Abstract
Bacteriophage PM2 presently is the only member of the Corticoviridae family. The virion consists of a protein-rich lipid vesicle, which is surrounded by an icosahedral protein capsid. The lipid vesicle encloses a supercoiled circular double-stranded DNA genome of 10,079 bp. PM2 belongs to the marine phage community and is known to infect two gram-negative Pseudoalteromonas species. In this study, we present a characterization of the PM2 genome made using the in vitro transposon insertion mutagenesis approach. Analysis of 101 insertion mutants yielded information on the essential and dispensable regions of the PM2 genome and led to the identification of several new genes. A number of lysis-deficient mutants as well as mutants displaying delayed- and/or incomplete-lysis phenotypes were identified. This enabled us to identify novel lysis-associated genes with no resemblance to those previously described from other bacteriophage systems. Nonessential genome regions are discussed in the context of PM2 genome evolution.
Studies of lipid-containing bacterial viruses have yielded considerable information on the structure and assembly of viral membranes (1, 7, 12, 35, 36, 42, 49). Crucial for this progress has been the availability of defined genetic systems (39, 47). Recently, we have initiated a structural characterization of phage PM2 as a model phage system (2, 26). Since no genetic tools are available in this system, we set out to probe the PM2 genome using the phage Mu-derived in vitro transposition mutagenesis technology, which represents an efficient alternative strategy for functional genome characterization (50).
PM2 is the first bacterial virus for which the presence of lipids as a structural component of the virion has been confirmed (9). PM2 was isolated along with its host bacterium Pseudoalteromonas espejiana BAL-31 from the coastal seawater of Chile (14, 15). The phage represented a new virus type, classified as Corticoviridae, and it is still the only known representative of this virus family. An alternative host for PM2, Pseudoalteromonas sp. strain ER72M2, was isolated from the East River in New York, N.Y. (33).
The PM2 virion consists of an icosahedrally organized proteinaceous capsid that surrounds a protein-rich lipid membrane, enclosing the highly supercoiled circular double-stranded DNA (dsDNA) genome (18, 26, 32, 38). The complete genome sequence revealed a 10,079-bp-long molecule with a GC content of 42.2% (16, 38). Replication of the PM2 genome takes place in the proximity of the cytoplasmic membrane (6) via a rolling-circle mechanism initiated by the phage-encoded replication initiation protein P12 (17, 38). The PM2 genome is organized into three operons: two early and one late. OEL is a leftward-transcribing early operon under the control of promoter P1207. This operon codes for two transcriptional repressors (proteins P15 and P16). P16 regulates the early rightward-transcribing operon (OER, under the control of promoter P1193), and protein P15 represses its own expression from promoter P1207 in addition to P1193. The late operon (OL, under the control of promoter P5321), which codes for almost all of the structural proteins, is positively regulated by the viral transcription factors P13 and P14 (37). The PM2 virion contains 10 virally encoded proteins. The protein coat is composed of P1 (a pentameric receptor binding protein) and P2 (a major capsid protein). All other proteins (P3 to P10) are associated with the lipid membrane (26, 32, 33, 38). Of the 21 open reading frames (ORFs) detected within the PM2 genome, 6 have not yet been confirmed to code for a protein (26).
Transposon mutagenesis can be used to map essential versus nonessential regions in viral genomes (8, 48, 50, 54). One of the most versatile methods is based on the bacteriophage Mu in vitro DNA transposition reaction, which can be used to disseminate custom-designed mini-Mu transposons into any DNA, including viral genomes. This strategy has been used successfully for the functional characterization of linear dsDNA genomes of bacteriophages PRD1 (50) and φYeO3-12 (31). A related strategy employing transposon ends has been applied to human immunodeficiency virus (34) and potato virus A (30) RNA genome characterization.
The in vitro transposition reaction catalyzed by the MuA transposase results in an essentially random distribution of transposons into the target DNA (19-21). A given transposon integrated into an essential gene or important noncoding region in the genome is expected to yield nonviable viruses. Alternatively, if the insertion occurs in a dispensable genomic region, the delivery of the viral genome into the host cell interior is expected to lead to successful virus propagation and progeny release. Theoretically, it also is possible to obtain transposon-containing virus clones with reduced viability.
To shed light on the functional organization of the phage PM2 genome, we used an in vitro DNA transposition-based insertion mutagenesis strategy. Our analysis revealed fully viable insertion mutants as well as mutants with an altered phenotype. The lack of insertions in many genomic regions revealed genes essential for phage viability. Analysis of mutant phenotypes identified two genes involved in host cell lysis.
MATERIALS AND METHODS
Virus and bacterial strains.
Phage PM2, Pseudoalteromonas espejiana BAL-31 (14, 15), and Pseudoalteromonas sp. strain ER72M2 cells were cultured in SB broth or on SB agar plates (33) at 28°C. Escherichia coli K-12 DH5α (Life Technologies) was used for transposon preparation as described previously (20, 50).
Production of PM2 DNA.
The virus was propagated, concentrated, and purified using rate zonal centrifugation as previously described (33). The virus zone was collected, and viruses were recovered by differential centrifugation (Beckman 50 Ti rotor, 18,000 rpm, 15 h, 5°C). The pellet was resuspended in MOPS buffer (20 mM morpholinepropanesulfonic acid [pH 7.4], 150 mM NaCl, 5 mM CaCl2), and DNA was released by the addition of sodium dodecyl sulfate (SDS) (2% final concentration, 50 min, 37°C). The mixture was phenol extracted (five times), followed by ether extraction (eight times). CsCl, water, and ethidium bromide were added to the resulting DNA fraction (to a 5-ml total volume; 4.5 g CsCl, 0.5 mg/ml ethidium bromide per tube). DNA was equilibrated by centrifugation (Sorvall TV865 rotor, 47,000 rpm, 17 h, 20°C). The DNA zone, corresponding to supercoiled molecules, was collected, and ethidium bromide was removed by isopropanol extraction (11 times). CsCl was removed by dialysis overnight at 4°C against TE (8.0) buffer (6 mM Tris-HCl [pH 8.0], 0.17 mM EDTA). Following dialysis, DNA was precipitated with ethanol, dried, and resuspended in water.
Mutagenesis of the PM2 genome using an in vitro-transposition-based approach.
