Abstract
Following UV irradiation, denaturing polyacrylamide gel electrophoresis and Northern blot hybridization revealed a cross-link in Peach latent mosaic viroid (PLMVd) plus-strand RNA. Primer extension and partial alkaline hydrolysis of the UV-irradiated PLMVd plus-strand RNA resulting from the hammerhead-mediated self-cleavage mapped the cross-link at U81 and at the 3′-terminal C289 (or at a very proximal nucleotide). Supporting this notion, in vitro-synthesized PLMVd plus-strand RNAs with short insertions/deletions at their 3′ termini failed to cross-link. Because U81 and C289 are conserved in PLMVd variants and because the initiation site of PLMVd minus-strand RNA maps at a short double-stranded motif containing C289, the UV-photo-cross-linkable element of tertiary structure may be functionally significant. A second cross-linked species similar in size and sequence to the monomeric circular PLMVd form, observed in some PLMVd variants, probably derives from UV-induced ligation of the two termini resulting from self-cleavage.
Elements of tertiary structure are crucial for RNA function (12, 18). Such elements exert their physiological role by promoting specific local RNA folding and/or facilitating binding of certain proteins. In viroids, which are small, non-protein-encoding, single-stranded, circular RNAs, able to replicate autonomously in some higher plants (11, 14, 29), several elements of tertiary structure have been reported. In vitro UV irradiation and enzymatic and chemical probing of purified preparations of Potato spindle tuber viroid (PSTVd) (10, 19)—the type species of the family of nuclear viroids (Pospiviroidae) characterized by the presence of a central conserved region (CCR) and the absence of hammerhead ribozymes (15)—have revealed that its CCR contains an element of local tertiary structure with high sequence and structural similarity to loop E of eukaryotic 5S rRNA (4, 17). There is evidence supporting the involvement of loop E in PSTVd replication (3), symptom expression (26), and host specificity (30).
In Peach latent mosaic viroid (PLMVd) (20)—which belongs to the family of chloroplastic viroids (Avsunviroidae) lacking a CCR but endowed with self-cleavage through hammerhead ribozymes embedded in the strands of both polarities (15)—in vitro nuclease mapping and oligonucleotide binding shift assays indicate the existence of a pseudoknot-like interaction between two hairpin loops of the proposed branched conformation (6). Moreover, inoculations with natural and artificial variants of another member of the family Avsunviroidae, Chrysanthemum chlorotic mottle viroid (24), to assess their biological properties and analysis of the genetic stability in the resulting progenies support the existence of a similar kissing-loop interaction in this viroid that is critical for its in vitro folding and in vivo viability (16). Also in the family Avsunviroidae, recent data show that modifications of loops 1 and 2 of natural hammerheads induce a severe reduction in their catalytic activity, indicating that these peripheral regions play a critical role in catalysis through tertiary interactions between some of their nucleotides that may favor the active site at the low magnesium concentration existing in vivo (8, 22). Here we present evidence for a UV-sensitive element of tertiary structure between two nucleotides located far apart in the primary structure of PLMVd and suggest a possible functional role for it.
Identification of a UV-induced cross-link in PLMVd-strand RNA.
