Abstract
Terminal repeat (TR) elements of Kaposi's sarcoma-associated herpesvirus (KSHV), the potential origin sites of KSHV replication, have been demonstrated to play important roles in viral replication and transcription and are most likely also critical for the segregation of the KSHV genome to daughter cells. To search for the cellular proteins potentially involved in KSHV genome maintenance, we performed affinity chromatography analysis, using KSHV TR DNA as the affinity ligand. Proteomic analysis was then carried out to identify the TR-interacting proteins. We identified a total of 123 proteins from both KSHV-positive and -negative cells, among which most were identified exclusively from KSHV-positive cells. These proteins were categorized as proliferation/cell cycle regulatory proteins, proteins involved in spliceosome components, such as heterogeneous nuclear ribonuclear proteins, the DEAD/H family, the switch/sucrose nonfermenting protein family, splicing factors, RNA binding proteins, transcription regulation proteins, replication factors, modifying enzymes, and a number of proteins that could not be broadly categorized. To support the proteomic results, the presence of four candidate proteins, ATR, BRG1, NPM1 and PARP-1, in the elutions was further characterized in this study. The binding and colocalization of these proteins with the TR were verified using chromatin immunoprecipitation and immunofluorescence in situ hybridization analysis. These newly identified TR binding proteins provide a number of clues and potential links to understanding the mechanisms regulating the replication, transcription, and genome maintenance of KSHV. This study will facilitate the generation and testing of new hypotheses to further our understanding of the mechanisms involved in KSHV persistence and its associated pathogenesis.
Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8, is a human gammaherpesvirus family member associated with Kaposi's sarcoma, body cavity-based lymphomas, and multicentric Castleman's disease (36). Typically, KSHV displays two modes of infection: latent infection, during which the viral genome persists in the host cell and no viral progeny are released, and lytic infection, during which the host cell is destroyed and viral progeny is produced (for a review, see reference 51). In the latent state, KSHV genomic DNA, which exists as a closed circular plasmid, appears to behave like host chromosomal DNA and is packaged onto nucleosomes with cellular histones (37, 42). During S phase, KSHV genomes are replicated once and are partitioned faithfully into daughter cells during the mitotic phase (20).
KSHV terminal repeat (TR) elements are multiple GC-rich, 801-bp DNA fragments at the terminus of the KSHV genome (25, 43). The viral TR is important for the tethering of viral genome to the host chromosomes and thus ensures efficient segregation of viral DNA upon mitosis (2, 9). Two latency-associated nuclear antigen (LANA) protein binding sites (LBS1 and LBS2) were located between nucleotides 571 and 610 in each TR sequence, and both binding sites contribute to ori activity, as determined by short-term replication assays (14, 15). An 89-bp highly GC-rich element is located upstream of LBS1/2, along with a 101-bp AT-rich stretch that is often found in origins of replication and believed to function in DNA unwinding. Both the GC-rich element and the LBS1/2 sequences are required for ori function, while the AT-rich element is dispensable (21). LANA is consistently expressed in KS lesions and crucial for viral maintenance in proliferating cells (1, 23). LANA not only modulates the transcription of viral and cellular genes but also recruits a number of molecules to regulate the replication of the viral episome and the segregation of the newly synthesized genome copies to daughter progeny nuclei by tethering to host chromosomes (15, 29, 44, 46). A simplified model suggests that LANA can mediate the tethering of the KSHV genome to specific components of the chromatin structure through the binding of its C terminus with the TR and association with components of the human chromatin at its N terminus, which includes linker histones and MeCP3 (3, 23).
The long-term persistence of a viral agent which includes KSHV depends on its interaction with are host cell. Its genome replication and viral gene transcription are typically dependent on the involvement of a number of cellular processes. The finding that KSHV genomic DNA as well as TR-containing plasmids is replicated only once during the cell cycle and that LANA has no detectable polymerase or helicase activity required for DNA replication strongly suggest that replication of the KSHV genome is dependent on cellular replication machinery (53). However, the mechanisms for the initiation and regulation of KSHV replication as well as the segregation of newly synthesized DNA copies to daughter cells are still largely unknown. The identification of the cellular molecules involved in KSHV replication, transcription control, and segregation may provide clues towards improved understanding of the life cycle and pathogenesis of KSHV. This study is designed to identify the cellular proteins binding to the TR, which is a critical component of the complex involved in viral tethering and genome replication.
MATERIALS AND METHODS
Cell lines and plasmid.
KSHV-negative cell line BJAB and KSHV-positive cell line BC-3 were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively). Both cell lines were grown at 37°C in a humidified environment supplemented with 5% CO2. pBSpuro was constructed by subcloning the puromycin resistance expression cassette from pBABEpuro into the SalI and ClaI sites of pBS (Stratagene, Inc., La Jolla, CA) containing multiple cloning sites (50). The complete TR unit of KSHV (801 bp) was excised from cosmid clone Z6 with restriction endonuclease NotI and ligated into pBSpuro at the NotI site to obtain pBSpuroA3. The resultant plasmid contains three copies of the TR.
Preparation of nuclear extracts.
To isolate nuclear proteins binding with TRs, nuclear extracts were prepared from the BJAB and BC-3 cell lines. Briefly, 100 × 106 cells were harvested and washed with cold phosphate-buffered saline (PBS), resuspended in buffer A (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of aprotinin/ml), incubated on ice for 1 h, and Dounce homogenized (25 to 30 strokes, observed under a microscope if necessary). Extracted nuclei were collected by centrifugation, washed once with buffer A, and then resuspended in buffer B (20 mM HEPES, 10% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 5 mM DTT, 0.5 mM PMSF, 10 μg of aprotinin/ml). After 30 min of incubation on ice, nuclear debris was removed by centrifugation at high speed. The supernatant containing the nuclear protein was added to an equal volume of buffer C (20 mM HEPES, 30% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 5 mM DTT, 0.5 mM PMSF, 10 μg of aprotinin/ml), and the mixture was snap frozen in aliquots for storage at −80°C. The protein concentration in the nuclear extracts was determined by a standard Bradford assay.