The standard in vitro-transposition reaction mixture (50 μl) contained 2.0 pmol (0.6 μg) LacZ′-Mu(NotI) transposon DNA (50), 0.3 pmol (2 μg) PM2 target DNA, 10.8 pmol (0.88 μg) MuA transposase (Finnzymes), 25 mM Tris-HCl [pH 8.0], 100 μg/ml bovine serum albumin (Sigma), 15% (wt/vol) glycerol, 0.05% (wt/vol) Triton X-100 (Boehringer Mannheim), 126 mM NaCl, and 10 mM MgCl2. The reaction was carried out for 30 min at 30°C. Reaction products were extracted once with phenol and twice with chloroform, precipitated with ethanol, and resuspended in TE (7.5) buffer (10 mM Tris [pH 7.5], 0.5 mM EDTA). To isolate transposon-containing genomes, the reaction products were separated initially on a preparative 0.8% agarose gel (SeaPlaque; Cambrex) at 4 V/cm (90 min). A zone corresponding to the open circular forms of PM2 DNA was then excised from the gel, and the gel slice was melted at 65°C (10 min). The DNA within the mixture was digested with KpnI (New England Biolabs) at 37°C overnight, and the resulting fragments were separated on a preparative 1.3% agarose gel (SeaPlaque) at 3 V/cm (180 min). A fragment corresponding to transposon-containing genomes linearized within the transposon sequence was excised, and DNA was isolated by electroelution (44), extracted three times with 1-butanol and twice with chloroform, precipitated with ethanol, and resuspended in 15 μl of TE (7.5) buffer. Insertion-mutated phage genomes were regenerated by recircularization with T4 DNA ligase (Promega) at a low DNA concentration (2 ng/μl). Ligation products were purified by extraction once with phenol and twice with chloroform, precipitated with ethanol, and resuspended in 9 μl of TE (7.5) buffer.
DNA transfer to Pseudoalteromonas strains.
Electroporation into P. espejiana BAL-31 cells was developed using information available for related bacteria (29). Overnight cultures of the bacteria were diluted 1/50 into 50 ml of fresh SB medium and grown at 28°C to an optical density at 600 nm of 0.8 to 0.9, corresponding to mid-logarithmic growth phase. The cells were collected by centrifugation (Sorvall SLA1500 rotor, 6,000 rpm, 4°C, 20 min), washed three times with ice-cold HSG buffer (7 mM HEPES [pH 7.0], 252 mM sucrose, 20% glycerol) (29), resuspended in 500 μl of the buffer (about 1011 CFU/ml), and stored at −80°C. Electroporation was carried out using the Bio-Rad Gene Pulser. Briefly, 40 μl of P. espejiana BAL-31 cells was mixed with 200 ng of mutagenized PM2 DNA; the mixture was kept on ice for 5 min and transferred into a chilled cuvette (0.2-cm electrode spacing). A single pulse of 12.5 kV/cm (1.9 kV, 200 Ω, 25 μF) was applied, and 950 μl of SBE medium (0.8% Difco nutrient broth, 200 mM NaCl, 100 mM MgSO4, 10 mM CaCl2, 10 mM KCl, 100 mM sucrose) was added. The sample was incubated for 50 min at 28°C, plated with SB soft agar and the host cells onto solid SB medium, and incubated overnight at 28°C.
Lysis assay.
P. espejiana BAL-31 cells were grown with aeration at 28°C to a cell density of ∼6 × 108 CFU/ml and infected with wild-type (wt) or mutant PM2 using a multiplicity of infection of ∼10. Absorbance (λ = 550 nm) was measured at different time points using a Selecta Clormic digital colorimeter (J. P. Selecta). The experiments were done in triplicate with the wt-PM2 infection as a control. The presence of the transposon in each mutant was confirmed by PCR from a single plaque as described previously (27). Briefly, plaques were resuspended in 0.5 ml of SB broth and left overnight at 4°C, after which the material was divided into two aliquots. One aliquot was saved for phage recovery, and the other was boiled for 5 min in the presence of 1% SDS to disrupt phage particles. A 1/10 dilution of this solution was used as a template in a PCR. The presence of the transposon was visualized by the mobility shift of a particular PCR fragment in a 1.1% agarose gel.
DNA isolation from mutant phages.
The clonality of each mutant phage was ensured by two successive single-plaque isolations. Mutant virus stocks were prepared by collecting the soft agar layer from confluent SB plates with the addition of SB broth (3 ml/plate), followed by incubation with aeration for 1 h at 28°C. The debris was removed by centrifugation (Sorvall SA-600 rotor, 14,000 rpm, 5°C, 30 min). The resultant virus stock was mixed with an equal volume of polyethylene glycol (PEG) solution (20% PEG 6000, 0.5 M NaCl) and incubated for 30 min on ice. The virus was collected by centrifugation (Sorvall SA-600 rotor, 14,000 rpm, 5°C, 30 min) and washed twice with cold PM2 buffer (20 mM Tris-HCl [pH 7.2], 100 mM NaCl, 5 mM CaCl2). The PEG pellet was resuspended in cell resuspension solution (Promega), and the phage DNA was isolated using the Wizard plus SV miniprep system (Promega).
Determination of transposon insertion sites.
The presence of the transposon DNA was confirmed initially by digestion of the viral genome with HpaI (New England Biolabs). The precise site of the transposon integration was determined by sequencing (Sequencing Laboratory, Institute of Biotechnology, University of Helsinki, Finland) with transposon-specific primers. Since the circular supercoiled PM2 genome was an inefficient template for sequencing, the genome was linearized with the restriction endonuclease PvuII (MBI Fermentas), which recognizes a unique site within the transposon.
RESULTS
Transposon mutagenesis of the PM2 genome.
Circular covalently closed genomic PM2 DNA was subjected to in vitro transposon insertion mutagenesis using the artificial mini-Mu LacZ′-Mu(NotI) transposon as a DNA donor (50). The transposon-containing PM2 genomes were separated from the wt-PM2 genomes by a strategy employing two successive rounds of preparative agarose gel electrophoresis (see Materials and Methods), which produced mutant clones with close to 100% efficiency. In addition, a method for DNA transfer into P. espejiana BAL-31 cells by electroporation was developed (see Materials and Methods), which yielded 5 × 104 to 1 × 105 PFU/μg PM2 DNA. We initially obtained 162 plaques (phage clones), and their clonality was verified by at least two successive single-plaque purifications.
