated sialic acid-deficient Lec-2 cells. The following sialyltransferases were used to add specific sialic acids to the surfaces of Lec-2 cells: α2,3(O)-sialyltransferase, α2,3(N)-sialyltransferase, and α2,6(N)-sialyltransferase. Resialylated Lec-2 cells were then transduced (MOI, 1,000) with either AAV1-luc or AAV6-luc (A), AAV4-luc (B), or AAV2-luc (C), and luciferase expression was measured 24 h posttransduction. RLU, relative light units.
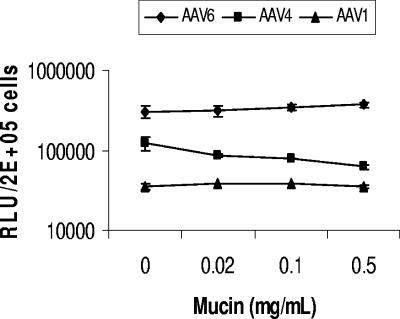
AAV1 and AAV6 transduction is not inhibited by mucin.
Mucin, which contains primarily O-linked oligosaccharides and which has previously been shown to inhibit gene transfer with AAV4 (45), was tested for inhibition of AAV1 and AAV6 transduction. As shown in Fig. 9, mucin inhibited AAV4 transduction in a dose-dependent manner. At 0.5-mg/ml mucin concentration, 50% inhibition of AAV4 transduction was observed. In contrast, but consistent with our data supporting a requirement for N-linked sialic acid, mucin did not inhibit AAV1 and AAV6 transduction.
FIG. 9.
AAV1 and AAV6 transduction is not inhibited by mucin. Particles (2 × 108) of AAV1-luc, AAV4-luc, or AAV6-luc were incubated with the indicated concentrations of mucin for 30 min at 20°C. Virus alone or virus plus mucin were added to Cos-7 cells growing in 24-well plates (2 × 105 cells/well) in equal volumes of DMEM, and cells were incubated for 1 h at 37°C. Cells were rinsed twice with DMEM and incubated at 37°C. Luciferase expression was tested 24 h later. RLU, relative light units.
Specificity of carbohydrate binding by AAV1 capsids on a glycan array.
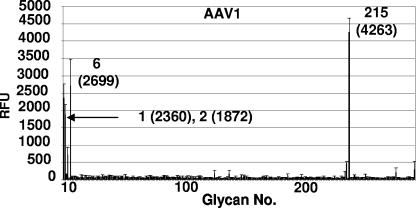
We utilized a glycan array, available through the CFG, to screen a library of 264 different synthetic and natural glycans, which represent the carbohydrates commonly found on cell surfaces, to test the AAV1 capsid binding specificity. This array, containing sialylated and nonsialylated sugars with different linkages and modifications, was constructed for identifying specific carbohydrate binding partners for proteins. The AAV1 capsids recognized only four glycans: NeuAcα2-3GalNAcβ1-4GlcNAcβ (sialylated di-N-acetyl-lactosamine; PA address 215), apo-transferrin (PA address 6), α1-acid glycoprotein (AGP) (PA address 1), and AGP-A (concanavalin A flowthrough; PA address 2) in the array (Fig. 10). The AGP and AGP-A (prepared as described in reference 37a), and apo-transferrin (Sigma-Aldrich) are natural purified glycoproteins that were covalently attached to the array. AGP contains five N-linked glycan sites that contain biantennary, triantennary, and tetra-antennary chains which contain terminal sialic acids linked either α2,3 or α2,6 to lactosamine (Galβ1-4GlcNAc) (37a). AGP-A is a form of AGP that contains a reduced amount of biantennary chains (37a). Apo-transferrin is known to be glycosylated with two biantennary N-linked glycan chains that contain α2,6 terminal sialic acids linked to lactosamine (4, 38). Fractions of glycans purified from AGP, AGP-A, and apo-transferrin can also be fucosylated to create the sLeX motif [NeuAcα2-3/6Galβ1-4(Fucα1-3)GlcNAc], which is a marker for cancer cells. The glycan array binding data provide independent support of AAV1 interaction with α2,3 and α2,6 trisaccharides.
FIG. 10.