TR column preparation and nuclear extracts binding.
pBSpuroA3 (containing three copies of the terminal repeat) was amplified and extracted. The entire 801-bp TR fragment was purified after the vector was digested with a NotI restriction enzyme (New England Biolabs, Beverly, MA) and resolved on a 1% agarose gel. The 801-bp DNA fragment was retrieved from the gel. About 1 mg of the purified fragment was incubated with CNBr-activated Sepharose (GE Healthcare, Milwaukee, WI) according to the manufacturer's protocol. The unbound reactors were inactivated, and the Sepharose was extensively washed with binding buffer and packed into an Econocolumn (Bio-Rad Laboratories, Inc., Hercules, CA), resulting in TR(+)Sepharose. Nonspecific DNA binding was absorbed with 125 μg of poly(dI-dC) (GE Healthcare, Milwaukee, WI).
The nuclear extracts were diluted 1:4 with buffer A and loaded to the TR Sepharose column or the control column. The elutant that passed through the TR Sepharose fraction was stored as the unbound fraction. After the column was washed with buffer A, the bound proteins were eluted with the same buffer containing sequentially increasing concentrations (50, 100, 300, 500, and 1,000 mM) of NaCl and finally washed with 2.5 M NaCl. Eluted proteins were denatured by boiling in sample buffer and then separated on a 15-by-15-cm 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Proteins were visualized by Coomassie blue staining.
Two 30-bp complementary oligonucleotides (corresponding to nine 30-base pair, high-GC-content repeats [83%] from oligonucleotide 24637 to 24907 of the KSHV genome) (6), 5′GATCCCGGCGCGCCACCCTCCCCGGAGGGG3′ and 5′GATCCCCCTCCGGGGAGGGTGGCGCGCCGG3′, were synthesized (Integrated DNA Technologies, Inc., Coralville, IA) as control DNA fragments. First, two oligonucleotides were annealed together and the double-strand DNA was purified with 15% PAGE gel. The 5′ terminal end of the purified DNA was then phosphorylated with T4 polynucleotide kinase and ligated with T4 DNA ligase (New England Biolabs, Beverly, MA). The DNA was resolved on a 1% agarose gel, and fragments between 400 and 1,000 bp were retrieved from the gel as a control DNA column with a repeated sequence similar to that of the KSHV TR column.
LCQ mass spectrometry (MS) analyses.
Distinct bands between 28 and >260 kDa from 300-, 500-, and 1,000-mM NaCl elutions from both cell lines were excised off the gel and subjected to LCQ proteomic analysis at the Proteomics Core Facility at the University of Pennsylvania School of Medicine. Proteins for each band with an LCQ score over 20 were reported.
Western blot analysis.
Electrophoresed proteins were blotted onto 0.45-μm nitrocellulose paper (Osmonics, Inc., Minnetonka, MN) at 100 V for 1 to 2 h. Blots were blocked with 5% milk in phosphate-buffered saline and washed three times with TBST buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween 20) before overnight incubation with rabbit anti-LANA antiserum or mouse anti-PARP (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and -NPM1 (Cell Signaling Technology, Inc., Danvers, MA) antibodies or rabbit anti-ATR and -BRG1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies at 4°C. Blots were washed three times with TBST and incubated with a 1:10,000 dilution of appropriate Alexa Fluor 680 or IRDye 800 secondary antibody (Molecular Probes, Carlsbad, CA). Membranes were scanned with an Odyssey infrared scanner (Li-Cor Biosciences, Lincoln, NE). Densitometric analysis was performed with Odyssey scanning software.
Immunofluorescence in situ hybridization analysis.
Immunofluorescence assay was performed as described previously (47). Briefly, BC-3 cells were fixed with 3% paraformaldehyde at room temperature for 10 min and permeabilized with 0.1% Triton in PBS for 5 min. Fixed cells were blocked with appropriate serum and then incubated with the specific rabbit antibodies for LANA or KSHV-positive human serum and antibody against one of the candidate proteins, ATR, BRG1, NPM1, or PARP-1. Slides were washed three times in PBS, followed by incubation with a 1:1,000 dilution of appropriate immunoglobulin-Alexa Fluor 488/647-conjugated secondary antibodies. Then cells were postfixed with 3% paraformaldehyde in PBS for 5 min, permeabilized in 0.1% Triton in PBS for 3 min, and treated with 0.1 M Tris-HCl (pH 7.0) for 2 min and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) twice for 2 min. Cells were dehydrated in 70, 80, 90, and 100% ethanol at 4°C for 2 min each and dried. Then cells were treated with 100 μg/ml RNase A for 45 min at 37°C, washed, denatured in denaturing solution (70% formamide in 2× SSC) at 70°C for 2 min, dehydrated, dried, and subjected to in situ hybridization. A Z6 cosmid containing the left end of the KSHV genome was used as a probe after being biotin labeled with a NEBlot Phototope kit (New England Biolabs, Beverly, MA) according to the manufacturer's instructions. Slides were hybridized with the denatured biotinylated Z6 cosmid probe overnight at 55°C. After hybridization, slides were washed in 2× SSC and 0.1× SSC for 15 min each at 55°C, followed by the detection of the KSHV genome with a streptavidin-conjugated Alexa Flour 594 (Molecular Probes, Carlsbad, CA). The slides were washed in PBS, counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and mounted with antifade solution. Slides were examined with an Olympus FluoView FV300 confocal microscope, and images were analyzed with FluoView software (Olympus, Inc., Melville, NY).
Chromatin immunoprecipitations.