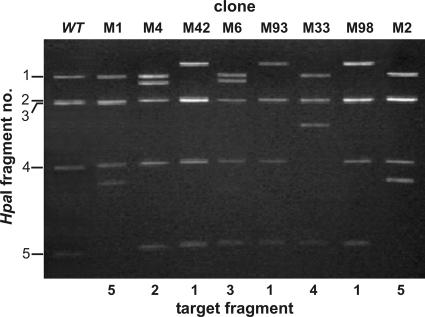
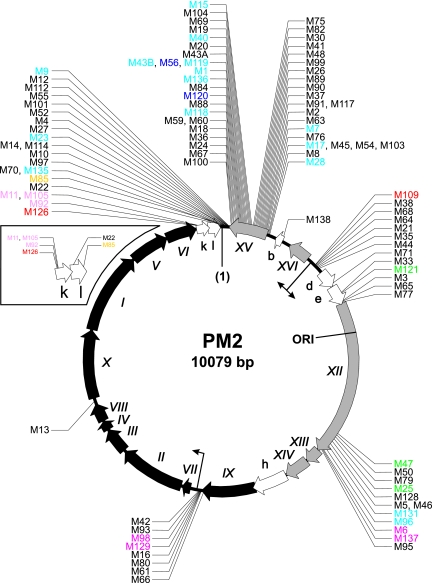
PM2 genomic DNA from 101 such clones was isolated, and the approximate location of the transposon was determined by restriction analysis with HpaI. Digestion of the wt-PM2 genome with HpaI yielded five fragments. LacZ′-Mu(NotI) transposon insertion into the genome increases the genome size by ∼460 bp. HpaI digestion of mutagenized PM2 genome clones showed that transposon insertions were tolerated at different genomic loci (Fig. 1). The exact location of each transposon insertion was determined by sequencing using a pair of transposon-specific primers to yield the sequence from each transposon end. In the 101 clones analyzed, 82 insertion sites appeared only once in our mutant collection, while the remaining 19 were mapped to eight different positions (Fig. 2). This indicates with a high probability that we had saturated all the nonessential PM2 genomic regions that could include transposon insertions.
FIG. 1.
Representative assortment of transposon insertion mutant phage genomes analyzed by HpaI digestion and agarose gel electrophoresis. Integration of a single LacZ′-Mu(NotI) transposon into a given restriction fragment adds 460 bp to the size of the fragment. Wild-type HpaI restriction fragment sizes are (fragment 1) 3,053 bp, (fragment 2) 2,393 bp, (fragment 3) 2,361 bp, (fragment 4) 1,451 bp, and (fragment 5) 819 bp.
FIG. 2.
Fine mapping of transposon integration sites within the PM2 genome. The nucleotides of the circular genome are numbered starting from the unique EcoRII restriction site (1) and proceeding clockwise (38). Genes encoding structural and nonstructural proteins (Roman numerals) and ORFs (lowercase letters) are visualized as black, gray, and white arrows, respectively, indicating the direction of transcription. The origin of replication (ORI) and the known promoters (bent arrows) are indicated. The inset depicts an enlarged view of ORFs k and l. The transposon mutants are indicated by their clone numbers. The exact locations are shown in Table 1. Color-coded mutants represent the phenotypical classes described in the legend to Fig. 4.
Transposon integration and repair of the transposition DNA intermediate.
A closer inspection of the actual target site sequences revealed that out of the 101 mutants for which the integration site was defined, 62 had intact transposon ends flanked by a 5-bp target site duplication that is considered a hallmark of Mu transposition (3, 28). Thirty-seven mutants had a normal transposon end at one end but retained the 4-bp transposon flank sequence at the other (discussed later). Of these mutants, 7 had a 1-bp target site duplication, 29 had no PM2 target sequence duplications/deletions, and 1 had a 1-bp target site deletion. Mutants M55 and M69 included larger deletions (456 bp and 148 bp, respectively), probably arising from the initial integration of two separate transposons into two different genomic locations and subsequent homologous recombination between these elements.
Identification of essential and nonessential regions of the PM2 genome.
A spectrum of transposon insertion sites mapped in a viral genome portrays a picture of the distribution of nonessential versus essential genomic loci. Transposon integration sites of 101 clones were determined in the PM2 genome, and a functional map was generated (Fig. 2).
More than half (64) of the insertions were located in the coding or putative coding regions of the PM2 genome. Only one insertion was detected in a gene encoding a structural protein. There was a transposon inserted at the far 3′ end of gene IX, encoding protein P9 (32), which contains putative ATP/GTP binding site motifs and is implicated in genome packaging (38, 49). Only the last C-terminal amino acid of the resulting mutant protein was predicted to have been replaced with a transposon-encoded fragment of 12 amino acids.
The remaining gene-associated transposon insertions were mapped to genes encoding nonstructural proteins, the majority of which reside within the two early operons OEL and OER. Transposons were tolerated within genes XV and XII as well as in ORFs b, d, e, k, and l (Fig. 2). Gene XV, which resides in the distal end of the early leftwards-transcribed operon OEL, has been shown to code for a repressor for the two early PM2 promoters (37). The entire OEL operon has been shown to be highly similar to the maintenance region of a Pseudoalteromonas plasmid, pAS28 (29, 38). Surprisingly, 39 integrations were distributed throughout the entire length of gene XV, indicating the nonessential nature of this gene. This result was supported by the observation that insertion mutants M55 and M69, with large deletions in the transposon target site (Table 1), also were located at the end of gene XV.
TABLE 1.