AAV1 capsid glycan specificity on glycan array. A total of 264 glycans were screened for binding to the capsids as described in Materials and Methods. The plot shows the average relative fluorescence (RFU) for the six addresses for each glycan (thick line) versus glycan number. The standard error measurement (SEM) in the fluorescence for the six addresses (thin line) is drawn at the top of each thick line. The four top hits (printed array addresses: 1, AGP; 2, AGP-A; 6, transferrin; 215, NeuAcα2-3GalNAcβ1-4GlcNAcβ) with acceptable SEMs are indicated, with their relative fluorescence levels given in parentheses. All glycans recognized contain a common motif: sialic acid linked to N-acetyl-lactosamine.
DISCUSSION
Sialic acid is the most abundant carbohydrate moiety on the cell surface, and as a result of this, a number of viruses use it as their primary attachment factor for productive infection (27). Among the 11 known AAV serotypes and hundreds of variants, AAV4 and AAV5 utilize α2,3 O-linked or α2,3 N-linked sialic acids for their binding and subsequent infection (19), respectively. In this report, we demonstrate that AAV1 and AAV6 use both α2,3 and α2,6 N-linked sialic acids for binding and infection. Using competition, genetic, inhibitor, and enzymatic reconstitution experiments we demonstrate that a glycoprotein(s) with N-linked α2,3 and/or α2,6 sialic acids serves as a receptor(s) for AAV1 and AAV6 transduction. In addition, through the CFG, we were able to test AAV1 on a unique glycan array and demonstrate specific binding to glycans containing terminal sialic acids attached α2,3 and α2,6 to the Galβ1-4GlcNAc motif (Fig. 10), with the sugars being N-linked in the glycoproteins recognized. The glycan array has been used to test the glycan binding profiles of a wide variety of proteins (3) and represents a novel approach for investigating virus-receptor interactions. The present study represents the first utilization of the printed array to analyze the glycan binding profile for whole intact virus capsids. Interestingly, an older version of the array, containing glycans immobilized as biotinylated glycosides on a 384-well streptavidin-coated plate, was used to screen the sugar binding specificity of the parvovirus minute virus of mice (MVM) capsids (M. Agbandje-McKenna, personal communication). This study was able to identify the sialic acid structures recognized by MVM, which were consistent with the oncotropic properties of this virus, in addition to the neurotropism displayed by the lymphotropic strain MVMi. While the data in our present study provided independent support for AAV1 binding to a 2,3 trisaccharide, we did not observe a similar interaction with the equivalent 2,6 trisaccharide. One simple possibility for this observation might reside in the fact that an α2,6 trisaccharide adopts a more “kinked” structure and thus might not protrude out enough from the printed glass slide to “reach” into a potential binding pocket on the AAV1 capsid compared to α2,3 trisaccharide, which would be more extended and long enough to access a receptor binding pocket. In the context of the two glycoproteins recognized in the array, AGP and apo-transferrin, the terminal α2,3/α2,6 trisaccharide motif is linked to several other sugars before they are N-linked to the protein, and thus the length of the chain is not likely to be limiting for binding. Ongoing X-ray crystallographic studies of AAV1 and these sugars should provide the detailed insight required to fully understand the determinants of the interactions between the capsid and the sugars.