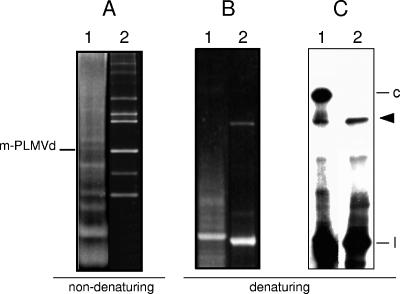
Nucleic acids were extracted and fractionated from PLMVd-infected peach leaves (20). In Northern blot analyses of PLMVd RNAs separated by double polyacrylamide gel electrophoresis (PAGE), first under nondenaturing and then under denaturing conditions (13) (Fig. 1), a band of variable intensity was observed below that of the monomeric circular PLMVd RNAs in the second gel (Fig. 1C, lane 1). To test the hypothesis that this band could arise from a UV-induced cross-linking of PLMVd RNA, a linearized plasmid containing a dimeric PLMVd-cDNA insert of variant esc5 (1)—identical to the second variant originally reported (20)—was used as template to generate plus-polarity transcripts. Following electrophoresis in a nondenaturing 5% polyacrylamide gel in which the PLMVd monomeric circular and linear RNAs have the same mobility (Fig. 1A), the segment containing these RNAs (delimited by the position of the monomeric linear form resulting from self-cleavage) was cut, irradiated for 5 to 30 min in a UV transilluminator, and laid on top of a second denaturing 5% polyacrylamide gel. Generation of the cross-linked PLMVd species was dependent on the UV dose but independent of the RNA concentration (data not shown), indicating that it results from an intramolecular interaction. These data also revealed that such interaction takes place within the monomeric positive-strand linear PLMVd RNA arising from self-cleavage (Fig. 1B and C, lanes 2), the most abundant form accumulating in vivo (5, 9). Moreover, the cross-linked species was also observed when PLMVd-infected leaves were UV irradiated prior to RNA extraction, indicating that the UV-sensitive element of tertiary structure exists also in vivo (data not shown). On the other hand, UV irradiation of a purified monomeric linear PLMVd minus-strand RNA resulting from in vitro self-cleavage failed to reveal a similar RNA species (data not shown), illustrating that cross-linking occurs only in the PLMVd plus strand.
FIG. 1.
Analysis of PLMVd RNAs by PAGE and Northern blot hybridization. RNAs from PLMVd-infected leaves (lanes 1) and in vitro transcription products of plus polarity from a linearized plasmid with a head-to-tail dimeric PLMVd cDNA insert of variant esc5 (lanes 2) were separated in a nondenaturing gel (A). After ethidium bromide staining, the segment containing the monomeric PLMVd RNAs (m-PLMVd) was excised, exposed to UV light, and applied to the top of a denaturing gel that was stained with ethidium bromide (B); the RNAs were then electrotransferred onto a nylon membrane and hybridized with a radioactive full-length PLMVd minus-strand riboprobe (C). In panels B and C, the arrowhead at the right indicates the position of the cross-linked PLMVd plus-strand RNA between the circular (c) and linear (l) forms. The autoradiogram corresponding to lane 1 in panel C was overexposed to make visible the cross-linked form.
Mapping the cross-link induced by UV irradiation of PLMVd plus-strand RNA.
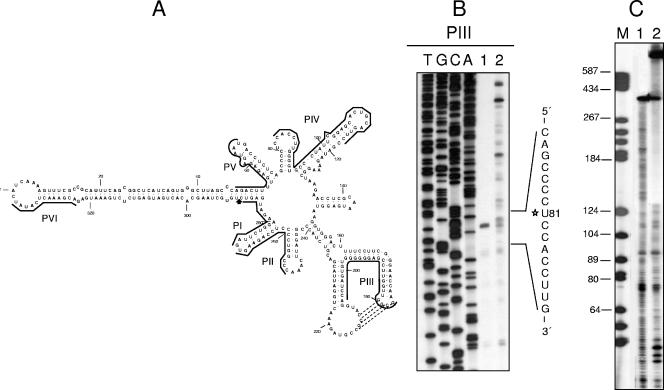
To identify the cross-linked nucleotides, monomeric linear PLMVd plus-strand RNAs of variant esc5 resulting from self-cleavage (UV irradiated and nonirradiated) were eluted and reverse transcribed with the minus-polarity primers PI to PVI using internal labeling and standard protocols (28) (Fig. 2A). In these experiments reverse transcriptase was expected to stop at the residue preceding the cross-link, as in stops caused by chemically modified nucleotides (21). With PIII, a cDNA doublet consistent with two consecutive strong stops at C80 and U81 was observed in the irradiated sample, most likely resulting from the extension arrest at C80 and the occasional addition of a nontemplate nucleotide (Fig. 2B, lane 1). No further extension products were detected in the irradiated sample (Fig. 2B, lane 1), in contrast to the situation found in the nonirradiated control (Fig. 2B, lane 2). The same results were obtained when primers PI, PII, and PIV were used to extend cDNAs on the cross-linked PLMVd form (data not shown), thus pointing to U81 as one of the residues involved in the cross-link. Reverse transcriptions with PV and PVI on both the cross-linked and non-cross-linked PLMVd plus-strand RNAs led to cDNAs that were extended up to the 5′ terminus of the linear monomeric PLMVd plus-strand RNA (data not shown). To complement this approach, both the cross-linked form and the nonirradiated control were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase and then subjected to partial alkaline hydrolysis. After fractionation of the products by PAGE in a sequencing gel, a gap in the ladder derived from the cross-linked form was observed beginning at the position corresponding to U81, which is located 129 nucleotides (nt) downstream of the 5′ end (Fig. 2C). These results, together with those of the primer extension assays, indicated that no residue upstream of U81 was involved in the PLMVd cross-link and also suggested that the second nucleotide implicated was very close to the PLMVd plus-strand self-cleavage site.