Locations and nature of LacZ′-Mu(NotI) transposon insertions
| Clone name | Plaque morphologyc | Flanking sequencea in:
|
Position in PM2 genomeb | Integration typed | LacZ′ orientation | Clone name | Plaque morphologyc | Flanking sequencea in:
|
Position in PM2 genomeb | Integration typed | LacZ′ orientatione | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSP210 | HSP211 | HSP210 | HSP211 | |||||||||||
| M1 | N + L | ACCGCGC | GATCGGT | 309 | C | + | M61 | N | AGTAGCT | TTAGATC | 5267 | C | − | |
| M2 | N + S | AATCAGT | TCAGTCA | 548 | A | + | M63 | N | GTCATAT | TCAGATC | 547 | C | − | |
| M3 | N + S | ATGCAAT | GATCTGT | 1789 | B | + | M64 | N | ACAGATC | GTAACCG | 1298 | C | + | |
| M4 | N + S | TATTGAA | TATATTG | 26 | A | − | M65 | N | TCGCTGG | GCTGGTG | 1801 | A | + | |
| M5 | N | AACAATG | GAAACAA | 3616 | A | − | M66 | N | TAACGTT | ACGTTTA | 5266 | A | + | |
| M6 | N + L | AAGTAGA | GTAGACG | 3647 | A | + | M67 | N | GTTTATC | TTGTTTA | 94 | A | − | |
| M7 | N + S | CATATTA | AGTGATC | 548 | C | − | M68 | N | CCCCGAC | GATCGTA | 1297 | C | + | |
| M8 | N + S | GCGGAAG | GCGCGGA | 597 | A | − | M69 | N | AAAGATC | CCAGTCT | 201/349 | E | + | |
| M9 | N + L | GCCGCAA | AGGCCGC | 56 | A | − | M70 | N | CGCCTGG | CCTGGGC | 6 | A | + | |
| M10 | N | GATCAAA | CCTGGGC | 8 | C | − | M71 | ST | GATCTCA | TTGCCAC | 1559 | C | − | |
| M11 | ST | TGACCGT | GATCTGT | 9827 | B | + | M75 | N | GATCTTA | CGCTGCT | 392 | B | − | |
| M12 | N | GCCGCAA | AAGGATC | 52 | C | − | M76 | N | CCTTTGA | TTTGAGT | 564 | A | + | |
| M13 | N | AAAGATA | TAAAAGA | 7034 | A | − | M77 | N | GTGAGCT | GTGTGAG | 1805 | A | − | |
| M14 | N + S | CTTGTAA | ACCTTGT | 17 | A | − | M79 | N | GGGTGAT | GTGATAG | 3570 | A | + | |
| M15 | N + L | CCGTTAA | CGCCGTT | 380 | A | − | M80 | N | GATCTAG | TTTAAGT | 5270 | B | − | |
| A16 | N | TAGCTAA | GCTAAAG | 5276 | A | + | M82 | N | GCTGCT- | GATCAAG | 392 | D | + | |
| M17 | N + S | CCATTGA | ATTGACA | 574 | A | + | M84 | N | GATCTTT | ACATGGC | 236 | C | − | |
| M18 | N | GCTGATC | AAAGAGT | 126 | C | + | M85 | NT | CCCTTGA | CTTGAAG | 10062 | A | + | |
| M19 | N | CCAGTCT | CCCCAGT | 355 | A | − | M88 | N | GATCAGC | CACTCGC | 182 | C | − | |
| M20 | N | AACCCGC | GATCCCC | 349 | C | + | M89 | N | CCATTGC | GATCGGC | 482 | C | + | |
| M21 | N | GACGATC | GCCAGAG | 1317 | C | + | M90 | N | GCGGCGC | TTGCGGC | 485 | A | − | |
| M22 | ND | ACAGAGA | TAACAGA | 9969 | A | − | M91 | N | CCTAAAA | GCCCTAA | 526 | A | − | |
| M23 | N + S | CCTGATC | TGTAATA | 14 | C | + | M92 | ST | TTGCAAG | GCAAGTA | 9813 | A | − | |
| M24 | N | GCCGTAT | TCGCCGT | 100 | A | − | M93 | N | AATTTAG | TTTAGAG | 5320 | A | + | |
| M25 | N + S | TTAAATC | GATTAAA | 3579 | A | − | M95 | N | AACTTAC | CTTACAC | 3698 | A | + | |
| M26 | N | GTTCGCT | TCGCTCT | 454 | A | + | M96 | N + S | AGGGGGG | TAAGATC | 3623 | B | − | |
| M27 | N | CTTGTAA | TGTAATA | 20 | A | + | M97 | N | CTGGGCA | GCCTGGG | 7 | A | − | |
| M28 | N + S | ACGATGA | TAACGAT | 609 | A | − | M98 | N + S | ACCTCAA | CTCAAAT | 5310 | A | + | |
| M30 | N | GCTGATC | GCTTAAG | 389 | C | + | M99 | N | TTCAGCA | TTTTCAG | 430 | A | − | |
| M33 | N | TGATTGC | GATCTCG | 1749 | C | + | M100 | N | GGCGCGT | CGCGTGT | 89 | A | + | |
| M35 | N + S | ACGCCAG | GATCAGT | 1321 | C | + | M101 | N | TTGAAAA | TATTGAA | 28 | A | − | |
| M36 | N | ATTACAA | TCATTAC | 110 | A | − | M103 | N | CCATTGA | ATTGACA | 574 | A | + | |
| M37 | N | CCCTAAA | AGCCCTA | 525 | A | − | M104 | N | TACCGCC | CCGCCGT | 377 | A | + | |
| M38 | N | GCTGGGC | TTGGATC | 1271 | C | − | M105 | ST | GATCTGT | TGACCGT | 9827 | B | − | |
| M40 | N + S | CCCAGTC | GCCCCAG | 354 | A | − | M109 | M | GGGCAAC | GATCCGC | 1255 | C | + | |
| M41 | N | GATCGGC | GCGCGGG | 412 | C | − | M112 | N + S | AAGGATC | GCCGCAA | 52 | C | + | |
| M42 | N | TTAGAGC | ATTTAGA | 5321 | A | − | M114 | N | CTTGTAA | ACCTTGT | 17 | A | − | |
| M43A | M | CCCCGAC | GATCAAA | 334 | C | + | M117 | N | CCTAAAA | GCCCTAA | 526 | A | − | |
| M43B | L | CCCCGAC | CCGACAA | 334 | A | + | M118 | N + S | ATCATTG | GATCCCA | 156 | C | + | |
| M44 | N | GATCAAC | CAGAGTG | 1325 | C | − | M119 | N + S | CCGACAA | CCCCGAC | 334 | A | − | |
| M45 | N | ATTGACA | CCATTGA | 574 | A | − | M120 | N + S | AACATGG | CATGGCT | 234 | A | + | |
| M46 | N | AACAATG | GAAACAA | 3616 | A | − | M121 | N + S | GCTCGGT | TTGCTCG | 1751 | A | − | |
| M47 | M | CCCCGCC | CCGCCAA | 3543 | A | + | M126 | N + S | TATGATC | TGCAAGT | 9806 | C | + | |
| M48 | N | TTGTTTT | GTTTGTT | 426 | A | − | M128 | N | TCCCTAA | CCTAAAG | 3599 | A | + | |
| M50 | N | GGTGATA | CGGGTGA | 3570 | A | − | M129 | L + M | AAGTAGC | GATCTAA | 5273 | C | + | |
| M52 | N | ATATTGA | ATTGAAA | 27 | A | + | M131 | N + L | TAAGATC | AGGGGGG | 3623 | B | + | |
| M54 | N | CCATTGA | ATTGACA | 574 | A | + | M135 | N + L | CCTGGGC | CGCCTGG | 6 | A | − | |
| M55 | N | GATCGCG | TATTGAA | 28/484 | E | − | M136 | N + M | TAGCGCA | GCGCACC | 250 | A | + | |
| M56 | M | CCCCGAC | CCGACAA | 334 | A | + | M137 | N + L | CAGCTTA | GCTTAAA | 3658 | A | + | |
| M59 | N | TTTTAAT | TTAATCA | 146 | A | + | M138 | N | GATCGTT | ATTTATA | 744 | C | − | |
| M60 | N | TTTTAAT | TTAATCA | 146 | A | + | ||||||||
Duplicated sequences are in bold, and a 1-bp target site deletion is marked by a dash.