By DNA sequence, AAV1 and AAV6 are closely related serotypes. AAV6 is regarded as a laboratory strain derived from recombination between AAV1 and AAV2, with its upstream sequence being identical to AAV2 and its downstream sequence similar to AAV1 (34, 46). Analysis of the capsid sequence of AAV1 and AAV6 shows only six amino acid differences in the capsid protein. Therefore, it is not surprising that AAV1 and AAV6 are almost identical in their serology (14, 16). Both AAV1 and AAV6 have been shown to transduce muscle very efficiently (2, 5), although a side-by-side comparison has not been reported. However, AAV1 and AAV6 do show different kinetics and efficiency of transduction in nonmuscle tissue such as liver (16), raising the questions whether they use the same receptor(s) and how these six different amino acids may affect transduction. In the present study, we showed that AAV1 competes with AAV6 transduction and vice versa in cultured cells, suggesting that AAV1 and AAV6 might use the same receptor or that they share some common receptors for transduction. Sialic acids are abundant on the surfaces of muscle cells, which may partly explain the high transduction efficiency of AAV1 and AAV6 vectors for this tissue. However, because AAV6 is more efficient than AAV1 for liver transduction (16), this raises the question of how this difference may exist if these serotypes share identical receptors. From the in vitro data we collected in this study, although sialic acid is important for both AAV1 and AAV6 transduction, it appears that AAV6 transduction is more dependent on sialic acid than AAV1 (Fig. 2A to C, 3A, 7C, and 8A). Since the amount of AAV6-specific binding to sialic acid is not higher than that of AAV1 (Fig. 2D and 3B), it is possible that AAV6 relies more on sialic acid or sialic acid-containing glycoproteins than AAV1 for cell entry and/or subsequent steps of infection. In addition, if the interaction between sialic acid and the AAV6 capsid is more efficient than that of AAV1 for cell entry, this might account for the increased transduction of AAV6 compared to AAV1 in liver cells. Recently, we have swapped each of the six divergent amino acids between AAV1 and AAV6 and have identified a subset responsible for efficient liver transduction (manuscript in preparation) although the mechanism is still unknown.
A recent report demonstrated that in immortalized and high-passage nonimmortalized human airway cells, AAV6 transduction, unlike AAV5 transduction, was insensitive to neuraminidase treatment (37). Based on our present study, we would have predicted otherwise. To help understand these differences, we repeated identical experiments on immortalized CF16 cells provided by Seiler et al. (37). In our hands, we demonstrated that both AAV5 and AAV6 transduction was reduced after neuraminidase treatment (data not shown). In addition, among all other cell lines tested, AAV6 transduction markedly drops following neuraminidase treatment (Fig. 2). These cell lines are derived from different origins, including Pro-5, HepG2, Cos-7 (Fig. 2), and HeLa (data not shown) cells, suggesting that this observation is not unique to airway cells. Our data do not support a sialic acid-independent pathway for AAV6 transduction. As the authors discussed in their publication, it is possible that the sialic acid was not completely removed on the CF16 cells after neuraminidase treatment in their study. To support this hypothesis, it has been reported that neuraminidase from Vibrio cholerae, which has a broad spectrum, prefers the cleavage of α2,3 sialic acid to α2,6 sialic acid (19; Sigma product information). From our studies, AAV6 uses both α2,3- and α2,6-linked sialic acids, while AAV5 prefers α2,3-linked sialic acid for transduction (19, 42). This would explain why transduction of CF16 cells with AAV5 decreases more than that with AAV6 after neuraminidase treatment in their study (37).
Unlike AAV4 and AAV5, which use α2,3 sialic acid, AAV1 and AAV6 can use both α2,3 and α2,6 sialic acids for efficient transduction. Phylogenetic analyses of primate AAV isolates suggest that the existing primate AAV serotypes originated from avian AAV (12). Based on similar analysis, AAV4 and AAV5 are divergent from the other AAV serotypes and thought to have branched off earlier from avian AAV. Thus it will be of interest to determine if avian AAV also uses α2,3 sialic acid as its primary receptor. While the origin of AAV4 is monkeys (15), AAV5, although originally isolated from humans (1), is rarely found in human populations (18). The existence of AAV1 and AAV6 in human tissues (12) suggests that these serotypes may have evolved to use α2,6 sialic acid as their receptor in order to adapt to infection of humans, since α2,6 sialic acid is the predominant linkage form in a variety of human tissues (11). Precedent for this type of adaptation can be seen today when one observes the conversion of avian influenza virus, which uses α2,3 sialic acid, to a virus which infects humans with a preference for α2,6 sialic acid (27).
Mucin, which contains mainly O-linked sialic acids, plays an important role in the host defense system against pathogens. A previous report has shown that mucin specifically blocks AAV4 transduction (45), which uses O-linked sialic acid as its receptor, but does not block AAV5 transduction. Consistent with this published study, our results demonstrated that transduction with AAV1 and AAV6 was not inhibited by mucin (Fig. 9). Since mucin is secreted by several tissues in humans, this result has important implications for clinical gene transfer. It is anticipated that human airway epithelial cells, the target cells for the treatment of cystic fibrosis, can be efficiently transduced by AAV1 and AAV6 vectors. Since α2,3 sialic acid is present on ciliated cells, while α2,6 sialic acid is present on both ciliated and nonciliated cells (23), it will be of interest to determine if AAV1 and AAV6 are able to transduce both types of epithelial cells. In summary, we have used a number of assays (competition, genetic, inhibitor, enzymatic reconstitution, and direct virus binding to a glycan array) to support glycoproteins with N-linked α2,3 and/or α2,6 sialic acid serving as the receptor(s) for AAV1 and AAV6 transduction. These observations continue to add to our knowledge of AAV receptor/virus interactions and should provide important insight when determining optimum use of these reagents for vectors in human gene transfer studies.