FIG. 2.
Mapping the UV-induced cross-link in PLMVd plus-strand RNA. (A) Predicted secondary structure of lowest free energy proposed for the PLMVd variant esc5 (1, 6, 20). The positions complementary to the six primers, PI to PVI, used in the reverse transcriptions are indicated with lines, and the positive-strand self-cleavage site is marked by an arrow. (B) Primer extensions with PIII in a 6% sequencing gel. Lanes 1 and 2 show the cDNAs synthesized by reverse transcription using as templates the cross-linked and non-cross-linked monomeric linear PLMVd plus-strand RNAs (resulting from in vitro self-cleavage of the corresponding dimeric transcript), respectively, of the PLMVd variant esc5. Sequencing ladders were obtained with the same primer and a recombinant plasmid containing a monomeric PLMVd insert. The 5′ terminus of the UV cross-linked RNA species is indicated with a star in the written sequence at the right. (C) Denaturing PAGE in a 6% sequencing gel of the products resulting from partial alkaline hydrolysis of 5′-end-labeled non-cross-linked (lane 1) and cross-linked (lane 2) monomeric linear PLMVd plus-strand RNAs. DNA markers, with their sizes (in nucleotides) indicated at the left, were applied to lane M.
Effects on cross-linking of insertions and deletions at the 3′ terminus of linear monomeric plus-strand PLMVd RNA.
To determine the influence on cross-linking of the 3′ terminus of the linear monomeric PLMVd plus-strand RNA, several insertions and deletions were introduced at this terminus in variant esc5. For this purpose, monomeric constructs were obtained by cloning the products from PCR amplifications using the esc5 dimeric plasmid and pairs of primers delimiting the 5′ and 3′ borders of the PLMVd plus-strand self-cleavage site. The 5′ primer was the same in all cases and was preceded by the T7 promoter. The 3′ primers carried proper restriction sites for in vitro synthesis of monomeric transcripts with 3′ termini identical in length to, or slightly modified from, that resulting from self-cleavage. Following UV irradiation, the PLMVd RNA control with the 3′ terminus at the same position as that resulting from self-cleavage (but with a 3′-OH instead of a 2′,3′ cyclic phosphodiester produced by the hammerhead-mediated self-cleavage) generated the cross-linked species (Fig. 3, lanes 1), confirming our previous results and excluding the requirement for a 2′,3′ cyclic phosphodiester for cross-linking. However, PLMVd RNAs carrying a 2-nt insertion and 1-, 4-, or 15-nt deletions at their 3′ termini failed to produce the cross-linked species (Fig. 3, lanes 2 to 5). Only the 15-nt deletion is predicted to cause a partial rearrangement of the PLMVd molecule, while the effect of the remaining mutations on UV cross-linking should be specifically attributed to the altered nucleotides. These results further support the idea that the nucleotide forming the cross-link with U81 is located in, or highly proximal to, the 3′ terminus resulting from the hammerhead-mediated self-cleavage.