For each immunoprecipitation, 2 × 107 BC-3 or BJAB cells were cross-linked by adding formaldehyde to a final concentration of 1% directly to the growth medium at 37°C for 10 min. The reaction was quenched by adding glycine at a final concentration of 0.125 M. Cells were washed once in ice-cold PBS containing protease inhibitor mini-mixture and 1 mM phenylmethylsulfonyl fluoride. Nuclei were pelleted at low speed, lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8) for 10 min on ice, and diluted to 1 ml in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, 16.7 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor mini-mixture). Chromatin was sonicated to ∼500-bp fragments and centrifuged at 13,000 rpm for 10 min at 4°C to remove debris. A portion (5%) of each sample was set aside to measure input DNA, and the remainder was diluted to 2 ml in dilution buffer and split into two 1-ml aliquots. Nonspecific background was precleared with 30 μl of salmon sperm DNA-protein A agarose beads and 1 μl of normal mouse immunoglobulin G for 1 h at 4°C with rotation. Supernatants were incubated with 2 ml of either anti-ATR, BRG1, NPM1, or PARP-1 antibody overnight at 4°C with rotation before immune complexes were collected with 30 μl salmon sperm DNA-protein A agarose for 1 h at 4°C. Beads were washed once each with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 150 mM NaCl), high-salt wash buffer (same as low salt but with 500 mM NaCl), and LiCl wash buffer (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris, pH 8.0) and then twice in TE (10 mM Tris, 1 mM EDTA) (pH 8.0). Pellets were resuspended in 150 μl of chromatin immunoprecipitation (ChIP) assay elution buffer (1% SDS, 0.1 M NaHCO3) and rotated at room temperature for 15 min. Samples were centrifuged, and eluates were removed. Elution was repeated one more time, and eluates were combined. Input DNA (5% of total sample previously set aside) was diluted to 300 μl in elution buffer, followed by reverse cross-linking of all the samples, with the addition of NaCl to a concentration of 0.3 M and 20 μg of RNase A for 5 to 6 h at 65°C. Proteins were removed from samples with 10 mM EDTA, 53 mM Tris-HCl, pH 6.5, and 50 μg proteinase K overnight at 37°C. Samples were extracted once with phenol, once with 1:1 phenol-chloroform, and once with chloroform and precipitated in alcohol. DNA samples were then amplified with primers (forward, 5′-GGGGGACCCCGGGCAGCGAG-3′, and reverse, 5′-GGCTCCCCCAAACAGGCTCA-3′) flanking TR nucleotides 677 to 766. pBSpuroA3 plasmid was amplified in each test as a positive control.
Immunoprecipitation.
For immunoprecipitation, 8 × 107 BJAB and BC-3 cells were lysed on ice with 1 ml of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, pH 8.0) supplemented with protease inhibitors (1 mM PMSF, 10 μg of pepstatin/ml, 10 μg of leupeptin/ml, and 10 μg of aprotinin/ml). The lysates were centrifuged at high speed to remove the cell debris. A control serum was used to preclear the lysate before it was incubated with specific antibody. Precleared lysates were then incubated with anti-ATR, NPM1, BRG1 or PARP-1 antibodies overnight at 4°C with rotation and further incubated with protein A/G Sepharose beads at 4°C for 1 h with rotation. The resulting immunoprecipitates were collected by centrifugation at 2,000 × g for 3 min at 4°C, and the pellets were washed four times with 1 ml of ice-cold RIPA buffer. The immunoprecipitated pellets were resuspended in 30 μl of 2× SDS protein sample buffer (62.5 mM Tris [pH 6.8], 40 mM DTT, 2% SDS, 0.025% bromophenol blue, and 10% glycerol) and then resolved using SDS-PAGE with an 8% polyacrylamide gel. The separated proteins were transferred to a nitrocellulose membrane. Western blot analysis was performed for the detection of LANA protein by the use of an anti-rabbit polyclonal antibody. Similarly, reverse immunoprecipitation with an anti-LANA polyclonal serum was performed for BJAB and BC-3 cells, which were probed for the detection of ATR, NPM1, BRG1, or PARP-1 coimmunoprecipitation with LANA.
RESULTS
Affinity purification of KSHV TR binding proteins.
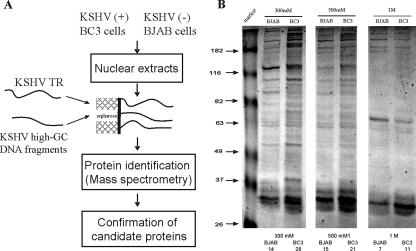
DNA binding proteins are involved in a large number of cellular processes, such as transcription, replication, and recombination. It is well known that the biological activities of virus are tightly associated with the activities of a number of cellular processes. Moreover, since the terminal repeat element of KSHV is a pivotal component required for the modulation of viral replication, transcription, and long-term maintenance of the virus in human cells (20, 30, 48), identification of the cellular proteins binding to TR would be critically important for a better understanding of the mechanisms involved in KSHV maintenance and pathogenesis. A proteomics technological approach is well suited for systematically studying the molecular anatomy of the TR and its DNA binding proteins. In this study, the 801-bp TR DNA was purified and bound to a simple and effective DNA affinity resin (22). Nuclear proteins isolated from KSHV-positive (BC-3) and KSHV-negative (BJAB) cell lines were isolated and subjected to the TR affinity column separately. TR binding proteins bound to the column were sequentially eluted with elution buffer containing an increased concentration of salt. The proteins eluting at high-salt concentrations (300 mM, 500 mM, and 1,000 mM NaCl) were collected and resolved with 8% SDS-PAGE and stained with Coomassie blue (Fig. 1A and B).
FIG. 1.
Proteomic study of TR binding proteins. (A) experimental design. KSHV TR element and 30-bp high-GC repeat control DNA were bound to an activated CNrB resin column. Nuclear extracts from KSHV-positive (+) (BC-3) or -negative (−) (BJAB) cells were incubated with the column, and proteins were eluted from the TR or control column, respectively. High-salt eluted materials were resolved on an SDS-PAGE gel. The visible bands from a TR DNA affinity column were collected and subjected to protein mass spectrometry. (B) Coomassie blue staining of TR binding proteins. Nuclear proteins from KSHV-positive (BC-3) and -negative (BJAB) cells bound to the TR affinity column were extensively washed. TR binding proteins were eluted with elution buffer containing increased concentrations of salt. The proteins eluted at high-salt concentrations (300 mM, 500 mM, and 1,000 mM NaCl) were resolved by SDS-PAGE (8%) and stained with Coomassie blue. Distinct bands were identified and excised for proteomic analysis (LCQ sequencing). In total, 96 bands were subject to proteomic analysis, of which 36 were from BJAB cells and 60 bands were from BC-3 cells.
A total of 96 clearly visible bands, ranging from larger than 260 kDa to 28 kDa, were obtained from both KSHV-positive and -negative cells from the above-mentioned three elutions after Coomassie blue staining (Fig. 1). A comparison of protein bands revealed that there were some specific differences seen in elutions from the KSHV-positive TR column for all the salt concentrations (Fig. 1B). In total, 24 additional bands were seen when combining all three elutions of KSHV-positive cells compared to those seen with elutions of KSHV-negative cells (60 and 36 bands, respectively) (Fig. 1). In addition, a small number of bands (7 bands in the BJAB lane and 11 bands in the BC-3 lane) were obtained at a very high-stringency elution condition of 1 M NaCl in both cell lines (Fig. 1B).
