Abstract
Mouse mammary tumor virus (MMTV) induces breast cancer with almost 100% efficiency in susceptible strains through insertional activation of protooncogenes, such as members of the wnt and fibroblast growth factor (fgf) families. We previously showed that expression of the MMTV envelope protein (Env) in normal immortalized mammary epithelial cells grown in three-dimensional cultures caused their morphological transformation, and that this phenotype depended on an immunoreceptor tyrosine-based activation motif (ITAM) present in Env and signaling through the Syk tyrosine kinase (E. Katz, M. H. Lareef, J. C. Rassa, S. M. Grande, L. B. King, J. Russo, S. R. Ross, and J. G. Monroe, J. Exp. Med. 201:431-439, 2005). Here, we examined the role of the Env protein in virus-induced mammary tumorigenesis in vivo. Similar to the effect seen in vitro, Env expression in the mammary glands of transgenic mice bearing either full-length wild-type provirus or only Env transgenes showed increased lobuloalveolar budding. Introduction of the ITAM mutation into the env of an infectious, replication-competent MMTV or into MMTV/murine leukemia virus pseudotypes had no effect on incorporation of Env into virus particles or on in vitro infectivity. Moreover, replication-competent MMTV bearing the ITAM mutation in Env infected lymphoid and mammary tissue at the same level as wild-type MMTV and was transmitted through milk. However, mammary tumor induction was greatly attenuated, and the pattern of oncogene activation was altered. Taken together, these studies indicate that the MMTV Env protein participates in mammary epithelial cell transformation in vivo and that this requires a functional ITAM in the envelope protein.
The nonacute transforming betaretrovirus mouse mammary tumor virus (MMTV), which induces mammary carcinomas in mice, has long been used as an in vivo model for the study of virus-induced cancer. The major route of MMTV infection in vivo is through milk, and mice can be freed of virus by foster nursing on uninfected mothers (42). MMTV-induced transformation is mediated by proviral integration near cellular oncogenes in mammary epithelial cells, and a number of different integration sites (int genes) have been implicated in this process, including the wnt1 and fgf3 oncogenes (3). Surprisingly, although integration near protooncogenes is critical to transformation, the latency and incidence of tumor induction after MMTV infection is similar to that of transgenic mice bearing wnt1, fgf3, or other oncogenes under the control of the MMTV long terminal repeat (LTR) (8, 38-40, 57). Moreover, like virus-infected mice, the transgenic mice develop only one to two clonal mammary tumors. Given that most, if not all, of the mammary cells of transgenic mice express the oncogene while oncogene activation in MMTV-infected mice occurs by stochastic integration of the provirus, this similarity in pathogenesis between the two models suggests that the virus itself contributes to tumorigenesis.
Indeed, recent evidence suggests that the MMTV envelope (Env) protein participates in mammary epithelial cell transformation. Like other retroviruses, the MMTV Env consists of two subunits generated by proteolytic processing of a polyprotein precursor to cell surface (SU) and transmembrane (TM) domains (59); both subunits are required for virus entry via the entry receptor, transferrin receptor 1 (TfR1) (50). However, the Env protein appears to have additional roles. Env expression in the absence of other viral proteins in immortalized normal mammary epithelial cells caused them to bear multiple hallmarks of cell transformation, including a depolarized acinar morphology and markers typical of epithelial-to-mesenchymal transition (26). This activity depended on a functional immunoreceptor tyrosine activation motif (ITAM) in the SU domain. ITAMs are highly conserved sequences found in receptors involved in the activation, proliferation, survival, and differentiation of hematopoietic cells such as B and T lymphocytes, mast cells, platelets, and natural killer cells (13, 37). The tyrosine residues found in the canonical motif D(xx)Yxx(L/I)x6-12Yxx(L/I) are necessary and sufficient for signaling, and it is well established that mutation of the two tyrosine residues to phenylalanine in ITAM-containing molecules abrogates signaling without altering protein biosynthesis and folding (24). After ligand binding, the two tyrosine residues in ITAMs are phosphorylated by protein tyrosine kinases and function as docking sites for SH2-containing signaling proteins, linking receptor-initiated signals to downstream cellular responses. Indeed, wild-type Env protein was shown to bind in pervanadate-treated cells, and mutation of the two tyrosine residues abrogated its abilities to bind Syk and to transform mammary epithelial cells (26). Morphological transformation was phenocopied in cells constitutively expressing the B-cell receptor ITAM and was reversed by treatment of cells expressing either the MMTV Env or B-cell receptor ITAM-containing proteins with Syk or Src family tyrosine kinase inhibitors (19, 26), indicating that signaling through the ITAM was critical to phenotype induction.
Regulation of signaling in hematopoietic cells is the result of a complex balance between Src and Syk family kinases and protein tyrosine phosphatases such as SHP-1 and SHP-2, which are located in the cytoplasm until they are recruited to regions of the plasma membrane occupied by ITAM-containing proteins by a second set of hematopoietic-restricted transmembrane proteins with cytoplasmic immunoreceptor tyrosine inhibitory motifs (13, 37). ITAM-containing proteins also generate signals independent of ligand triggering; these “tonic” signals require only that the ITAM-containing proteins be present in the plasma membrane (1). Expression of ITAM-containing plasma membrane proteins in cells where inhibitory regulatory proteins are not present could lead to deregulated, uncontrolled signaling, and thus expression of the MMTV Env in infected mammary epithelial cells would play a role in virus-induced breast cancer.
Here, we examined whether the Env ITAM motif affected MMTV infection in vivo and show that it plays a critical role in virus-induced breast cancer.
MATERIALS AND METHODS
Plasmids.
The wild-type pEnv (16) and pEnv-Y1Y2 plasmids (26) as well as HP (53) and LEL (14) constructs were all previously described. Site-directed mutagenesis to introduce the ITAM mutation into the HP plasmid to generate HP-Y1Y2 and mutation into pEnv was performed using the QuickChange XL Site-Directed Mutagenesis kit (Stratagene, Inc.) as previously described (26); the sequence of the mutagenic oligonucleotides are available upon request.
Cells.
293T human kidney epithelial cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and penicillin-streptomycin (50 mg/ml). TRH3 cells (293T cells stably expressing mouse TfR1 [64]) and NMuMG (normal murine mammary gland; ATCC CRL-1636) cells were grown in the same medium supplemented with Geneticin (100 mg/ml) or 10 mg/ml insulin, respectively. NMuMG cells stably expressing the HP and HP-Y1Y2 molecular clones were generated by cotransfection with a plasmid containing the neomycin resistance gene (pSV2-neo) (55) followed by selection in medium containing Geneticin (100 μg/ml).
Virus preparation, infection, and Western blotting.