FIG. 3.
Analysis by denaturing PAGE of UV-irradiated monomeric linear PLMVd plus-strand RNAs with insertions and deletions at their 3′ terminus. Linear RNAs synthesized in vitro by transcription of different constructs derived from PLMVd variant esc5 were loaded on a nondenaturing gel, UV irradiated, and electrophoresed on a second denaturing gel (shown here) that was stained with ethidium bromide (A); the RNAs were then electrotransferred onto a nylon membrane and hybridized with a radioactive full-length PLMVd minus-strand riboprobe (B). Lanes 1, linear monomeric PLMVd plus-strand RNA with the 3′ terminus at the same position as that resulting from self-cleavage (but with a 3′-OH instead of a 2′,3′ cyclic phosphodiester). Lanes 2, the same RNA carrying a 2-nt insertion at the 3′ terminus. Lanes 3, 4, and 5, the same RNA with a 1-, 4-, or 15-nt deletion at the 3′ terminus, respectively. The arrowhead indicates the position of the PLMVd cross-linked form, and the region containing the non-cross-linked RNAs is delimited by bars. The overexposure of the Northern blot shown in panel B did not reveal the cross-linked species in any of the mutated RNAs.
Effects of UV irradiation on cross-linking of the PLMVd RNAs from different variants.
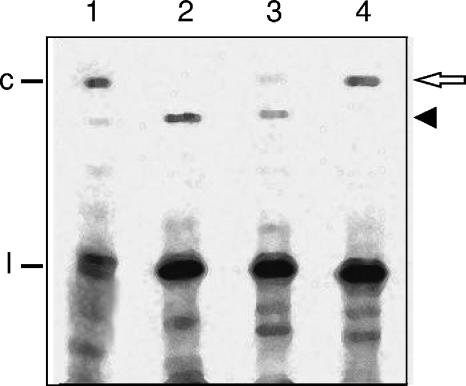
In a further attempt to uncover additional factors influencing the UV-induced cross-linking, the linear monomeric plus-strand RNAs resulting from self-cleavage of the three natural infectious PLMVd variants, esc5, ls1, and gds19 (accession numbers AJ005307, AJ005314, and AJ005301, respectively) (1), were UV irradiated and analyzed as described previously. Variants ls1 and gds19 differ in 19 nucleotide positions from each other and in 21 and 23 nucleotide positions, respectively, from variant esc5, some of which are located near the 3′ terminus of the linear monomeric plus-strand RNA resulting from self-cleavage. The PLMVd cross-linked species initially detected was generated with high, intermediate, and low yield in esc5, ls1, and gds19 variants, respectively (Fig. 4, lanes 2, 3, and 4), and was also visible in an RNA extract from PLMVd-infected leaves (Fig. 4, lane 1). However, a second PLMVd cross-linked species with an electrophoretic mobility in the denaturing gel indistinguishable from that of the PLMVd monomeric circular form was observed, with its relative yield in the three variants displaying an inverse trend with respect to the other cross-linked species (Fig. 4, lanes 2, 3, and 4). Elution of the cross-linked species with low mobility from PLMVd variant gds19, reverse transcription-PCR amplification with the overlapping primers PIII (Fig. 2A) and PVII [5′-d(TGTGATCCAGGTACCGCCGTAGAAACT-3′), homologous to positions 198 to 224], cloning, and sequencing yielded a cDNA with the complete viroid sequence except the nucleotides preceding and following the self-cleavage site that were deleted. This latter observation strongly supports the existence of an atypical linkage in or very close to the self-cleavage site. Therefore, following UV irradiation in vitro, the linear monomeric plus-strand RNAs of certain PLMVd variants are more prone to form one of two alternative cross-link species. In this context, nucleotides UGAG (positions 284 to 287), near the 3′ terminus of the monomeric linear PLMVd plus-strand RNAs resulting from self-cleavage, are complementary to nucleotides CUUG (positions 86 to 89) located in the same loop as U81 and might help bring positions 81 and 289 close enough to be UV cross-linked. Variants gds19 and ls1 have the change U284A, which would weaken the potential interaction, thus accounting for the reduced yield of the initially detected cross-linked species in these variants.