Transposon insertion sites are counted from the first change in the PM2 genome sequence (GenBank accession no. AF155037).
L, large; M, minute; N, normal; S, small; T, turbid; ND, not determined.
A, normal 5-bp target site duplication; B, GATC in one end and a 1-bp target site duplication; C, GATC in one end and no duplication; D, GATC in one end and a 1-bp target site deletion; E, large deletion in target.
The + denotes the orientation of the LacZ′ gene (5′ to 3′), and − denotes the other orientation.
Gene XII, the largest in the PM2 genome, codes for the 634-amino-acid replication initiation protein P12 (38) and is supposed to be essential for successful phage propagation. Consequently, ∼91% of the gene did not tolerate insertions. Twelve transposon insertions were found at the 3′ end of the gene (Fig. 2). Accordingly, 58 C-terminal amino acids in P12 could be replaced by a transposon-derived peptide fragment (M47) without observable effects on phage propagation (phenotypic changes caused by transposon insertions are discussed below).
The two short ORFs b and d each contained one transposon insertion. ORF e included five transposon insertions, of which two were detected in the central region and three in the 3′ distal region of the ORF. ORF h, the fourth putative coding region in the early operons for which a protein product has not yet been confirmed, did not tolerate any transposon insertions (Fig. 2). The end of the late PM2 operon OL contains two short, overlapping ORFs, k and l. We isolated four and two clones, respectively, with insertions in those ORFs (see the inset in Fig. 2).
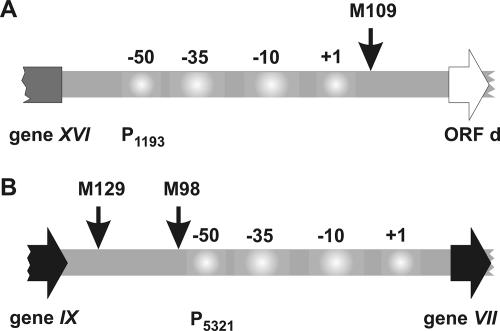
The remaining 37 clones had transposons inserted into the intergenic regions of the PM2 genome (Fig. 2). Twenty-two integrations were in the noncoding regions flanking gene XV. Fifteen insertions were between gene XV and ORF l, and seven insertion mutations were detected between gene XV and ORF b. Interestingly, seven insertions were located in the downstream region of the P1193 promoter, which controls the expression of the OER operon and is located between gene XVI and ORF d. One mutant (M109) showed a clearly altered phenotype (discussed below) and had the transposon inserted near the +1 position of promoter P1193 (Fig. 3A). In addition, there were seven insertions in the intergenic region between genes IX and VII, where the late PM2 promoter P5321 is located. All these insertions occurred in the upstream region of the promoter. Nevertheless, in the cases of mutants M98 and M129, insertions had an effect on the phage phenotype (Fig. 3B). Transposon insertions in the downstream regions of the P1207 and P5321 promoters were not detected. Only one clone (M13) had the transposon inserted in the noncoding region inside the late operon OL, between genes VIII and X.
FIG. 3.
Transposon integrations into promoter regions of phage PM2. (A) Promoter P1193 controls the early rightwards-transcribed operon OER. (B) Promoter P5321 controls the late operon OL. The numbers refer to the promoter consensus sequences. The arrows point to the locations of the transposon integrations that affected the wild-type phenotype.
Phenotypic changes in PM2 transposon insertion mutants and identification of lysis functions.
We analyzed insertion-mutagenized PM2 clones using a plaque assay. Thirty-nine out of 101 clones displayed plaque morphologies deviating from that of wt PM2 (Table 1). Among these, 29 clones formed plaques that were heterogeneous in size: normal and small (20 clones), normal and minute (1 clone), normal and large (7 clones), and large and minute (1 clone). Clones retained the plaque size heterogeneity even if one type was used in single-plaque isolation. The remaining 10 clones produced plaques that were homogenous in size but morphologically different from those formed by the wt phage: small and turbid plaques (four clones), large plaques (one clone), turbid plaques of normal size (one clone), and minute plaques (four clones).
These thirty-nine mutant viruses were analyzed further in a lysis assay. It was possible to prepare virus stocks and analyze 30 such clones. The remaining mutants (nine) either had lost the transposon (M4, M14, M71, and M112) or were inactivated during storage and failed to form a plaque (M2, M3, M8, M35, and M43A). The mutants analyzed were divided into seven types according to the profiles of their lysis curves (Fig. 2 and 4). Nearly half (15 clones) of the mutants analyzed displayed the wt lysis curve (Fig. 4, type 1), even though the plaque morphologies differed from the wt. Two mutants in this phenotypic class had the transposon inserted at the 3′ end of gene XII. In the remaining 13 PM2 clones, insertions occurred either in gene XV or in the noncoding regions flanking this gene. In addition, two clones that had an ∼5-min-shorter infectious cycle than the wt (type 2) had the transposon inserted in gene XV (Fig. 2).
FIG. 4.
Lysis curves of representative mutants that have different phenotypes. P. espejiana BAL-31 cells were grown to a cell density of 6 × 108 CFU/ml and infected with wt PM2 or an appropriate PM2 mutant using a multiplicity of infection of ∼10. Type 1, cells grew like cells infected with the wt (cyan); type 2, cells started to lyse ∼5 min earlier than the wt-infected cells (blue); type 3, cells started to lyse ∼5 min later than the wt-infected cells (green); type 4, cells started to lyse ∼10 min later than the wt-infected cells (magenta); type 5, cells showed delayed and incomplete lysis (red); type 6, cells were lysis deficient (light violet); type 7, cells showed abnormal lysis (yellow). p.i., postinfection.
Cells infected with transposon mutant types 3 and 4 lysed, respectively, 5 and 10 min later than the cells infected with wt PM2 (Fig. 4). The type 3 mutants had the insertions inside gene XII or ORF e, while transposon insertions in the type 4 members were mapped either to gene XII or to the noncoding region between genes IX and VII.