Identification of the proteins associated with the TR DNA elements.
To identify TR binding proteins, all 96 protein bands were excised from the gel and subjected to liquid chromatography-tandem mass spectrometry analysis at the Proteomics Core Facility at the University of Pennsylvania School of Medicine.
In this study, the search engine Mascot was used for protein identification by searching MS data against primary sequence databases (39). Mascot uses a statistical scoring algorithm, the MOWSE score, to calculate the matching scores that represent the level of confidence for the identification. According to its calculation, significance thresholds differ between peptides in the search. Therefore, in our results both MOWSE scores and significance thresholds were considered. MS spectra were also manually inspected to ensure that the identifications were reasonable and of high confidence. In some cases, the score of a single peptide was low (lower than the threshold), but correlation with other identified peptides from the same protein and peptide mass mapping results led to increased confidence in the identification. The theoretical molecular weights of the identified proteins were typically well matched to their correlated positions on the gel, also supporting the MS results and the level of confidence.
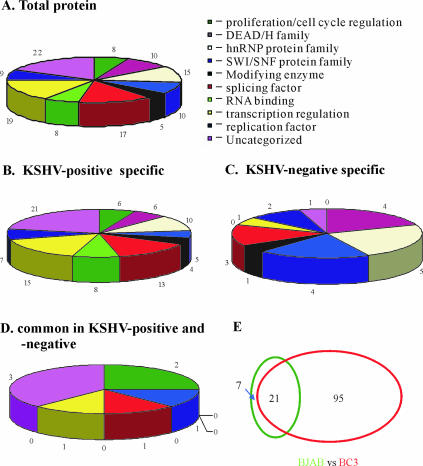
The results of the analysis identified 123 proteins from the 96 excised bands (Fig. 2). No protein was identified in three of the gel bands (Tables 1 to 3). Table 1 shows the protein identification results categorized in KSHV-positive and -negative cells and the corresponding molecular masses of proteins eluted at 300 mM. Two additional detailed tables (Tables 2 and 3) show protein identification results of the bands eluted at 500 mM and 1 M, respectively. Notably, more than one protein was identified in most bands, suggesting that the most visible bands were a mixture of multiple proteins in the one-dimensional gel slice. A comparison of the proteins identified indicated that some were found in more than one band, corresponding to different molecular masses, such as BAF250a, splicing factor prp8, NPM1, BRG1/SMARCA4 isoform 2, U5 200-kDa snRNP-specific protein, and the switch/sucrose nonfermenting (SWI/SNF) complex 155-kDa subunit. This may reflect potential posttranslational modifications of a protein altering its molecular mass (e.g., glycosylation and ribosylation, alternative splice forms, or partial degradation of a protein); the identities of these modifications are beyond the scope of this initial study.
FIG. 2.
Profiling of TR binding proteins. The total proteins eluted from both the KSHV-positive and -negative column cells (A), proteins specific from KSHV-positive (B) and -negative columns (C), and proteins common in both columns (D) were categorized into nine categories according to the functions of the proteins. The profiling of these TR binding proteins is summarized in panels A to E.
TABLE 1.
KSHV TR binding proteins eluted with 300 mM NaCl from BC3 and BJAB cellsa
| Molecular mass (kDa) | Description of TR binding proteins eluted from cell typeb
|
|
|---|---|---|
| BJAB | BC3 | |
| 260 | DNPK1 | DNPK1 |
| 240 | N | DNA-PKcs; BRG1-associated factor 250a; nucleophosmin 1; ATR kinase |
| 235 | N | U5 200-kDa protein; RNA polymerase II largest subunit; Ki-67; nucleophosmin 1 |
| 230 | N | Topo II beta; DNA topoisomerase (ATP-hydrolyzing) (EC 5.99.1.3) alpha |
| 220 | BRG1 | Nuclear DNA helicase II; splicing factor 3b, subunit 2 |
| 205 | N | BRG1; SNF2-alpha |
| 195 | SWI/SNF complex 155 | N |
| 185 | SWI/SNF related, subfamily c, member 2; DNA topoisomerase alpha | BRG1-associated factor 170 |
| 180 | N | Scaffold attachment factor B; l-histidine decarboxylase; SWI/SNF complex 155-kDa subunit |
| 175 | NA | Splicing factor 3b, subunit 1; nuclear DNA helicase II |
| 128 | PARP-1; ADPRT; splicing factor 3a, subunit 1 | PARP-1; ATP-dependent RNA helicase |
| 120 | Proliferating cell nuclear protein P120; DEAD box polypeptide 24; PARP-1; 5′ to 3′ exoribonuclease 2 | 5′ to 3′ exoribonuclease |
| 110 | DNA topoisomerase I, similar to splicing factor proline/glutamine rich; gamma interferon-inducible protein 16 | DNA topoisomerase I; mitochondrial topoisomerase I; Cdc5-related protein, similar to splicing factor praline/glutamine rich; gizzard PTB-associated splicing factor |
| 105 | N | RNA helicase Gu; UBF-1 |
| 95 | DEAD box-1; GTF3C4 protein | DEAD box protein RB |
| 85 | N | hnRNP R; DEAD/H box polypeptide 18; FUSE binding protein |
| 82 | N | DEAD/H box-3, Y linked; CAP-Rf; hnRNP Q2; PABP-2 |
| 75 | N | hnRNP R; KU70; heat shock 70-kDa protein 8 isoform 1; DEAD box polypeptide 17 isoform 1; |
| 70 | N | N |
| 62 | DEAD/H box polypeptide 3 | hnRNP K; DEAD/H box polypeptide 5 |
| 60 | N | hnRNP K isoform A; SWI/SNF complex 60-kDa subunit |
| 56 | N | hnRNP K; PRP4; dyskerin; HSPC117; PAI-RBP1 |
| 55 | N | HSPC117; PAI-RBP1 protein; hnRNP K; mRNA (guanine-7) methyltransferase |
| 52 | DNA topoisomerase I | Paraneoplastic antigen like 5; hnRNP H1; CD2 antigen (cytoplasmic tail) binding protein |
| 48 | N | BAF53; HIV-1 Rev binding protein 2; nucleophosmin 1; translation initiation factor eIF-4A2 homolog; RNA binding protein AUF1; PDIP46 protein; hnRNP F; DAZ-associated protein 1 isoform B; DEAD box protein |
| 39 | N | hnRPC; nucleophosmin 1; poly(rC) binding protein 2 isoform B; HNRAB protein; NF-AT 45-kDa protein |
| 38 | BA18I14.2.2 | N |
| 35 | NA | FBRNP; splicing factor U2AF 35-kDa subunit |
| 31 | hnRNP A2; hnRNP A1; hnRNP H3; casein alphaS1 | hnRNP R; DEAD/H box polypeptide 18; FUSE binding protein |
| 30 | N | hnRPA1B2 protein; hnRNP A1 |
| 29 | N | hnRNP A1; hnRNP A2; hnRNP A3; alternative splicing factor ASF-2; fibulin 1 isoform D; fibrillarin; dynein, axonemal, heavy polypeptide; bZIP-enhancing factor; histone H1c |
eIF, eukaryotic initiation factor; HIV-1, human immunodeficiency virus type 1; N, no bands shown; NA, data not available; PABP-2, poly(A) binding protein 2; PTB, polypyrimidine tract binding protein; RB, retinoblastoma; UBF, upstream binding factor.