Acknowledgments
We thank Matthew Hirsch and Aravind Asokan for critically reading the manuscript. We also acknowledge Brittney Whitaker-Gurda for technical support, Colin Parrish for providing AAV1 anticapsid antibodies, and Sergei Zolotukhin for providing the baculovirus construct expressing the VP proteins of AAV1. We also thank Richard Alvarez, Director of Core H, and Angela Lee for their help and advice.
This work was supported by research grants from NIH (HL051818, HL069973, and GM059299) and a Brain Tumor Research Award from The Goldhirsh Foundation to R. Jude Samulski, research grants from NIH (HL59412 and HL51811) to Mavis Agbandje-McKenna, and the Beckman Foundation of a fellowship to Edward Miller. Zhijian Wu is a recipient of a Judith Graham Poole postdoctoral fellowship from the National Hemophilia Foundation. The glycan array analysis was conducted by the Protein-Carbohydrate Interaction Core H of The Consortium for Functional Glycomics, funded by National Institute of General Medical Sciences grant GM62116.
REFERENCES
- 1.Bantel-Schaal, U., and H. zur Hausen. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52-63. [DOI] [PubMed] [Google Scholar]
- 2.Blankinship, M. J., P. Gregorevic, J. M. Allen, S. Q. Harper, H. Harper, C. L. Halbert, D. A. Miller, and J. S. Chamberlain. 2004. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol. Ther. 10:671-678. [DOI] [PubMed] [Google Scholar]
- 3.Blixt, O., S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong, and J. C. Paulson. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 101:17033-17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonsdorff, L. V., H. Tolo, E. Lindeberg, T. Nyman, A. Harju, and J. Parkkinen. 2001. Development of a pharmaceutical apotransferrin product for iron binding therapy. Biologicals 29:27-37. [DOI] [PubMed] [Google Scholar]
- 5.Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., M. Kapturczak, S. A. Loiler, S. Zolotukhin, O. Y. Glushakova, K. M. Madsen, R. J. Samulski, W. W. Hauswirth, M. Campbell-Thompson, K. I. Berns, T. R. Flotte, M. A. Atkinson, C. C. Tisher, and A. Agarwal. 2005. Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Hum. Gene Ther. 16:235-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2005. AAV hybrid serotypes: improved vectors for gene delivery. Curr. Gene Ther. 5:299-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMattia, M., L. Govindasamy, H. C. Levy, B. Gurda-Whitaker, A. Kalina, E. Kohlbrenner, J. A. Chiorini, R. McKenna, N. Muzyczka, S. Zolotukhin, and M. Agbandje-McKenna. 2005. Production, purification, crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 5. Acta Crystallogr. F. Struct. Biol. Crystalliz. Comm. 61:917-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, W., L. Zhang, Z. Yan, and J. F. Engelhardt. 2005. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 12:873-880. [DOI] [PubMed] [Google Scholar]
- 10.Di Pasquale, G., B. L. Davidson, C. S. Stein, I. Martins, D. Scudiero, A. Monks, and J. A. Chiorini. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 9:1306-1312. [DOI] [PubMed] [Google Scholar]
- 11.Gagneux, P., M. Cheriyan, N. Hurtado-Ziola, E. C. van der Linden, D. Anderson, H. McClure, A. Varki, and N. M. Varki. 2003. Human-specific regulation of alpha 2-6-linked sialic acids. J. Biol. Chem. 278:48245-48250. [DOI] [PubMed] [Google Scholar]
- 12.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, G., L. H. Vandenberghe, and J. M. Wilson. 2005. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 5:285-297. [DOI] [PubMed] [Google Scholar]
- 14.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, D., and M. A. Kay. 2003. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 3:281-304. [DOI] [PubMed] [Google Scholar]
- 16.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, J. Allen, R. Surosky, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 102:2412-2419. [DOI] [PubMed] [Google Scholar]