FIG. 4.
Analysis by denaturing PAGE and Northern blot hybridization of the UV-irradiated monomeric linear PLMVd plus-strand RNAs from different PLMVd variants. The RNAs were generated by self-cleavage of in vitro-synthesized dimeric transcripts, loaded on a nondenaturing gel, exposed to UV light, electrophoresed on a denaturing gel, and electrotransferred onto a nylon membrane that was hybridized with a radioactive PLMVd minus-strand riboprobe. Lane 1, RNAs extracted from PLMVd-infected leaves. Lanes 2, 3, and 4, monomeric linear RNAs corresponding to PLMVd variants esc5, ls1, and gds19, respectively. The solid arrowhead marks the position of the PLMVd cross-linked species initially detected and the open arrow that of an additional PLMVd cross-linked species. The locations of circular (c) and linear (l) PLMVd RNAs are indicated at the left.
Conclusions.
A new PLMVd RNA species, migrating between the circular and the linear forms of the viroid, has been detected after UV irradiation of RNA preparations from PLMVd-infected tissue and of the linear monomeric PLMVd plus-strand RNA from certain variants. Primer extension assays have identified U81, located in a loop of the predicted branched PLMVd secondary structure (1, 6, 20), as one of the cross-linked nucleotides. Interestingly, no changes have been reported in this position despite the high sequence heterogeneity reported in PLMVd (1, 2, 20, 23, 25, 27), thus supporting a biological relevance for U81. Although the second nucleotide involved in the UV-sensitive interaction has not been unequivocally determined, linear monomeric PLMVd plus-strand RNAs carrying small insertions or deletions at their 3′ terminus were not competent for cross-linking, strongly suggesting that the conserved 3′-terminal C289 arising from self-cleavage, or a very close one, is the other nucleotide implicated. This adds a different element of tertiary structure to the pseudoknot interaction between two kissing loops characterized previously in vitro (6). Assignment of a function to the UV-photo-cross-linkable element here identified would be premature at this stage. However, the recent finding that the transcription initiation site of PLMVd minus-strand RNA maps at a double-stranded motif of 6 to 7 bp containing the conserved C289 that precedes the self-cleavage site (9) suggests a possible role in initiation, although other possibilities cannot be excluded. The second cross-linked RNA species similar in size and sequence to the monomeric circular PLMVd form, observed in some PLMVd variants, might derive from a UV-induced ligation of the two termini resulting from self-cleavage. This reaction, reminiscent of that promoted by incubation in a high magnesium concentration leading to a 2′,5′ phosphodiester bond (7), would be facilitated by these variants being particularly liable to adopt in vitro a conformation like the thermodynamically most stable conformation wherein the two termini lie in physical proximity. Lack of the two nucleotides flanking the self-cleavage site in the reverse transcription-PCR-amplified product from the second UV-induced adduct is consistent with an atypical bond in this position.
Acknowledgments
This work has been partially supported by grant BFU2005-06808/BMC from the Ministerio de Educación y Ciencia of Spain (to R.F. and J.-A.D.). The laboratories of F.D. and R.F. have been jointly supported by the CNR-CSIC project 2004IT0028.