The remaining mutants had defects in cell lysis. Insertion mutations in the noncoding region between gene XVI and ORF d resulted in a delayed and incomplete lysis phenotype (type 5). Interestingly, transposon insertions in ORF k abolished the lysis of the infected cells (type 6) or lysis was delayed and only partial (type 5). Furthermore, ORF l, which is adjacent to ORF k (Fig. 2), also seems to participate in cell lysis events, as the ORF l insertion mutant displayed an abnormal lysis phenotype (type 7).
DISCUSSION
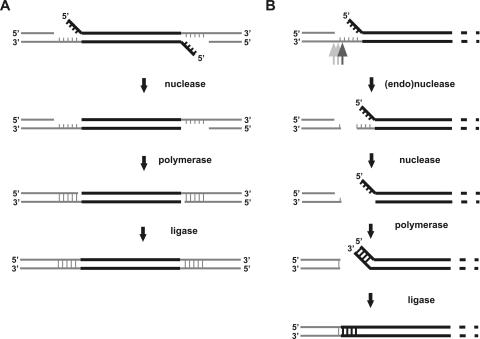
In this study, we analyzed the dsDNA genome of phage PM2 using in vitro transposon insertion mutagenesis. In addition to functionally characterizing the viral genome, we obtained data pointing to an alternative and, to our knowledge, previously unreported pathway to repair the transposition reaction intermediate. The 5-base-pair target site duplication that normally ensues from Mu transposon integration (3, 28) is a result of the target site being cut in a staggered manner by a transposase and the subsequent replication/repair of the transposition DNA intermediate by the host machinery (46). In E. coli, this type of end repair appears to be uniform (19, 20, 40, 51). However, other host replication machineries might, at least occasionally, repair the transposon ends differently, producing alternative transposon-target site junctions. Figure 5 outlines a feasible model for this alternative transposon DNA intermediate processing observed in P. espejiana BAL-31. Interestingly, even though this alternative processing was relatively common (37/101 mutants [∼37%]), no mutants with two alternatively processed transposon ends were detected, suggesting that simultaneous alternative processing of the two ends may not be possible.
FIG. 5.
Model of transposition DNA intermediate processing. Although the processing enzymes are unknown, nuclease (endo- or exonuclease), DNA polymerase, and ligase activities must be involved. (A) Standard processing. Transposon integration generates a DNA intermediate that contains 5-nucleotide (nt) single-stranded regions and 4-nt noncomplementary overhangs. The 4-nt overhangs are removed by a nuclease. Single-stranded regions are filled in by DNA polymerase, and the nicks are sealed by ligase. The process generates a 5-bp target site duplication (i.e., the same 5-bp sequence flanks the transposon DNA as a direct repeat). (B) Alternative processing. The single-stranded region initially is cleaved by a nuclease. The ensuing DNA ends are then processed by exonuclease and polymerase. Finally, the ends are joined by ligase. Within the joint, the 4-nt overhang sequence from the transposition intermediate (5′-GATC) will be incorporated, and the site of initial cleavage determines the exact number of genome-derived base pairs. The most probable cleavage site is depicted by a dark-gray arrow, with other possibilities shown in lighter gray.
The saturation mutagenesis of the circular highly supercoiled PM2 genome indicated that at least 460 bp (length of the transposon used in this study) can be additionally packaged into the PM2 virion without detectable adverse effects. Although the upper packaging capacity of the PM2 particle is currently unknown, it is evident that the available volume in the proteinaceous PM2 membrane vesicle is not fully occupied by the wt genome. This also is indicated by the mean DNA packaging density of 0.35 bp nm−3 (26).
A total of 101 insertion mutations were mapped in the PM2 genome, which allowed us to identify essential and dispensable regions of the viral genome. In addition, it was possible to reveal the functions of several PM2 genes. All but one of the genes encoding structural proteins are organized into the late operon OL (37) and are tightly packed (Fig. 2). None of these genes tolerated transposon insertions, indicating stringent constraints for the virion assembly process, as has been observed in other viral systems (4, 5). The genes encoding the regulatory proteins P13, P14, and P16, as well as ORF h, also were sensitive to change. The lack of mutations in ORF h indicates that, with a high probability, it is an authentic protein-coding gene.
Gene XII, coding for the replication initiation protein P12, tolerated insertions only in the 3′ distal region of the gene, resulting in a 5- to 10-min delay in lysis. Either the resultant truncated protein initiates DNA replication less efficiently, leading to a delay in the virus life cycle, or transposon insertions upstream of the genes encoding transcription activators P13 and P14 (37) could result in delayed expression of these transcriptional factors, possibly due to premature termination of transcription, consequently postponing the activation of the late operon. The latter interpretation is supported by the observation that the strongest phenotype (longest delay in lysis) is caused by the insertions (M6, M137) that are closest to the start of gene XIII.
Similarly, infection with mutant M109 resulted in delayed and incomplete lysis. The clone had an insertion in the noncoding region between gene XVI and ORF d, near the +1 position of the P1193 promoter (Fig. 3A). Such an insertion probably has a negative effect on the RNAP-promoter interaction and, consequently, on the expression of the early rightwards-transcribing operon, which contains genes needed for viral DNA replication and further activation of the late viral functions. In addition, a 10-min delay in lysis was observed with clones M98 and M129, which had the transposon inserted upstream of the late promoter P5321 (Fig. 3B). However, the lysis kinetics were comparable to those of the wt and not incomplete as in the case of M109. It is possible that the delay in lysis might be the result of the retarded expression of the lysis genes that reside in the late operon OL (see below).
Mutations in ORFs k and l resulted in almost complete lysis deficiency and/or in turbid plaques, respectively (Fig. 4), suggesting that these two ORFs are involved in the release of PM2 from the infected cell. ORFs k and l reside next to each other. All dsDNA bacteriophages characterized so far use a holin-endolysin system to lyse the cell at the end of the infectious cycle (52). Therefore, ORF k, which has been predicted to have a membrane-spanning domain (38), would represent a holin gene and the soluble product of ORF l might be, consequently, involved in disruption of the peptidoglycan layer. It is notable that the gene coding for the holin protein P35 of the lipid-containing phage PRD1 also tolerated transposon insertions (43, 50). It is evident from the sequence data that the protein products of the genes involved in PM2 cell lysis greatly deviate from the corresponding proteins described for other phages (53). The roles of ORFs k and l in the process of PM2 release currently are being investigated.