MMTV Env-pseudotyped murine leukemia virus (MLV) recombinant virions containing the β-galactosidase gene under the control of the murine leukemia virus long terminal repeat (54) were prepared in 293T cells and used for infection as previously described (16). Infection was quantified by in situ staining of lacZ-positive colonies; the Lac-forming units/milliliter of each virus preparation was determined. HP and HP-Y1Y2 virions were harvested from the supernatants of NMuMG transfectants (induced with dexamethasone [0.5 μM] overnight) by ultracentrifugation at 32,000 rpm for 2 h in an SW50.1 rotor. Equal amounts of virions or cell lysates were resolved on 10% denaturing polyacrylamide gels. The proteins were transferred to nitrocellulose membrane and probed with polyclonal goat anti-MMTV antiserum as previously described (9).
Infection of mice with HP and HP-Y1Y2 molecular clones.
Three- to 4-week-old BALB/c female mice purchased from the National Cancer Institute were injected with 107 NMuMG cells expressing the HP and HP-Y1Y2 molecular constructs, as described previously (53). All injected females were bred with BALB/c males to generate pedigrees of HP- and HP-Y1Y2-infected mice. MMTV-infected mice were palpated weekly starting at 5 months of age and sacrificed when tumors were less that 1 cm in diameter. Kaplan-Meier survival curves were computed for age at tumor onset. The log-rank statistic was used to test for a significant difference between the two viruses. All mice were housed according to the policies of the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Transgenic mice.
HP transgenic mice in the C3H/HeN background were previously described (14). The LEL transgenic mice were made by the Transgenic and Chimeric Mouse Facility of the University of Pennsylvania School of Medicine. Both HP and LEL strains were carried by breeding heterozygous transgenic males to C3H/HeN females, obtained from the Animal Program of the National Cancer Institute. MMTV-Wnt1 transgenic mice were obtained from the Jackson Laboratory and were maintained by breeding with FVB females.
Histopathology.
C3H/HeN, HP, LEL, and MMTV-Wnt1 transgenic mice of similar ages, as indicated, were injected intraperitoneally with bromodeoxyuridine (BrdU) (1 mg/g body weight) 2 h prior to sacrifice. Mammary gland 3 was used for whole-mount/carmine red staining (49). Photographs of the whole mounts were analyzed for the average number of buds and branches per given area in a blinded fashion. Mammary gland 4 was fixed in formaldehyde, and paraffin-embedded sections were used to detect BrdU incorporation using mouse anti-BrdU antibodies (BD Biosciences) (30). The slides were analyzed by the Transgenic Laboratory at the University of California at Davis.
PCR to detect integrated exogenous viral DNA.
To detect newly integrated copies of exogenous MMTV (C3H), DNA isolated from spleens and thymi of mice infected by milk-borne HP or HP-Y1Y2 virus was amplified by semiquantitative PCR using HP LTR primers, as previously described (15). Simultaneously, the DNA was amplified with universal MMTV primers that amplify the endogenous viruses present in the mouse gene. Both sets of amplification products were analyzed on 1.5% agarose gels, blotted to nitrocellulose, and hybridized with an LTR-specific probe. The gels were subjected to PhosphorImager analysis (Molecular Dynamics STORM scanner 860), and the bands were quantified using ImageQuant 5.2 (Molecular Dynamics). Data are presented as the relative amount of HP-specific DNA normalized to the endogenous MMTV DNA.
RNase protection assays.
RNase T1 protection assays were performed as previously described (14) using a probe specific for MMTV (C3H) viral transcripts (17) and for mouse β-actin (Ambion, Inc.). Twenty micrograms of total RNA isolated from spleen, 40 μg from virgin mammary glands, and 5 μg from the milk of mice infected by milk-borne HP or HP-Y1Y2 virus were used. The gels were dried and scanned with a Molecular Dynamics Storm PhosphorImager and analyzed with Molecular Dynamics ImageQuant 5.2 software. Data are presented as relative expression of MMTV normalized to β-actin.
RT-PCR to detect common integration site expression.
Total RNA was isolated from tumors using the Trizol Reagent (Invitrogen, Inc.) according to the manufacturer's instructions. Two micrograms of total RNA was used to generate cDNA using Superscript II Reverse Transcriptase (Invitrogen, Inc.) according to the manufacturer's instructions. Radioactive PCR was performed using Platinum Taq Polymerase, 1.0 mCi [α-32P]ATP, and 4 pmol of the following primers: wnt1 forward, CCACCTCTTCGGCAAGATCGTCAA; wnt1 reverse, GTGGCATTTGCACTCTTGGCGCAT; fgf3 forward, GGAGATTACTGCGGTGGAAGTGGG; fgf3 reverse, CTTTTGTGTGCGGCGGGTCTTGAA; ActB forward, CTCCTATCGTGAGTCCGTTCCGCT; ActB reverse, TGGATGGCTACGTACATGGCTGGG. The amplification conditions for all PCRs (except for that of the primers) consisted of initial denaturation at 94°C for 2 min, denaturation at 94°C for 30 s, annealing at 55°C or 50°C for 30 s, and extension at 72°C for 30 s for 35 cycles. The amplification conditions for the ActB primers consisted of initial denaturation at 94°C for 2 min, denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s for 21 cycles. The reverse transcription-PCR (RT-PCR) products were resolved on 5% acrylamide gels, dried, and scanned with a Molecular Dynamics Storm PhosphorImager; all samples with intact RNA, as determined by a visible mouse β-actin signal, were scored for wnt1 and fgf3 expression. Pearson's chi-square statistic or two-sided Fisher's exact test was used to test whether the proportions of tumors that expressed each gene were the same for HP- and YIY2-infected mice; Fisher's exact test was used when the chi-square test was not appropriate due to small samples sizes.
T-cell deletion.
Peripheral blood lymphocytes isolated at the times indicated were stained with fluorescein isothiocyanate-conjugated anti-Vβ14 and phycoerythrin-conjugated anti-CD4 antibodies (BD Biosciences, Inc.) and analyzed by flow cytometry. Cells were acquired on a FACScan cytometer (Becton Dickinson, Mountainview, CA) and analyzed using CellQuest software (Becton Dickinson Immunocytometry Systems).
Statistics.
Statistical analyses were done using SAS/STAT software, version 9, of the SAS System for Windows.
RESULTS
Transgenic mice that express full-length MMTV or MMTV Env alone have altered mammary tissue morphology.