The numbers of proteins eluted from BJAB and BC3 cells were 14 and 28, respectively.
TABLE 3.
KSHV TR binding proteins eluted with 1 M NaCl in BC3 and BJAB cellsa
| Molecular mass (kDa) | Description of TR binding proteins eluted from cell typeb
|
|
|---|---|---|
| BJAB | BC3 | |
| 240 | BRG1-associated factor 250a | BRG1-associated factor 250a |
| 235 | N | Splicing factor Prp8 |
| 220 | SNF2-alpha; SNF2-like 4 | SMARCA4 isoform 2 |
| 185 | SWI/SNF related, subfamily c, member 2 | SWI/SNF related, subfamily c, member 2 |
| 175 | Cytokeratin 1 | Cytokeratin 1 |
| 70 | N | Fusion derived from t(12;16) malignant liposarcoma |
| 62 | Keratin 2a | Cytokeratin 1 |
| 60 | hnRNP K; splicing factor U2AF large chain, human | hnRNP H1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin d2; splicing factor U2AF |
| 35 | hnRNP A2/B1 isoform B1; dynein, axonemal, heavy polypeptide 8 | hnRNP A2; hnRNP A1; hnRNP H3 isoform A; hnRNP A3 |
| 31 | hnRNP A2/B1 isoform B1; hnRNP H3 isoform A; TRRAP | hnRNP A2/B1 isoform B; hnRNP A3; hnRNP H3 isoform A |
| 30 | N | hnRNP A2/B1 isoform B; dynein, axonemal, heavy polypeptide 8; histone acetyltransferase MOZ2; envelope glycoprotein (HIV-1) |
HIV, human immunodeficiency virus type 1; N, no bands shown.
The numbers of proteins eluted from BJAB and BC3 cells were 7 and 11, respectively.
TABLE 2.
KSHV TR binding proteins eluted with 500 mM NaCl in BC3 and BJAB cellsa
| Molecular mass (kDa) | Description of TR binding proteins eluted from cell typeb
|
|
|---|---|---|
| BJAB | BC3 | |
| 260 | N | Splicing factor Prp8; nucleophosmin 1 |
| 240 | BRG1-associated factor 250a | BRG1-associated factor 250a; nucleophosmin 1 |
| 235 | BRG1-associated factor 250a; splicing factor Prp8 | Splicing factor Prp8; U5 snRNP-specific protein, 200 kDa |
| 230 | U5 snRNP 200 kDa; BRG1-associated factor 250a; SMARCA4 isoform 2 | U5 snRNP-specific protein, 200 kDa |
| 220 | SMARCA4 isoform 2; SNF2-alpha | SMARCA4 isoform 2; SNF2-alpha; PDCD 11 |
| 185 | SWI/SNF complex 170-kDa subunit; BRG1-associated factor 170 | SWI/SNF related, subfamily c |
| 175 | SWI/SNF complex 155-kDa subunit; splicing factor 3b, subunit 1, 155 kDa | SWI/SNF complex 155-kDa subunit |
| 128 | PARP-1; E1B 55-kDa-associated protein 5 | PARP-1 |
| 110 | DNA topoisomerase I | DNA topoisomerase I; DEAD box polypeptide 23, similar to splicing factor proline/glutamine rich; E1B 55-kDa-associated protein 5 |
| 95 | DEAD box-1, similar to Ewing's sarcoma breakpoint region 1 | DEAD box-1, similar to Ewing's sarcoma breakpoint region 1; histone H1c |
| 82 | DEAD 3 | NA |
| 70 | N | DEAD box-5; CARF; FUS/TLS protein |
| 62 | hnRNP K isoform 1; cellular senescence-inhibited gene protein; splicing factor U2AF large chain, human | hnRNP isoform K |
| 60 | Splicing factor U2AF large chain, human | hnRNP K; U4/U6.U5 tri-snRNP-associated 65-kDa protein; splicing factor U2AF large chain, human; splicing factor SF3a60; dynain |
| 56 | N | hnRNP; hypothetical protein DKFZp434I1614.1 |
| 55 | Cytokeratin 1 | RNA binding protein 56; RNA polymerase II; HSPC117; ATR kinase; nuclear matrix protein NMP200 related to splicing factor PRP19 |
| 39 | N | hnRNP H1; paraneoplastic antigen like 5; Ecto-ATPase |
| 35 | hnRNP K isoform A2; hnRNP A3 | FBRNP; hnRNP A3; HNRPC; EndoA′ cytokeratin |
| 31 | hnRNP A2; hnRNP A3; hnRNP H3; hnRNP core protein A1 | hnRNP A2/B1 isoform B1; hnRNP core protein A1; hnRNP H3 isoform A; fibrillarin; histone H1d |
| 30 | N | hnRNP A1; hnRNP A2; hnRNP A3; hnRNP H3; dynein, axonemal, heavy polypeptide 8 |
| 29 | N | hnRNP A2; RPS2 |
N, no bands shown; NA, data not available.
The numbers of proteins eluted from BJAB and BC3 cells were 15 and 21, respectively.