- 17.Halbert, C. L., J. M. Allen, and A. D. Miller. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildinger, M., A. Auricchio, G. Gao, L. Wang, N. Chirmule, and J. M. Wilson. 2001. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J. Virol. 75:6199-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashiwakura, Y., K. Tamayose, K. Iwabuchi, Y. Hirai, T. Shimada, K. Matsumoto, T. Nakamura, M. Watanabe, K. Oshimi, and H. Daida. 2005. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J. Virol. 79:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovacs, P., M. Pinter, and G. Csaba. 2000. Effect of glucosphingolipid synthesis inhibitor (PPMP and PDMP) treatment on Tetrahymena pyriformis: data on the evolution of the signaling system. Cell Biochem. Funct. 18:269-280. [DOI] [PubMed] [Google Scholar]
- 23.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori, S., L. Wang, T. Takeuchi, and T. Kanda. 2004. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330:375-383. [DOI] [PubMed] [Google Scholar]
- 25.Nakai, H., S. Fuess, T. A. Storm, S. Muramatsu, Y. Nara, and M. A. Kay. 2005. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 79:214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nokhbeh, M. R., S. Hazra, D. A. Alexander, A. Khan, M. McAllister, E. J. Suuronen, M. Griffith, and K. Dimock. 2005. Enterovirus 70 binds to different glycoconjugates containing α2,3-linked sialic acid on different cell lines. J. Virol. 79:7087-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olofsson S., and T. Bergstrom. 2005. Glycoconjugate glycans as viral receptors. Ann. Med. 37:154-172. [DOI] [PubMed] [Google Scholar]
- 28.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 79:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 31.Rabinowitz, J. E., D. E. Bowles, S. M. Faust, J. G. Ledford, S. E. Cunningham, and R. J. Samulski. 2004. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J. Virol. 78:4421-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabinowitz, J. E., W. Xiao, and R. J. Samulski. 1999. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265:274-285. [DOI] [PubMed] [Google Scholar]
- 34.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, M., E. Grot, P. Cervenka, S. Wainer, C. Buck, and J. A. Chiorini. 2006. Identification and characterization of novel adeno-associated virus isolates in ATCC virus stocks. J. Virol. 80:5082-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, M., and J. A. Chiorini. 2006. Gangliosides are essential for bovine adeno-associated virus entry. J. Virol. 80:5516-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seiler, M. P., A. D. Miller, J. Zabner, and C. L. Halbert. 2006. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum. Gene Ther. 17:10-19. [DOI] [PubMed] [Google Scholar]
- 37a.Shiyan, S. D., and N. V. Bovin. 1997. Carbohydrate composition and immunomodulatory activity of different glycoforms of alpha1-acid glycoprotein. Glycoconj. J. 14:631-638. [DOI] [PubMed] [Google Scholar]
- 38.Spik, G., B. Bayard, B. Fournet, G. Strecker, S. Bouquelet, and J. Montreuil. 1975. Studies on glycoconjugates. LXIV. Complete structure of two carbohydrate units of human serotransferrin. FEBS Lett. 50:296-299. [DOI] [PubMed] [Google Scholar]
- 39.Stanley, P., V. Caillibot, and L. Siminovitch. 1978. Specific changes in the oligosaccharide moieties of VSV grown in different lectin-resistant CHO cells. Cell 13:515-526. [DOI] [PubMed] [Google Scholar]
- 40.Stanley, P., S. Narasimhan, L. Siminovitch, and H. Schachter. 1975. Chinese hamster ovary cells selected for resistance to the cytotoxicity of phytohemagglutinin are deficient in a UDP-N-acetylglucosamine-glycoprotein N-acetylglucosaminyltransferase activity. Proc. Natl. Acad. Sci. USA 72:3323-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. αVβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 42.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 78:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 45.Walters, R. W., J. M. Pilewski, J. A. Chiorini, and J. Zabner. 2002. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 277:23709-23713. [DOI] [PubMed] [Google Scholar]
- 46.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]