We first determined whether the Env gene affected mammary cell differentiation in vivo. A number of years ago we created HP transgenic mice in the C3H/HeN background. The HP MMTV molecular clone is not tumorigenic in C3H/HeN mice (14, 21), and thus transgenic mice in the C3H/HeN background develop tumors at very low incidence (<1%) and with long latency (>400 days) (data not shown). To determine whether MMTV altered mammary gland morphology when expressed as a transgene, histological analysis of HP mammary gland whole mounts was performed (Fig. 1). Shown are the virgin mammary glands of a 2-month-old transgenic mouse and a nontransgenic littermate from a mating between a heterozygous transgenic male and a nontransgenic female (no virus was acquired through milk). These analyses revealed a dramatic increase in the number of lobuloalveolar buds in HP transgenic mice compared to their nontransgenic littermates (compare C3H/HeN to HP in Fig. 1). This difference in lobuloalveolar differentiation persisted in older mice (data not shown).
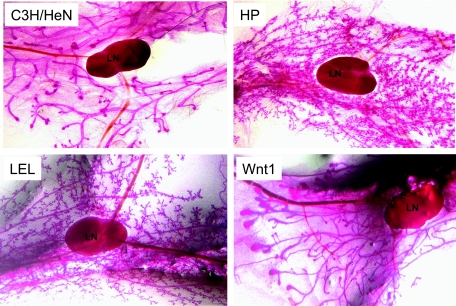
FIG. 1.
Whole-mount analysis of mammary glands from 2-month-old virgin C3H/HeN, MMTV-Wnt1, LEL, and HP mice. All pictures were taken at ×4 magnification; the lymph nodes (LN) are included in each picture for comparison.
To confirm that these morphological changes were due to Env expression, we created two independent strains of transgenic mice that expressed only the Env gene under the control of the MMTV LTR (Fig. 1) (14). Both strains of LEL mice showed the typical pattern of MMTV transgene expression in lymphoid, salivary gland, prostate/seminal vesicle, and mammary tissue (5) (data not shown). The virgin mammary glands of the LEL strains showed the same morphological changes as did those from the HP mice (Fig. 1). Thus, the MMTV Env protein has the ability to alter the morphology of mammary tissue in vivo as well as in vitro (26). LEL and HP transgenic mice consistently showed this phenotype; approximately 17% of nontransgenic mice (2 of 12) also demonstrated increased alveolar budding of unknown etiology (data not shown). To determine if Env expression caused changes in mammary gland morphology similar to those induced by a known oncogene activated by MMTV insertion, we also examined the mammary glands of age-matched, virgin MMTV-Wnt1 transgenic mice (57). In contrast to the LEL or HP mice, MMTV-Wnt1 transgenic mice did not show increased numbers of buds but instead had much larger end buds (Fig. 1).
We also examined BrdU incorporation in the mammary tissues of the HP and LEL transgenic mice to determine if transgene expression affected mammary cell proliferation. In both cases, there was no increase in the number of BrDU-positive cells (data not shown); in contrast, MMTV-Wnt1 transgenic mice showed a significant increase in mammary epithelial cell proliferation, as previously described (30). Thus, Env expression in mammary epithelial cells in vivo appeared to alter their morphology without affecting cell proliferation. This is similar to the phenotype we described for in vitro studies, where Env expression affected mammary cell differentiation but not proliferation (26).
Mutation of the SU ITAM does not affect pseudovirus infection or virion production in cultured cells.
It is well established that mutation of the two Y residues to F in cellular or viral ITAMs preserves protein structure but abrogates signaling by such proteins (24). Indeed, we showed that introduction of the mutations Yaa418, aa428→F (Y1Y2) into the wild-type MMTV envelope expression plasmid pEnv did not affect protein expression or its ability to traffic to the cell surface (26). However, YXXL/I motifs are often used for protein sorting, and given its sequence similarity to the ITAM, its mutation could affect its incorporation into virions and thereby reduce infectivity. To test this, we prepared MLV pseudotypes in 293T cells (16) bearing the wild-type and mutant Env proteins. The producer 293T cells all expressed similar levels of the different Env proteins (data not shown). The pseudoviruses were then used to infect NMuMG or TRH3 cells (293T cells stably expressing the mouse transferrin receptor) (50). The Env mutant pseudoviruses infected both NMuMG and TRH3 cells with efficiencies similar to those of the virus bearing the wild-type Env (Fig. 2A, shown for TRH3 cells). A construct bearing a Yaa418→I mutation was also tested; although expression of this mutant Env was similar to that of the wild type, this change dramatically reduced virus titers (data not shown), indicating that structural conservation of this region is important for infection.
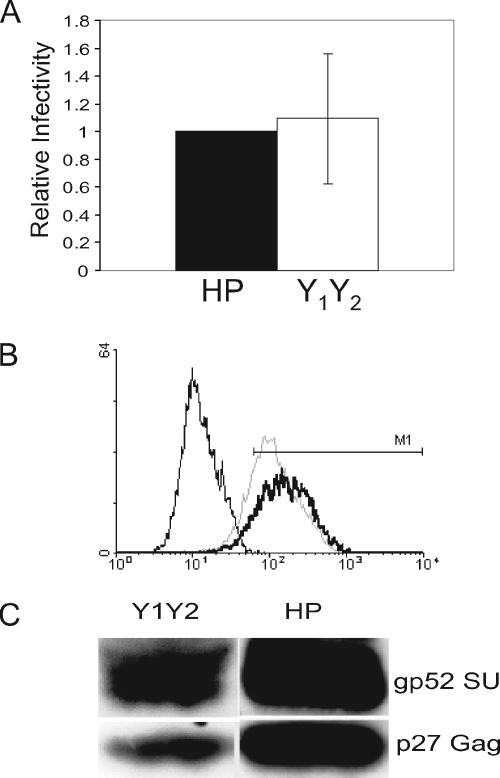
FIG. 2.
Mutation of the Y residues in the SU ITAM does not alter pseudovirus infectivity or Env incorporation into virions. (A) Pseudovirus titers on TRH3 cells of the particles bearing wild-type and mutant Env proteins. Shown is the average of three independent experiments (infectivity of pseudoviruses bearing the mutant Env normalized to the wild-type Env). (B) Fluorescence-activated cell sorting analysis of HP- (dark line) and HP-Y1Y2- (dotted line) transfected NMuMG cells using anti-MMTV Env antisera; unstained cells are shown by the thin line. The mean fluorescense intensity of cells within the M1 window was 183 for HP and 142 for HP-Y1Y2. Western blot analysis of virus particles isolated from HP- and HP-Y1Y2-transfected NMuMG cell supernatants.
We also introduced these same Y→F mutations into an infectious MMTV molecular clone, HP (53), and stably transfected these constructs into NMuMG cells; the cells expressed similar amounts of Env on their surface, indicating that the protein was processed (Fig. 2B). Supernatants were harvested from these cultures and pelleted by high-speed centrifugation. The virions and cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis using polyclonal anti-MMTV or SU antisera. Env levels on virions were proportional to the amount of capsid proteins (Fig. 2C), indicating that mutation of the Y residues did not affect its incorporation into virions.