As indicated above, the identification results also showed a trend of more proteins in low-salt elution and KSHV-positive BC-3 cells than in KSHV-negative cells (compare Tables 1, 2, and 3 and Fig. 3A). The 123 polypeptides identified were grouped into nine specific categories, and one group was uncategorized as the proteins did not fit into any selected groups (Fig. 2A). In KSHV-positive BC-3 cells, 116 proteins were identified from 59 gel bands (no polypeptides were identified in 1 of the bands) from all three elutions and 95 proteins were seen exclusively in KSHV-positive BC-3 cells (Fig. 2B). However, in KSHV-negative BJAB cells, 28 proteins were identified from 34 gel slices (no proteins were identified in 2 of the bands) from all three elutions, with only 7 proteins seen exclusively in KSHV-negative BJAB cells (Fig. 2C and D). A comprehensive list, including accession numbers of all proteins identified from all three elutions of KSHV-positive and -negative cell lines, are presented in Table 4 and are schematically shown in Fig. 2E.
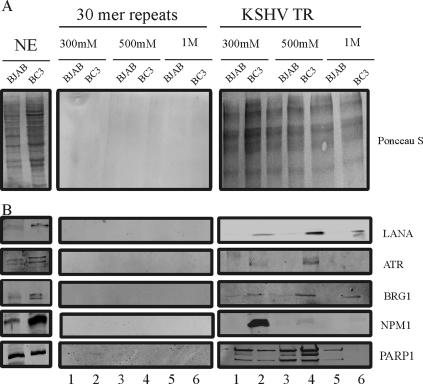
FIG. 3.
Specific binding of candidate proteins to TR DNA affinity column. (A) Ponceau S staining of nuclear extracts and TR binding proteins identified at different salt concentrations. Nuclear extracts (NE) and the high-salt-eluted proteins from both TR as well as control DNA column were resolved on an 8% SDS-PAGE gel and stained with Ponceau S. In comparison with the protein bands obtained from the TR column, fewer bands were shown in control column. (B) The presence of candidate proteins LANA, ATR, BRG1, NPM1, and PARP-1 was detected in nuclear extracts, with faint signals for LANA in BJAB due to overflow from the BC-3 NE lane. These proteins were eluted from the TR column lane but not the control column.
TABLE 4.
KSHV TR binding proteins identified in BC3 and BJAB cellsaa
| Molecular mass (kDa) | Description of TR binding proteins eluted from cell type
|
|
|---|---|---|
| BJAB | BC3 | |
| 260 | DNPK1 (AAC50210) | DNPK1; splicing factor Prp8 (CK610786); nucleophosmin 1 (NP_002511) |
| 240 | BRG1-associated factor 250a (AF231056) | DNA-PKcs (P78527); BRG1-associated factor 250a; nucleophosmin 1; ATR (NP_001175) |
| 235 | BRG1-associated factor 250a; splicing factor Prp8 | U5 220-kDa protein (NP_006436); RNA polymerase II (NP_631961); Ki-67 (NP_002408); nucleophosmin 1; splicing factor Prp8; U5 snRNP-specific protein, 200 kDa |
| 230 | U5 snRNP 200 kDa; BRG1-associated factor 250a; BRG1 | Topo II beta (NP_033435); DNA topoisomerase (ATP-hydrolyzing) (EC 5.99.1.3) alpha (A40493); U5 snRNP-specific protein, 200 kDa (O75643) |
| 220 | BRG1 (AAG24790) | Nuclear DNA helicase II; splicing factor 3b, subunit 2 (AAH00401); SNF2-alpha (P51531); PDCD 11 (NM_014976); BRG1 |
| 195 | SWI/SNF complex 155 (AAC50693) | BRG1; SNF2-alpha |
| 185 | SWI/SNF-related subfamily c, member 2; DNA topoisomerase alpha (CAA76313); SWI/SNF complex 170-kDa subunit | BRG1-associated factor 170 (NP_620706); SWI/SNF related, subfamily c |
| 180 | N | Scaffold attachment factor B (AAC18697); l-histidine decarboxylase (D16583); SWI/SNF complex 155-kDa subunit |
| 175 | SWI/SNF complex 155-kDa subunit (XP_343571); splicing factor 3b, subunit 1; cytokeratin 1 | Splicing factor 3b, subunit 1 (XP_343571); nuclear DNA helicase II(NP_776461); SWI/SNF complex 155-kDa subunit; cytokeratin 1 (P04264) |
| 128 | PARP-1 (P09874); ADPRT (NP_037195); splicing factor 3a, subunit 1 (AAH29753); E1B 55-kDa-associated protein 5 (AAH02564) | PARP-1; ATP-dependent RNA helicase (Q08211) |
| 120 | Proliferating cell nuclear protein P120 (AAA36398); DEAD box polypeptide 24 (NP_065147); PARP-1; 5′-3′ exoribonuclease 2 (NP_036387) | 5′-3′ exoribonuclease (NP_036387); DEAD box polypeptide 23 (AAH02366), similar to splicing factor proline/glutamine rich (AAH04534); E1B 55-kDa-associated protein 5 |
| 110 | DNA topoisomerase I (NP_003277), similar to splicing factor proline/glutamine rich; gamma interferon-inducible protein 16 (AB208989) | DNA topoisomerase I; mitochondrial topoisomerase I (NP_443195); Cdc5-related protein (NP_001244), similar to splicing factor praline/glutamine rich (AAH04534); gizzard PTB-associated splicing factor (AAC59935) |
| 105 | N | RNA helicase Gu (PC6010); UBF-1 |
| 95 | DEAD box-1 (NP_004930); GTF3C4 protein (BC094774) | DEAD box protein RB (NP_004930), similar to Ewing's sarcoma breakpoint region 1 (AAH11048) |
| 85 | N | hnRNP R (AAH01449); DEAD/H box polypeptide 18 (AAH01238); FUSE binding protein (AAC50892) |
| 82 | DEAD 3 | DEAD/H box-3, Y linked (NP_004651); CAP-Rf (NM_001356); hnRNP Q2 (AAK59704); PABP-2 (Q15097) |
| 75 | N | HnRNP R (XP_001541); KU70 (NP_001460); heat shock 70-kDa protein 8 isoform 1 (NP_006588); DEAD box polypeptide 17 isoform 1 (NP_006377) |
| 70 | N | DEAD box-5; CARF (NM_017632); FUS/TLS protein; fusion derived from t(12;16) malignant liposarcoma (AAH26062) |
| 62 | DEAD/H box polypeptide 3 (NP_001347); hnRNP K isoform 1; cellular senescence-inhibited gene protein (AAN46298); splicing factor U2AF large chain, human (S20250); keratin 2a (NP_000414) | Cytokeratin 1; hnRNP K (NP_079555); DEAD/H box polypeptide 5 (NP_004387) |