REFERENCES
- 1.Ambrós, S., C. Hernández, J. C. Desvignes, and R. Flores. 1998. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasispecies. J. Virol. 72:7397-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrós, S., C. Hernández, and R. Flores. 1999. Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J. Gen. Virol. 80:2239-2252. [DOI] [PubMed] [Google Scholar]
- 3.Baumstark, T., A. R. W. Schröder, and D. Riesner. 1997. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branch, A. D., B. J. Benenfeld, and H. D. Robertson. 1985. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. USA 82:6590-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussière, F., J. Lehoux, D. A. Thompson, L. J. Skrzeczkowski, and J. P. Perreault. 1999. Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J. Virol. 73:6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussière, F., J. Ouellet, F. Côté, D. Lévesque, and J. P. Perreault. 2000. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Côté, F., and J. P. Perreault. 1997. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol. 273:533-543. [DOI] [PubMed] [Google Scholar]
- 8.De la Peña, M., S. Gago, and R. Flores. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22:5561-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgado, S., E. Martínez de Alba, C. Hernández, and R. Flores. 2005. A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 79:12934-12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diener, T. O. 1972. Potato spindle tuber viroid VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 50:606-609. [DOI] [PubMed] [Google Scholar]
- 11.Diener, T. O. 2003. Discovering viroids—a personal perspective. Nat. Rev. Microbiol. 1:75-80. [DOI] [PubMed] [Google Scholar]
- 12.Doudna, J. A. 2000. Structural genomics of RNA. Nat. Struct. Biol. 7(Suppl.):954-956. [DOI] [PubMed] [Google Scholar]
- 13.Flores, R., N. Durán-Vila, V. Pallás, and J. S. Semancik. 1985. Detection of viroid and viroid-like RNAs from grapevine. J. Gen. Virol. 66:2095-2102. [Google Scholar]
- 14.Flores, R., C. Hernández, A. E. Martínez de Alba, J. A. Daròs, and F. Di Serio. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 15.Flores, R., J. W. Randles, M. Bar-Joseph, R. A. Owens, and T. O. Diener. 2005. Viroidae, p. 1145-1159. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and A. L. Ball (ed.), Virus taxonomy, eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 16.Gago, S., M. De la Peña, and R. Flores. 2005. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA 11:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gast, F. U., D. Kempe, R. L. Spieker, and H. L. Sänger. 1996. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J. Mol. Biol. 262:652-670. [DOI] [PubMed] [Google Scholar]
- 18.Gesteland, R. F., T. R. Cech, and J. F. Atkins. 2005. The RNA world, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Gross, H. J., H. Domdey, C. Lossow, P. Jank, M. Raba, H. Alberty, and H. L. Sänger. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273:203-208. [DOI] [PubMed] [Google Scholar]
- 20.Hernández, C., and R. Flores. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 89:3711-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue, T., and T. R. Cech. 1985. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. USA 82:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khvorova, A., A. Lescoute, E. Westhof, and S. D. Jayasena. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10:708-712. [DOI] [PubMed] [Google Scholar]
- 23.Malfitano, M., F. Di Serio, L. Covelli, A. Ragozzino, C. Hernández, and R. Flores. 2003. Peach latent mosaic viroid variants inducing peach calico contain a characteristic insertion that is responsible for this symptomatology. Virology 313:492-501. [DOI] [PubMed] [Google Scholar]
- 24.Navarro, B., and R. Flores. 1997. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 94:11262-11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelchat, M., D. Levesque, J. Ouellet, S. Laurendeau, S. Levesque, J. Lehoux, D. A. Thompson, K. C. Eastwell, L. J. Skrzeczkowski, and J. P. Perreault. 2000. Sequencing of peach latent mosaic viroid variants from nine North American peach cultivars shows that this RNA folds into a complex secondary structure. Virology 271:37-45. [DOI] [PubMed] [Google Scholar]
- 26.Qi, Y., and B. Ding. 2003. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15:1360-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodio, M. E., S. Delgado, R. Flores, and F. Di Serio. 2006. Variants of peach latent mosaic viroid inducing peach calico: uneven distribution in infected plants and requirements of the insertion containing the pathogenicity determinant. J. Gen. Virol. 87:231-240. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, H., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Tabler, M., and M. Tsagris. 2004. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 9:339-348. [DOI] [PubMed] [Google Scholar]
- 30.Wassenegger, M., R. L. Spieker, S. Thalmeir, F. U. Gast, L. Riedel, and H. L. Sänger. 1996. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226:191-197. [DOI] [PubMed] [Google Scholar]