Thus, mutation of the tyrosine residues did not have any significant effect on viral protein expression, stability, or incorporation into virions. Moreover, because the pseudovirus titers of mutants were similar to those of the wild type, the ITAM mutations did not appear to affect receptor binding or entry.
Mutation of the tyrosine residues in the SU ITAM does not affect initial in vivo infection of lymphocytes.
During milk-borne MMTV transmission, virus infection is initially established in lymphocytes resident in the Peyer's patches and mesenteric lymph nodes. These infected lymphocytes then traffic to peripheral lymphoid organs and eventually to the pubescent mammary gland (10, 15). To determine whether the ITAM mutation altered the ability of MMTVs to infect lymphocytes, we inoculated 10 3-week-old BALB/c females with NMuMG cells expressing the HP wild type or HP-Y1Y2 mutant molecular clones. This well-established method for infecting mice with recombinant MMTVs allows efficient infection of both lymphocytes and mammary cells undergoing cell division at puberty (53, 62). When the transfected cells are from a different species (i.e., rat) or inbred background (NMuMG are NAMRU strain), they are immunologically rejected, but this occurs after transfer of the virus to the host.
We first examined Sag-cognate T-cell deletion in the inoculated animals for evidence of virus infection. MMTV-infected mice show deletion of CD4+ T cells that recognize the virally encoded Sag protein and the kinetics and extent of deletion reflect virus load (18); the HP-encoded Sag causes deletion of Vβ14+ T cells. At both 6 and 16 weeks after infection, there was no difference in Vβ14+ T-cell levels (BALB-HP and BALB-HP-Y1Y2; shown for the 16-week time point in Table 1).
TABLE 1.
Sag-mediated deletion of cognate T cells in wild-type- and ITAM mutant-infected micea
| Mouse and infection route | Vβ14+/CD4+ content (%) | n | Age (wk) |
|---|---|---|---|
| Uninfected | |||
| BALB/c | 8.0 ± 0.67 | 7 | NA |
| Injected | |||
| BALB-HP | 4.57 ± 1.2 | 10 | 16 |
| BALB-HP-Y1Y2 | 5.08 ± 1.1 | 10 | 16 |
| Milk borne | |||
| BALB f. BALB-HP | 3.59 ± 0.68 | 3 | 23 |
| BALB f. BALB-HP-Y1Y2 | 4.36 ± 0.23 | 3 | 23 |
Mice were bled at the indicated age, and the percentages of Vβ14+/CD4+ T cells were determined by fluorescence-activated cell sorter analysis. f., foster-nursed; NA, not applicable.
We next tested whether milk-borne transmission of MMTV and Sag-mediated T-cell deletion was affected by the ITAM mutation. The HP- and HP-Y1Y2-virus-injected mice were mated, and Vβ14+ T-cell deletion in their offspring was examined. The extent of deletion of Sag-cognate T cells was identical in both sets of mice (shown for 23-week-old BALB f. BALB-HP and BALB f. BALB-HP-Y1Y2 mice; Table 1).
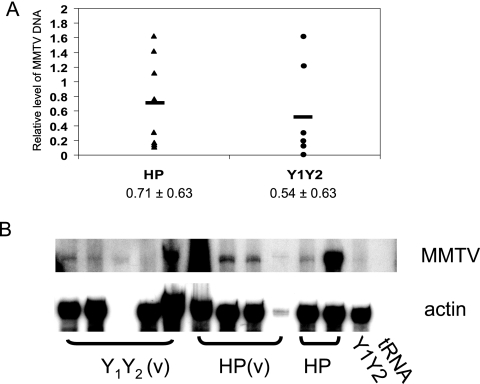
We then sacrificed groups of age-matched virgin mice infected by milk-borne MMTV and examined their spleens for evidence of infection by semiquantitative PCR for integrated virus DNA (Fig. 3A) and for MMTV-specific transcripts by RNase protection assays (Fig. 3B). Although there was some variation in the level of infection between individual mice infected with either virus, the average infection level of the HP-Y1Y2-infected mice was equivalent to that of the wild-type HP-infected mice. Thus, both lymphocyte infection and, importantly, milk-borne transmission appeared not to be impaired by mutation of the MMTV Env ITAM.
FIG. 3.
Infection of lymphoid tissue. A. Provirus integration was determined by semiquantitative PCR with DNA isolated from the spleens of age-matched mice infected by milk-borne HP or HP-Y1Y2 virus. Presented are the relative expression levels in individual mice (see Materials and Methods); shown below are the average values for each group. B. RNA analysis. RNA isolated from the spleens was subjected to RNase protection analysis as previously described (17). Shown are the results from splenic RNA isolated from virgin [HP(v) and Y1Y2(v)] and multiparous (HP and Y1Y2) females.
Mutation of the SU ITAM does not affect mammary gland infection.
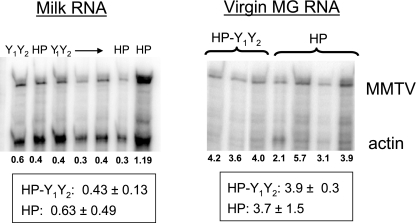
The primary source of milk-borne MMTV is mammary epithelial cells, which produce and secrete virus during lactation. As described in the preceding section, milk-borne transmission of the mutant HP-Y1Y2 virus was not impaired, indicating that mammary gland infection was also not affected. To directly test this, we examined virgin mammary gland virus expression of MMTV RNA as well as the levels of milk-borne virus produced by lactating HP and HP-Y1Y2 females infected through milk, using an RNase protection assay specific for the HP virus (Fig. 4). The mammary glands of the wild-type and mutant mice expressed similar levels of MMTV RNA and produced equivalent amounts of virus, indicating that the mutation had little or no effect on mammary gland infection.
FIG. 4.
Mammary gland infection. RNA isolated from the virus fraction of milk or from virgin mammary tissue of age-matched mice infected by milk-borne HP or HP-Y1Y2 virus was subjected to RNase protection analysis, as described in the legend to Fig. 3. Shown below each lane is the MMTV signal normalized to β-actin; the boxed values are the average normalized MMTV signal for each group of infected mice. MG, mammary gland.
The ITAM mutant virus is attenuated for tumor induction.