| 60 | hnRNP K; splicing factor U2AF large chain, human | hnRNP K isoform A; SWI/SNF complex 60-kDa subunit (AAC50696); U4/U6.U5 tri-snRNP-associated 65-kDa protein (AAK49524); splicing factor U2AF large chain, human; splicing factor SF3a60 (CAA57388); dynein, axonemal, heavy polypeptide 8 (NP_001362); hnRNP H1 (NP_005511) |
| 56 | N | hnRNP K; PRP4 (XP_131444); dyskerin (CAB51168); HSPC117; PAI-RBP1 PAI-RBP1 (AAH02488); hypothetical protein DKFZp434I1614.1 (T46344) |
| 55 | Cytokeratin 1 | HSPC117 (BC016707); PAI-RBP1 protein; hnRNP K; mRNA (guanine-7) methyltransferase (AB022605) |
| 52 | DNA topoisomerase I | Paraneoplastic antigen-like 5; hnRNP H1; CD2 antigen (cytoplasmic tail) binding protein (NP_006101) |
| 48 | N | BAF53 (CR533529); HIV-1 Rev binding protein 2 (AAH16778); nucleophosmin 1; translation initiation factor eIF-4A2 homolog (S45142); RNA binding protein AUF1 (A54601); PDIP46 protein (AAH01488); hnRNP F (AAH16736); DAZ-associated protein 1 isoform B (NP_061832); DEAD box protein (CAC14786) |
| 39 | BA18I14.2.2 (CAC08398) | hnRPC (AAC61695); nucleophosmin 1; poly(rC) binding protein 2 isoform B (NP_035995); HNRAB protein (NP_006749); NF-AT 45-kDa protein (BG625087); hnRNP H1; paraneoplastic antigen like 5 (BC101111); Ecto-ATPase (XP_302537) |
| 35 | hnRNP K isoform A2; hnRNP A3; dynein, axonemal, heavy polypeptide 8 | FBRNP (AA408025); splicing factor U2AF 35-kDa subunit; hnRNP A3; HNRPC; EndoA′ cytokeratin (AAA37551); hnRNP H3 isoform A |
| 31 | hnRNP A2 (NP_112533); hnRNP A1 (P09651); hnRNP H3 (XP_165561); casein alphaS1; hnRNP core protein A1 | hnRNP R; DEAD/H box polypeptide 18; FUSE binding protein; hnRNP A2/B1 isoform B1 (NP_112533); hnRNP core protein A1 (P09651); hnRNP H3 isoform A (NP_036339); fibrillarin (NP_001427); histone H1d (NP_005310); hnRNP A3 (XP_165561) |
| 30 | N | hnRPA1B2 protein; hnRNP A1; hnRNP A2; hnRNP A3; hnRNP H3; dynein, axonemal, heavy polypeptide 8; histone acetyltransferase MOZ2 (AF217500); envelope glycoprotein (HIV-1) (AAK85226) |
| 29 | N | hnRNP A1; hnRNP A2; hnRNP A3; alternative splicing factor ASF-2 (B40040); fibulin 1 isoform D (CO779977); fibrillarin (NP_001427); dynein, axonemal, heavy polypeptide; bZIP-enhancing factor (NP_005773); histone H1c (NP_005311); hnRNP A2; RPS2 (AB082925) |
eIF, eukaryotic initiation factor; HIV-1, human immunodeficiency virus type 1; N, no bands shown; PABP-2, poly(A) binding protein 2; PTB, polypyrimidine tract binding protein; RB, retinoblastoma; UBF-1, upstream binding factor 1.
The identified proteins were grouped in 10 categories based on their functions, which are shown in Fig. 2, and a detailed list is provided in Table 5. The categories include the following: proliferation/cell cycle regulatory proteins, proteins involved in spliceosome components, such as heterogeneous nuclear ribonuclear proteins, the DEAD/H family, the SWI/SNF protein family, splicing factors, RNA binding proteins, transcription regulation proteins, replication factors, modifying enzymes, and a number of proteins that did not specially fit into any of the above-listed groups based on known function and so remained uncategorized.
TABLE 5.
Category of KSHV TR binding proteinsa
| Category | Description of KSHV TR binding protein for cell type
|
||
|---|---|---|---|
| BJAB specific | Common | BC3 specific | |
| Proliferation/cell cycle regulation | Proliferating cell nuclear protein P120; cellular senescence-inhibited gene protein | Ki-67; PDCD 11; proto-oncogene c-ros-1 protein precursor; Cdc5-related protein; putative serine-rich protein, CARF; CD2 antigen (cytoplasmic tail) binding protein 2 | |
| DEAD/H family | DEAD/H box-1, -3, -9, and -23 | DEAD/H box-5, -17, -18, and -4, RB, polypeptide 21 (Gu) | |
| hnRNP protein family | hnRNP protein family (U, K, A2, A3, and H3) | hnRNP protein family (R, L, H1, F, A1, core protein A1, Q2, hnRNB, and FBRNP), Similar to hnRNP U | |
| SWI/SNF protein family | SNF2-like 4 | SNF60; SNF2-alpha; BAF250a; SWI/SNF complex 155 | SNFc2; SNFe1; SNF2d2; SMARCA4 isoform 2; possible global transcription activator SNF2L2 |
| Modifying enzyme | PARP-1 | HSPC117; ATR kinase; DNA-PK; kinesin family member 1B isoform alpha | |
| Splicing factor | Splicing factor 3a, subunit 1 | U5 200-kDa protein; splicing factor 3b, subunit 155 kDa; splicing factor prp8 | U5 snRNP-specific protein; U4/U6.U5 tri-snRNP-associated 65-kDa protein; splicing factor, arginine/serine-rich 8 isoform 1; precursor mRNA processing protein; splicing factor 3b, subunit 2; splicing factor SF3a60; alternative splicing factor ASF-2; splicing factor U2AF 35 kDa; alternative splicing factor ASF-3; nuclear matrix protein NMP200 related to splicing factor PRP19; gizzard PTB-associated splicing factor; PRP4 pre-mRNA processing factor 4 homolog; U3 small nucleolar interacting protein |
| RNA binding proteins | Poly(rC) binding protein 2 isoform B; PAI-RBP1 protein; putative RNA binding protein KOC; AUF1; poly(A) binding protein 2; TIA-1 protein; DAZ-associated protein 1 isoform B; scaffold attachment factor B | ||
| Transcription regulation proteins | GTF3C4 protein/general transcription factor 3C polypeptide 4; gamma interferon-inducible protein 16; TRRAP | 5′-3′ exoribonuclease 2 | mRNA (guanine-7) methyltransferase; bZIP-enhancing factor; RNA polymerase II largest subunit; Dhm1-like protein; nucleolar transcription factor 1 (UBF-1); FUS/TLS protein; FUSE binding protein; translation initiation factor eIF-4A2 homolog, human; transcription factor NF-AT 45K; ATP-dependent RNA helicase A; TBP-associated factor, RNA polymerase II; U3 small nucleolar interacting protein; dyskerin; BAF53; fibrillarin |