Another striking observation about the PM2 genome is its mosaic organization. There are only a few genes in the PM2 genome that have significant similarities to those in the sequence databases. Those few share similarity with sequences from very different origins. Transcription activator P14 has a zinc finger motif similar to the transcription factors of eukaryal and archaeal origins (37). P12 was shown to have similarity to the replication initiation proteins of bacterial plasmids as well as bacteriophages of the Myoviridae (P2, HP1, 186) and Microviridae (φX174, G4) families (38). We observed here that the structural virion protein P5 shares significant sequence similarity with a number of proteins from different phyla of bacteria, Methylococcus capsulatus prophage MuMc02, and bacteriophages of the Siphoviridae (D3112) and Podoviridae (Epsilon15, K1F) families (Fig. 6). Moreover, the entire OEL operon is homologous to the Pseudoalteromonas plasmid maintenance region (29, 37, 38). Consequently, it has been proposed (37) that this entire operon is a moron (24), an autonomous transcriptional unit captured by the phage via horizontal gene transfer that provides selective benefits to the phage.
FIG. 6.
Sequence alignment of the structural protein P5 of phage PM2 with similar sequences present in GenBank. Amino acid residues of P5 that are conserved in at least one other protein are shaded gray. A predicted transmembrane helix in the PM2 protein is boxed. P5-PM2, structural protein P5 of bacteriophage PM2 (accession no. NP_049909); Seq 1, Gp21 of bacteriophage D3112 (accession no. NP_938228); Seq 2, putative structural protein P5 of Methylococcus capsulatus prophage MuMc02 (accession no. AAU90959); Seq 3, Orf36 of Photorhabdus luminescens (accession no. AAO18061); Seq 4, conserved hypothetical phage protein of Bacteroides fragilis NCTC 9343 (accession no. CAH07979); Seq 5, conserved hypothetical protein of Desulfovibrio vulgaris subsp. vulgaris strain Hildenborough (accession no. AAS95986). The alignment was constructed using CLUSTALW (11).
Interestingly, gene XV, which resides in the OEL operon and was shown convincingly to code for a repressor of the early viral promoters (37), tolerated transposon insertions throughout its entire length. Moreover, noncoding regions flanking this gene also were densely packed with transposon insertions (Fig. 2). There are two possible explanations—the gene may be either a new addition to the genome or not required under the conditions used or both. The second possibility is supported by the fact that some insertions (M1, M2, M7, M15, M40, M43A, M43B, M56, M118, M119, M120, and M136) resulted in altered plaque morphology, indicating that gene XV has a measurable effect on virus production. It also has been shown that dispensable genes 1.3 and 3.5 of phage φYeO3-12 are involved in extending the host range to Yersinia enterocolitica serotype O:3 (31). Likewise, nonessential genes of T4-type phages were suggested to be important for adaptation to a particular lifestyle (13).
The current understanding of bacteriophage genome evolution has emerged mostly from the comparative analyses of tailed dsDNA phage (Caudovirales) genomic sequences (10, 22, 23, 41), while the relatedness of tailless icosahedral dsDNA phages is less well understood (45). Recent genome sequencing of six PRD1-like phages intriguingly revealed a high degree of conservation and a lack of any detectable morons in the Tectiviridae family, presenting a marked contrast to the mosaic genomes of the tailed dsDNA bacteriophages (45).
It has been suggested that horizontal gene transfer at high frequencies occurs presumably only between phages occupying the same ecological niche. However, a low-frequency horizontal exchange across a wide phylogenetic range of hosts was assumed to take place in the course of the so-called “random walk through phylogenetic space” (22, 25). The mosaicism of the PM2 genome supports this idea, further pointing to the presence of a common genetic pool accessible not only for the tailed bacteriophages but also for those of more remote types and those infecting hosts occupying diverse ecological niches.
Acknowledgments
We thank Lars Paulin for helpful advice on sequencing the highly supercoiled PM2 DNA molecules. Sara Ollila, Petri Papponen, Pirjo Rahkola, and Tanja Ruokoranta are thanked for skillful technical assistance.
The work was funded by a Finnish Technology Agency (TEKES) NeoBio Program (2002-2005) grant (H.S.), Academy of Finland grants 202874 (H.S.) and 1201964 (J.K.H.B.), and Finnish Center of Excellence Program 2006-2011 grants 1213467 and 1213992 (D.H.B.).
REFERENCES
- 1.Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. [DOI] [PubMed] [Google Scholar]
- 2.Abrescia, N. G., H. M. Kivelä, J. M. Grimes, J. K. Bamford, D. H. Bamford, and D. I. Stuart. 2005. Preliminary crystallographic analysis of the major capsid protein P2 of the lipid-containing bacteriophage PM2. Acta Crystallogr. Sect. F 61:762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allet, B. 1979. Mu insertion duplicates a 5 base pair sequence at the host inserted site. Cell 16:123-129. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 5.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 6.Brewer, G. J. 1978. Membrane-localized replication of bacteriophage PM2. Virology 84:242-245. [DOI] [PubMed] [Google Scholar]
- 7.Brewer, G. J. 1980. Control of membrane morphogenesis in bacteriophage. Int. Rev. Cytol. 68:53-96. [DOI] [PubMed] [Google Scholar]
- 8.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 9.Camerini-Otero, R. D., and R. M. Franklin. 1972. Structure and synthesis of a lipid-containing bacteriophage. XII. The fatty acids and lipid content of bacteriophage PM2. Virology 49:385-393. [DOI] [PubMed] [Google Scholar]
- 10.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8:451-458. [DOI] [PubMed] [Google Scholar]
- 11.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cockburn, J. J., N. G. Abrescia, J. M. Grimes, G. C. Sutton, J. M. Diprose, J. M. Benevides, G. J. Thomas, Jr., J. K. Bamford, D. H. Bamford, and D. I. Stuart. 2004. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432:122-125. [DOI] [PubMed] [Google Scholar]
- 13.Desplats, C., and H. M. Krisch. 2003. The diversity and evolution of the T4-type bacteriophages. Res. Microbiol. 154:259-267. [DOI] [PubMed] [Google Scholar]
- 14.Espejo, R. T., and E. S. Canelo. 1968. Properties and characterization of the host bacterium of bacteriophage PM2. J. Bacteriol. 95:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espejo, R. T., and E. S. Canelo. 1968. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology 34:738-747. [DOI] [PubMed] [Google Scholar]