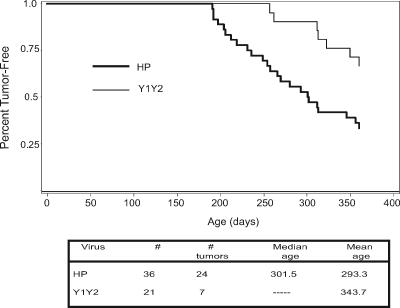
We next determined whether Env-mediated signaling through the ITAM was important for MMTV-induced mammary tumorigenesis. Mice that received milk-borne virus with the wild-type or ITAM mutant Envs were force bred and monitored for tumor formation. Compared to mice that received the wild-type HP virus, mice infected with the HP-Y1Y2 mutant virus had a much longer time to mean tumor formation (delayed by almost 2 months) (Fig. 5). Moreover, by the age of 365 days, when only 33% of the HP-infected mice were tumor free, 52% of the HP-Y1Y2-infected mice still had not developed tumors.
FIG. 5.
Mammary tumor incidence induced by HP and HP-Y1Y2 viruses. Mice infected by milk-borne HP (n = 36) or HP-Y1Y2 (n = 21) virus were force bred and monitored for mammary tumors by weekly palpation. Shown are Kaplan-Meier survival curves that were computed for age at tumor onset. The P value for the test in which these two curves are significantly different (null hypothesis of equality) was 0.0066. Shown in the table is the mean and median time to mammary tumor development for the two groups; numbers of mice infected with the mutant virus that had developed tumors at 1 year were insufficient to compute the median.
Retroviruses commonly undergo recombination when RNAs from different viruses are copackaged into the same particle, and we previously showed that exogenous MMTV recombines with endogenous proviruses expressed in the mammary gland (17). Although we used BALB/c mice that do not express any full-length endogenous proviruses in mammary tissue for these tumorigenesis studies, it was possible that reversion of the ITAM mutant viruses to wild type occurred through recombination with endogenous proviruses expressed in lymphoid tissue. We therefore made cDNA from the RNAs of four independent HP-Y1Y2-induced tumors (one from a virus-injected mouse, the others from three offspring of different infected mothers) and amplified the ITAM region of env using a 3′ primer specific for the HP LTR in conjunction with a 5′ primer that hybridizes to all MMTV env genes. The sequences of the tumor cDNAs showed that all had retained the Y1Y2 mutation (data not shown). Thus, the ITAM mutants were capable of mammary tumor induction, albeit with greatly delayed kinetics.
ITAM mutant viruses show altered oncogene activation.
It is well established that MMTV causes tumors by integrating next to protooncogenes, such as wnt1 and fgf3, thereby turning on their expression in mammary epithelial cells. We therefore examined expression of these two oncogenes to determine whether the integration site frequency was similar in the wild-type- and ITAM mutant-induced tumors. The wild-type HP virus turned on expression of wnt1 and fgf3 in 56% and 78% of tumors, respectively, similar to published data (25) (Table 2). In contrast, tumors induced by the HP-Y1Y2 virus showed lower turn on of both oncogenes (29% and 43%, respectively), with a statistically significant difference in the activation of expression of fgf3. Moreover, there was a statistically significant difference in the percentage of tumors in which both oncogenes were activated. Only one tumor (7%) induced by the HP-Y1Y2 virus, in contrast to eight tumors (44%) induced by the wild-type HP virus, fell into this category. Finally, 36% versus 11% of HP-Y1Y2- and HP-induced tumors, respectively, showed activation of neither gene, although because of the small number of analyzed tumors, these differences are not statistically significant. These data suggest that the ITAM mutant causes transformation by activating different protooncogenes than wild-type MMTV.
TABLE 2.
Expression analysis of MMTV common integration site genes in wild-type- and ITAM mutant-induced tumorsa
| Virus | wnt1 total | wnt1 alone | fgf3 total | fgf3 alone | wnt1+fgf3+ | wnt1 and fgf3 negative | Time to tumor (days) |
|---|---|---|---|---|---|---|---|
| HPb | |||||||
| 1 | + | − | + | − | + | − | 188 |
| 2 | + | − | + | − | + | − | 188 |
| 3 | + | − | + | − | + | − | 194 |
| 4 | + | − | + | − | + | − | 195 |
| 5 | + | − | + | − | + | − | 240 |
| 6 | − | − | + | + | − | − | 251 |
| 7 | − | − | + | + | − | − | 251 |
| 8 | − | − | + | + | − | − | 265 |
| 9 | + | − | + | − | + | − | 270 |
| 10 | − | − | + | + | − | − | 270 |
| 11 | + | − | + | − | + | − | 284 |
| 12 | + | + | − | − | − | − | 313 |
| 13 | − | − | + | + | − | − | 317 |
| 14 | − | − | − | − | − | + | 337 |
| 15 | + | − | + | − | + | − | 351 |
| 16 | − | − | − | − | − | + | 405 |
| 17 | − | − | + | + | − | − | 455 |
| 18 | + | + | − | − | − | − | UN |
| Y1Y2c | |||||||
| 1 | − | − | + | + | − | − | 304 |
| 2 | + | − | + | − | + | − | 313 |
| 3 | − | − | + | + | − | − | 317 |
| 4 | − | − | − | − | − | + | 390 |
| 5 | + | + | − | − | − | − | 390 |
| 6 | − | − | − | − | − | + | 405 |
| 7 | − | − | + | + | − | − | 405 |
| 8 | − | − | − | − | − | + | 412 |
| 9 | + | + | − | − | − | − | 422 |
| 10 | − | − | + | + | − | − | 424 |
| 11 | − | − | + | + | − | − | 443 |
| 12 | − | − | − | − | − | + | 470 |
| 13 | + | + | − | − | − | − | 470 |
| 14 | − | − | − | − | − | + | 525 |
Tumors were analyzed for expression of the indicated oncogenes by RT-PCR as described in Materials and Methods. UN, unknown.
The expression turn-on percentages for HP viruses were the following: wnt1 total, 55.6; wnt1 alone, 11.1; fgf3 total, 77.8; fgf3 alone, 44.4; wnt1 and fgf3 positive, 33.3; wnt1 and fgf3 negative, 11.1. The expression turn-on percentages for Y1Y2 viruses were the following: wnt1 total, 28.6; wnt1 alone, 21.4; fgf3 total, 42.9; fgf3 alone, 35.7; wnt1 and fgf3 positive, 6.7; wnt1 and fgf3 negative, 35.7.
P values for HP versus Y1Y2 viruses were the following: wnt1 total, 0.127 (Pearson's chi-square statistic); wnt1 alone, 0.631 (two-sided Fisher's exact test); fgf3 total, 0.043 (Pearson's chi-square statistic); fgf3 alone, 1.000 (two-sided Fisher's exact test); wnt1 and fgf3 positive, 0.044 (Pearson's chi-square statistic); wnt1 and fgf3 negative, 0.195 (two-sided Fisher's exact test).
DISCUSSION
Because the focus over the past 25 years has been on retroviral integration as the mechanism of tumor induction, including recent high-throughput analyses of integration sites (31, 35, 63), much less is known about the early steps in nonacute transforming retrovirus-induced cancers. Studies on the betaretrovirus Jaagsietke sheep retrovirus, which is related to MMTV and causes lung adenocarcinomas, have shown that its Env protein behaves as an oncogene (32, 48, 61), although it is also possible that insertional mutagenesis is required for transformation (7). Additionally, it is well established that murine leukemia viruses have effects on hematopoiesis long before proviral integration into specific oncogenes occurs, although the mechanism by which most of these virus-induced changes occur is not known (43, 52). MMTV, which has an easily definable target (the mammary epithelial cell) and whose natural pathway of infection is readily studied in its natural host, provides us with a tool to help in understanding these early events in transformation. We show here that an early step in MMTV-induced mammary tumorigenesis likely requires signaling through an ITAM in the Env protein that alters mammary gland differentiation/morphology.
The ITAM sequences in Env are located in the SU region of the protein, generally believed to be outside the cell membrane. However, there is evidence that the MMTV SU can exist as a transmembrane protein (45) and that it resides in lipid rafts (data not shown). Moreover, we previously demonstrated that Env interaction with Syk kinase, a cytoplasmic protein, depended on the ITAM phosphotyrosines (26), indicating that this motif resides in the cytoplasm in some Env conformations. These data predict that the MMTV Env, like other membrane proteins found in Newcastle disease virus (33), the adenovirus E3-6.7K protein (36), and hepatitis B virus (47), can assume multiple conformations and that these may serve different biological functions.
MMTV induces tumors through insertion site activation, and the wnt1 oncogene that is commonly activated in mammary tumors was the first integration site to be cloned (44). Although it is clear the MMTV requires insertional activation for full-blown tumorigenesis, previous studies had indicated that lobuloalveolar differentiation was significantly increased in MMTV-infected mice and that this effect was separable from transformation (56). In our previous studies (26) and the work presented here, we clearly show that the virus itself has profound effects on mammary cell morphology and that this is most likely due to signaling through an ITAM motif in the Env protein, since abrogation of signaling by mutating the required tyrosines to phenylalanine alters both its ability to morphologically transform cells (26) and its ability to induce tumors. Moreover, our previous data demonstrated that an identical morphological transformation of mammary epithelial cells occurred when cells expressed either the MMTV Env or a nonviral ITAM and was reversed by inhibitors of Syk or Src family kinases, known downstream effectors of ITAM signaling (19, 26).
The results presented here also show that the Env ITAM plays a role in MMTV-mediated mammary tumorigenesis. Although ITAM negative viruses induced tumors, the time to tumor formation was greatly increased and the rate of incidence was decreased, at least within the 1-year period examined here. Moreover, examination of the common integration sites (wnt1 and fgf3) in these HP-Y1Y2-induced tumors demonstrated a lower frequency of activation, indicating that there may be selection for activation of alternate oncogenes/pathways. This potential change in the selection of different integration sites further indicates that Env ITAM plays a role in mammary gland transformation.
A number of oncogenic viruses, some with tropism for nonhematopoietic cells, encode ITAM-containing plasma membrane-associated proteins that play a role in their ability to transform cells. These include Epstein-Barr virus LMP2A (12, 34), Kaposi's sarcoma virus K1 (28, 29, 46), and bovine leukemia virus gp30 (2, 4, 60). The spacing between the aspartate and tyrosine residues in the MMTV Env and other viral motifs [DYxx(L/I)x6-12Yxx(L/I)] usually differs from that found in cellular ITAMs, although the significance of this difference is not currently understood. Most, if not all, of these proteins are believed to signal in a ligand-independent manner.
There are also cellular proteins expressed in nonhematopoietic cells that contain ITAM repeats, such as members of the ezrin/moesin family (51, 58) and tamalin, a scaffold protein that associates with glutamate receptors in neuronal cells (20). Moreover, a bioinformatics search has revealed between 48 and 368 additional cellular proteins that contain potential ITAMs (11). PECAM-1, the endothelial adhesion receptor, was also thought to contain an ITAM, although more recent data suggests that it functions as an immunoreceptor tyrosine inhibitory motif (23). Interestingly, mammary tissue expression of PECAM-1 in transgenic mice led to attenuated mammary gland development (22). Recent work has also shown that Syk, a downstream target of ITAM signaling, is expressed in normal human breast cells but not in invasive breast carcinoma and that it acts as a growth suppressor when introduced into the latter (6). In addition, mice with targeted mutation of Cbl, a negative regulator of Syk, develop mammary gland hyperplasia, although this could also be due to its effects on epidermal growth factor receptor signaling (41). Thus, our recent finding that MMTV uses ITAM-containing protein in its transformation pathway of nonhematopoietic cells such as mammary epithelia has the potential to lead to the discovery of a new mode of breast cancer induction and, as a result, novel treatment paradigms.
MMTV must require the same types of functions supplied by the human immunodeficiency virus and human T-cell leukemia type 1 accessory genes for in vivo infection, such as cell activation and immune evasion. The MMTV Env may have acquired an ITAM to accomplish some of these functions. For example, ITAM signaling could contribute to increased virus load, as is thought to be in the case for the Kaposi's sarcoma-associated herpesvirus K1 protein in B cells, through affecting cellular metabolic processes (27). Alternatively, it may allow persistence of infected cells for longer time periods or, because Env expression seems to increase lobuloalveolar development, create a larger number of virus-shedding cells, thereby increasing MMTV transmission. Although we showed that viruses bearing Env proteins with the ITAM mutations were not apparently attenuated for infection and appeared to be transmitted at levels similar to those of the wild-type virus in vivo, these studies looked at transmission through only two generations. Future experiments will examine whether the ITAM mutation affects viral fitness through subsequent generations.
Acknowledgments
We thank John Rassa for providing the pENV-Yaa418→I construct and members of the laboratory for helpful discussions.
This work was supported by Public Health Services grant R01 CA73746 and the University of Pennsylvania Research Foundation.