- 16.Espejo, R. T., E. S. Canelo, and R. L. Sinsheimer. 1969. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc. Natl. Acad. Sci. USA 63:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espejo, R. T., E. S. Canelo, and R. L. Sinsheimer. 1971. Replication of bacteriophage PM2 deoxyribonucleic acid: a closed circular double-stranded molecule. J. Mol. Biol. 56:597-621. [DOI] [PubMed] [Google Scholar]
- 18.Gray, H. B., Jr., W. B. Upholt, and J. Vinograd. 1971. A buoyant method for the determination of the superhelix density of closed circular DNA. J. Mol. Biol. 62:1-19. [DOI] [PubMed] [Google Scholar]
- 19.Haapa, S., S. Suomalainen, S. Eerikäinen, M. Airaksinen, L. Paulin, and H. Savilahti. 1999. An efficient DNA sequencing strategy based on the bacteriophage mu in vitro DNA transposition reaction. Genome Res. 9:308-315. [PMC free article] [PubMed] [Google Scholar]
- 20.Haapa, S., S. Taira, E. Heikkinen, and H. Savilahti. 1999. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 27:2777-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haapa-Paananen, S., H. Rita, and H. Savilahti. 2002. DNA transposition of bacteriophage Mu. A quantitative analysis of target site selection in vitro. J. Biol. Chem. 277:2843-2851. [DOI] [PubMed] [Google Scholar]
- 22.Hendrix, R. W. 2002. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61:471-480. [DOI] [PubMed] [Google Scholar]
- 23.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 25.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huiskonen, J. T., H. M. Kivelä, D. H. Bamford, and S. J. Butcher. 2004. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol. 11:850-856. [DOI] [PubMed] [Google Scholar]
- 27.Huiskonen, J. T., L. Laakkonen, M. Toropainen, M. Sarvas, D. H. Bamford, and J. K. Bamford. 2003. Probing the ability of the coat and vertex protein of the membrane-containing bacteriophage PRD1 to display a meningococcal epitope. Virology 310:267-279. [DOI] [PubMed] [Google Scholar]
- 28.Kahmann, R., and D. Kamp. 1979. Nucleotide sequences of the attachment sites of bacteriophage Mu DNA. Nature 280:247-250. [DOI] [PubMed] [Google Scholar]
- 29.Kato, J., J. Amie, Y. Murata, A. Kuroda, A. Mitsutani, and H. Ohtake. 1998. Development of a genetic transformation system for an alga-lysing bacterium. Appl. Environ. Microbiol. 64:2061-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kekarainen, T., H. Savilahti, and J. P. Valkonen. 2002. Functional genomics on potato virus A: virus genome-wide map of sites essential for virus propagation. Genome Res. 12:584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiljunen, S., H. Vilen, M. Pajunen, H. Savilahti, and M. Skurnik. 2005. Nonessential genes of phage φYeO3-12 include genes involved in adaptation to growth on Yersinia enterocolitica serotype O:3. J. Bacteriol. 187:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kivelä, H. M., N. Kalkkinen, and D. H. Bamford. 2002. Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J. Virol. 76:8169-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kivelä, H. M., R. H. Männistö, N. Kalkkinen, and D. H. Bamford. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:364-374. [DOI] [PubMed] [Google Scholar]
- 34.Laurent, L. C., M. N. Olsen, R. A. Crowley, H. Savilahti, and P. O. Brown. 2000. Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting. J. Virol. 74:2760-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurinavičius, S., R. Käkelä, D. H. Bamford, and P. Somerharju. 2004. The origin of phospholipids of the enveloped bacteriophage phi6. Virology 326:182-190. [DOI] [PubMed] [Google Scholar]
- 36.Laurinmäki, P. A., J. T. Huiskonen, D. H. Bamford, and S. J. Butcher. 2005. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 13:1819-1828. [DOI] [PubMed] [Google Scholar]
- 37.Männistö, R. H., A. M. Grahn, D. H. Bamford, and J. K. H. Bamford. 2003. Transcription of bacteriophage PM2 involves phage-encoded regulators of heterologous origin. J. Bacteriol. 185:3278-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Männistö, R. H., H. M. Kivelä, L. Paulin, D. H. Bamford, and J. K. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355-363. [DOI] [PubMed] [Google Scholar]
- 39.Mindich, L., D. Bamford, C. Goldthwaite, M. Laverty, and G. Mackenzie. 1982. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J. Virol. 44:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuuchi, M., and K. Mizuuchi. 1993. Target site selection in transposition of phage Mu. Cold Spring Harbor Symp. Quant. Biol. 58:515-523. [DOI] [PubMed] [Google Scholar]
- 41.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 42.Poranen, M. M., R. Tuma, and D. H. Bamford. 2005. Assembly of double-stranded RNA bacteriophages. Adv. Virus Res. 64:15-43. [DOI] [PubMed] [Google Scholar]
- 43.Rydman, P. S., and D. H. Bamford. 2003. Identification and mutational analysis of bacteriophage PRD1 holin protein P35. J. Bacteriol. 185:3795-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 45.Saren, A. M., J. J. Ravantti, S. D. Benson, R. M. Burnett, L. Paulin, D. H. Bamford, and J. K. Bamford. 2005. A snapshot of viral evolution from genome analysis of the tectiviridae family. J. Mol. Biol. 350:427-440. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro, J. A. 1979. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. USA 76:1933-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinclair, J. F., J. Cohen, and L. Mindich. 1976. The isolation of suppressible nonsense mutants of bacteriophage phi6. Virology 75:198-208. [PubMed] [Google Scholar]
- 48.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strömsten, N. J., D. H. Bamford, and J. K. Bamford. 2005. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J. Mol. Biol. 348:617-629. [DOI] [PubMed] [Google Scholar]
- 50.Vilen, H., J. M. Aalto, A. Kassinen, L. Paulin, and H. Savilahti. 2003. A direct transposon insertion tool for modification and functional analysis of viral genomes. J. Virol. 77:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vilen, H., S. Eerikäinen, J. Tornberg, M. S. Airaksinen, and H. Savilahti. 2001. Construction of gene-targeting vectors: a rapid Mu in vitro DNA transposition-based strategy generating null, potentially hypomorphic, and conditional alleles. Transgenic Res. 10:69-80. [DOI] [PubMed] [Google Scholar]
- 52.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 53.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhan, X., M. Lee, G. Abenes, I. Von Reis, C. Kittinunvorakoon, P. Ross-Macdonald, M. Snyder, and F. Liu. 2000. Mutagenesis of murine cytomegalovirus using a Tn3-based transposon. Virology 266:264-274. [DOI] [PubMed] [Google Scholar]