REFERENCES
- 1.Bannish, G., E. M. Fuentes-Panana, J. C. Cambier, W. S. Pear, and J. G. Monroe. 2001. Ligand-independent signaling functions for the B lymphocyte antigen receptor and their role in positive selection during B-lymphopoiesis. J. Exp. Med. 194:1583-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaufils, P., D. Choquet, R. Mamoun, and B. Malissen. 1993. The (XXL/I)2 motif found in the cytoplasmic segments of the bovine leukemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 12:5105-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callahan, R., and G. H. Smith. 2000. MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene 19:992-1001. [DOI] [PubMed] [Google Scholar]
- 4.Cantor, G. H. 1996. A potential proline-rich motif upstream of the immunoreceptor tyrosine-based activation motif in bovine leukemia virus gp30, Epstein-Barr virus LMP2A, herpesvirus papio LMP2A, and African horsesickness virus VP7. Virology 220:265-266. [DOI] [PubMed] [Google Scholar]
- 5.Choi, Y. C., D. H. Henrard, I. Lee, and S. R. Ross. 1987. The mouse mammary tumor virus long terminal repeat directs expression in epithelial and lymphoid cells of different tissues in transgenic mice. J. Virol. 61:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coopman, P. J. P., M. T. H. Do, M. Barth, E. T. Bowden, A. J. Hayes, E. Basyuk, J. K. Blancato, P. R. Vezza, S. W. McLeskey, P. H. Mangeat, and S. C. Mueller. 2000. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 406:742-747. [DOI] [PubMed] [Google Scholar]
- 7.Cousens, C., J. V. Bishop, A. W. Philbey, C. A. Gill, M. Palmarini, J. O. Carlson, J. C. DeMartini, and J. M. Sharp. 2004. Analysis of integration sites of Jaagsiekte sheep retrovirus in ovine pulmonary adenocarcinoma. J. Virol. 78:8506-8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson, C., R. Smith, S. Brookes, and G. Peters. 1984. Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell 37:529-536. [DOI] [PubMed] [Google Scholar]
- 9.Dzuris, J. L., W. Zhu, T. V. Golovkina, and S. R. Ross. 1999. Lack of receptor interference by endogenous expression of the mouse mammary tumor virus envelope protein. Virology 263:418-426. [DOI] [PubMed] [Google Scholar]
- 10.Finke, D., and H. Acha-Orbea. 2001. Differential migration of in vivo primed B and T lymphocytes to lymphoid and non-lymphoid organs. Eur. J. Immunol. 31:2603-2611. [DOI] [PubMed] [Google Scholar]
- 11.Fodor, S., Z. Jakus, and A. Mocsai. 2006. ITAM-based signaling beyond the adaptive immune response. Immunol. Lett. 104:29-37. [DOI] [PubMed] [Google Scholar]
- 12.Fruehling, S., and R. Longnecker. 1997. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BGC-mediated signal transduction. Virology 235:241-251. [DOI] [PubMed] [Google Scholar]
- 13.Gergely, J., I. Pecht, and G. Sarmay. 1999. Immunoreceptor tyrosine-based inhibition motif-bearing receptors regulate the immunoreceptor tyrosine-based activation motif-induced activation of immune competent cells. Immunol. Lett. 68:3-15. [DOI] [PubMed] [Google Scholar]
- 14.Golovkina, T. V., A. Chervonsky, J. A. Prescott, C. A. Janeway, and S. R. Ross. 1994. The mouse mammary tumor virus envelope gene product is required for superantigen presentation to T cells. J. Exp. Med. 179:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovkina, T. V., J. P. Dudley, and S. R. Ross. 1998. Superantigen activity is need for mouse mammary tumor virus spread within the mammary gland. J. Immunol. 161:2375-2382. [PubMed] [Google Scholar]
- 16.Golovkina, T. V., J. L. Dzuris, B. van den Hoogen, A. B. Jaffe, P. C. Wright, S. M. Cofer, and S. R. Ross. 1998. A novel membrane protein is a mouse mammary tumor virus receptor. J. Virol. 72:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovkina, T. V., A. B. Jaffe, and S. R. Ross. 1994. Coexpression of exogenous and endogenous mouse mammary tumor virus RNA in vivo results in viral recombination and broadens the virus host range. J. Virol. 68:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golovkina, T. V., J. A. Prescott, and S. R. Ross. 1993. Mouse mammary tumor virus-induced tumorigenesis in sag transgenic mice: a laboratory model of natural selection. J. Virol. 67:7690-7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grande, S. M., E. Katz, J. E. Crowley, M. S. Bernardini, S. R. Ross, and J. G. Monroe. 2006. Cellular ITAM-containing proteins are oncoproteins in nonhematopoietic cells. Oncogene 25:2748-2757. [DOI] [PubMed] [Google Scholar]
- 20.Hirose, M., J. Kitano, Y. Nakajima, K. Moriyoshi, S. Yanagi, H. Yamamura, T. Muto, H. Jingami, and S. Nakanishi. 2004. Phosphorylation and recruitment of syk by immunoreceptor tyrosine-based activation motif-based phosphorylation of tamalin. J. Biol. Chem. 279:32308-32315. [DOI] [PubMed] [Google Scholar]
- 21.Hook, L. M., Y. Agafonova, S. R. Ross, S. J. Turner, and T. V. Golovkina. 2000. Genetics of mouse mammary tumor virus-induced mammary tumors: linkage of tumor induction to the gag gene. J. Virol. 74:8876-8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilan, N., L. Cheung, S. Miller, A. Mohsenin, A. Tucker, and J. A. Madri. 2001. PECAM-1 is a modulator of STAT family member phosphorylation and localization: lessons from a transgenic mouse. Dev. Biol. 232:219-232. [DOI] [PubMed] [Google Scholar]
- 23.Ilan, N., and J. A. Madri. 2003. PECAM-1: old friend, new partners. Curr. Opin. Cell Biol. 15:515-524. [DOI] [PubMed] [Google Scholar]
- 24.Isakov, N. 1998. ITAMs: immunoregulatory scaffolds that link immunoreceptors to their intracellular signaling pathways. Recept. Channels 5:243-253. [PubMed] [Google Scholar]
- 25.Jiang, Z., and G. M. Shackleford. 1999. Mouse mammary tumor virus carrying a bacterial supF gene has wild-type pathogenicity and enables rapid isolation of proviral integration sites. J. Virol. 73:9810-9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz, E., M. H. Lareef, J. C. Rassa, S. M. Grande, L. B. King, J. Russo, S. R. Ross, and J. G. Monroe. 2005. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J. Exp. Med. 201:431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagunoff, M., D. M. Lukac, and D. Ganem. 2001. Immunoreceptor tyrosine-based activation motif-dependent signaling by Kaposi's sarcoma-associated herpesvirus K1 protein: effects on lytic viral replication. J. Virol. 75:5891-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagunoff, M., R. Majeti, A. Weiss, and D. Ganem. 1999. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 96:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, H., J. Guo, M. Li, J. K. Choi, M. DeMaria, M. Rosenzweig, and J. U. Jung. 1998. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol. Cell. Biol. 18:5219-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y., B. Welm, K. Podsypanina, S. Huang, M. Chamorro, X. Zhang, T. Rowlands, M. Egeblad, P. Cowin, Z. Werb, L. K. Tan, J. M. Rosen, and H. E. Varmus. 2003. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc. Natl. Acad. Sci. USA 100:15853-15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund, J. M., L. Alexopoulou, A. Sato, M. Karow, N. C. Adams, N. W. Gale, A. Iwasaki, and R. A. Flavell. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 101:5598-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by Jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 98:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGinnes, L. W., J. N. Reitter, K. Gravel, and T. G. Morrison. 2003. Evidence for mixed membrane topology of the Newcastle disease virus fusion protein. J. Virol. 77:1951-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merchant, M., R. G. Caldwell, and R. Longnecker. 2000. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J. Virol. 74:9115-9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikkers, H., J. Allen, P. Knipscheer, L. Romeijn, A. Hart, E. Vink, A. Berns, and L. Romeyn. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 32:153-159. [DOI] [PubMed] [Google Scholar]
- 36.Moise, A. R., J. R. Grant, R. Lippé, R. Gabathuler, and W. A. Jefferies. 2004. The adenovirus E3-6.7K protein adopts diverse membrane topologies following posttranslational translocation. J. Virol. 78:454-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monroe, J. G. 2006. ITAM-mediated tonic signaling through pre-BCR and BCR complexes. Nat. Immunol. 6:283-294. [DOI] [PubMed] [Google Scholar]
- 38.Moore, D. H., C. A. Long, A. B. Vaidya, J. B. Sheffield, A. S. Dion, and E. Y. Lasfargues. 1979. Mammary tumor viruses. Adv. Cancer Res. 29:347-418. [DOI] [PubMed] [Google Scholar]
- 39.Moore, R., G. Casey, S. Brookes, M. Dixon, G. Peters, and C. Dickson. 1986. Sequence, topography and protein coding potential of mouse int2: a putative oncogene activated by mouse mammary tumour virus. EMBO J. 5:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, W. J., F. S. Lee, C. Dickson, G. Peters, P. Pattengale, and P. Leder. 1990. The int-2 gene product acts as an epithelial growth factor in transgenic mice. EMBO J. 9:907-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, M. A., R. G. Schnall, D. J. Venter, L. Barnett, I. Bertoncello, C. B. Thien, W. Y. Langdon, and D. D. Bowtell. 1998. Tissue hyperplasia and enhanced T-cell signaling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell. Biol. 18:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nandi, S., and C. M. McGrath. 1973. Mammary neoplasia in mice. Adv. Cancer Res. 17:353-414. [Google Scholar]
- 43.Nathanson, N. 2002. Viral pathogenesis and immunity. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 44.Nusse, R., and H. E. Varmus. 1982. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31:99-109. [DOI] [PubMed] [Google Scholar]
- 45.Platt, E. J., and G. L. Firestone. 1991. Expression of mouse mammary tumor virus glycoprotein truncations defines roles for the transmembrane domain and ectodomain hydrophobic region in constitutive exocytic trafficking and proteolytic processing. J. Biol. Chem. 266:19384-19395. [PubMed] [Google Scholar]
- 46.Prakash, O., O. R. Swamy, X. Peng, Z.-Y. Tang, L. Li, J. E. Larson, J. C. Cohen, J. Gill, G. Farr, S. Wang, and F. Sarmaniego. 2005. Activation of src kinase lyn by Kaposi sarcoma-associated herpesvirus K1 protein: implications for lymphomagenesis. Blood 105:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for Jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasmussen, S. B., L. J. T. Young, and G. H. Smith. 2000. Preparing mammary gland whole mounts from mice, p. 75-85. In M. M. Ip and B. B. Asch (ed.), Methods in mammary gland biology and breast cancer research. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 50.Ross, S. R., J. J. Schofield, C. J. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 99:12386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozsnyay, Z., G. Sarmay, M. Zoller, and J. Gergely. 1996. Membrane-bound ezrin is involved in B-cell receptor-mediated signaling: potential role of an ITAM-like ezrin motif. Immunol. Lett. 54:163-169. [DOI] [PubMed] [Google Scholar]
- 52.Rulli, K., J. Lenz, and L. S. Levy. 2002. Disruption of hematopoiesis and thymopoiesis in the early premalignant stages of infection with SL3-3 murine leukemia virus. J. Virol. 76:2363-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shackleford, G. M., and H. E. Varmus. 1988. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc. Natl. Acad. Sci. USA 85:9655-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under the control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 56.Squartini, F., F. Basolo, and M. Bistocchi. 1983. Lobuloalveolar differentiation and tumorigenesis: two separate activities of mouse mammary tumor virus. Cancer Res. 43:5879-5882. [PubMed] [Google Scholar]
- 57.Tsukamoto, A. S., R. Grosschedl, R. C. Guzman, T. Parslow, and H. E. Varmus. 1988. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55:619-625. [DOI] [PubMed] [Google Scholar]
- 58.Urzainqui, A., J. M. Serrador, F. Viedma, M. Yanez-Mo, A. Rodriguez, A. L. Corbi, J. L. Alonso-Lebrero, A. Luque, M. Deckert, J. Vazquez, and F. Sanchez-Madrid. 2002. ITAM-based interaction of ERM proteins with Syk mediates signaling by the leukocyte adhesion receptor PSGL-1. Immunity 17:401-412. [DOI] [PubMed] [Google Scholar]
- 59.Weiss, R., N. Teich, H. Varmus, and J. Coffin. 1984. RNA tumor viruses. Molecular biology of tumor viruses. Cold Spring Harbor Laboratory Press, .
- 60.Willems, L., J. S. Gatot, M. Mammerickx, D. Portetelle, A. Burny, P. Kerkhofs, and R. Kettmann. 1995. The YXXL signaling motifs of the bovine leukemia virus transmembrane protein are required for in vivo infection and maintenance of high viral loads. J. Virol. 69:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wootton, S. K., C. L. Halbert, and A. D. Miller. 2005. Sheep retrovirus structural protein induces lung tumours. Nature 434:904-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wrona, T., and J. P. Dudley. 1996. Major histocompatibility complex class II I-E independent transmission of C3H mouse mammary tumor virus. J. Virol. 70:1246-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, L., Z. Shen, L. Guo, B. Fodera, A. Keogh, R. Joplin, B. O'Donnell, J. Aitken, W. Carman, J. Neuberger, and A. Mason. 2003. Does a betaretrovirus infection trigger primary biliary cirrhosis? Proc. Natl. Acad. Sci. USA 100:8454-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., J. C. Rassa, E. M. deObaldia, L. Albritton, and S. R. Ross. 2003. Identification of the mouse mammary tumor virus envelope receptor-binding domain. J. Virol. 77:10468-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]