| Replication factor | E1B 55-kDa-associated protein; DNA topoisomerase I | PDIP46 protein; l-histidine decarboxylase; E1B 55-kDa-associated protein; DNA topoisomerase (ATP-hydrolyzing) (EC 5.99.1.3) alpha; DNA topoisomerase II; histone H1c; histone acetyltransferase MOZ2 | |
| Uncategorized | Dynein | Heat shock 70-kDa protein 8 isoform 1; beta spectrin; clathrin heavy chain 1; HIV-1 Rev binding protein 2; envelope glycoprotein (HIV-1); ecto-ATPase (ectonucleoside triphosphate diphosphohydrolase 2 isoform 1); nucleophosmin 1; myoblast antigen 24.1D5; similar to Caenorhabditis elegans hypothetical 55.2-kDa protein F16A11.2; KIAA 1934 protein; protein for IMAGE:3938975; thyroid hormone receptor interactor 12; CLTC protein; protein for MGC:9466, similar to Ewing's sarcoma breakpoint region 1; hypothetical protein DKFZp434I1614.1; fusion derived from t(12;16) malignant liposarcoma; centaurin beta 5; pyruvate kinase, liver, and RBC type; pyruvate kinase type L; similar to splicing factor proline/glutamine rich; fusion derived from t(12;16) malignant liposarcoma | |
eIF, eukaryotic initiation factor; HIV-1, human immunodeficiency virus type 1; PTB, polypyrimidine tract binding protein; RB, retinoblastoma; RBC, red blood cell; TBP, TATA box binding protein; UBF-1, upstream binding factor 1.
Confirmation of individual proteins binding to the TR DNA by Western blot analysis.
To confirm the specificity of the TR binding proteins, 400-to-1,000 bp DNA fragments composed of 30-bp double-stranded DNA repeats were used as the control DNA column. The repeats are noncoding sequences of the KSHV genome and have similar GC contents (approximately 83%) to TR. The eluted proteins from the control DNA column and TR element column were resolved on 8% SDS-PAGE and transferred to a nitrocellulose membrane. After Ponceau S staining, significantly fewer bands were seen, to an almost undetectable level, on the membranes of control DNA compared to those for the TR element elutions (Fig. 3A). After probing with specific antibodies against the candidate proteins PARP-1, ATR, NPM1, and BRG1, the presence of these proteins was predominantly detected in the TR column but not in the control DNA column (Fig. 3B). In addition, because KSHV LANA is demonstrated to bind to the TR, the presence of LANA protein was also verified in elutions from the TR column (Fig. 3B, top panel). The data showed that LANA mostly eluted from the column at 500 mM NaCl, an extent similar to those of ATR and PARP-1 (Fig. 3, lane 4). BRG1 was seen to be tightly associated with the TR in BC-3 but had the same lower level of signal as that in the elution from BJAB cells which are KSHV negative (Fig. 3, lanes 2, 4, and 6). Interestingly, NPM1/B23 associated predominately with TR from BC-3 extracts and was mostly eluted at 300 mM NaCl, with no detectable signals at 500 mM and 1 M salt elutions (Fig. 3, lanes 2 and 4). Thus, ATR, BRG1, NPM1/B23, and PARP-1 all associated with the TR DNA element predominantly in the presence of KSHV latent antigens which include LANA (Fig. 2, lanes 2, 4, and 6). Although PARP-1 associated with the TR from BC-3 extracts with the highest signal at 500 mM elution, some signals were also seen in BJAB extracts at 300 and 500 mM and 1 M elutions, suggesting that PARP-1 is associated with the TR element independently of the presence of the KSHV latent antigen, including LANA.
Physical interaction between LANA candidate proteins and KSHV TR element.
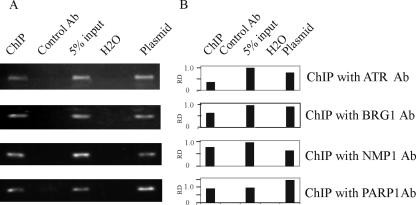
To determine whether the candidate proteins can bind to the TR DNA element, we examined the interaction between candidate proteins and TR in BC-3 cells by ChIP assay. The cross-linked chromatin from 50 million cells was immunoprecipitated with specific antibodies against PARP-1, ATR, NPM1, and BRG1, and a specific sequence was amplified using primers that amplified a 90-bp amplicon within the TR element. The results of the interaction between candidate proteins and the TR in the KSHV-positive cells are shown by ChIP analysis (Fig. 4). ChIP analysis showed that the four candidate proteins, PARP-1, ATR, NPM1, and BRG1, associated with the TR DNA element, with no detectable signals seen with the match antibody control (Fig. 4). These results suggest that these proteins associated with the TR elements and that it is possible that their signals also include association with LANA bound to the TR element, although they may also be able to be independent in their association with the TR element.
FIG. 4.
Physical interactions between candidate proteins and TR. The interaction between candidate proteins PARP-1, ATR, NPM1, BRG1, and TR in BC-3 cells were confirmed by ChIP assay. For each immunoprecipitation, 2 × 107 BC-3 or BJAB cells were cross-linked using formaldehyde. Chromatin was sonicated to ∼500-bp fragments and precleared. After incubation with anti-ATR, BRG1, NPM1, or PARP-1 antibodies (Abs), immune complexes were collected with salmon sperm DNA-protein A/G Sepharose. With extensive washing, chromatin was reverse cross-linked to purify bound DNA. The region of TR was amplified with primers flanking nucleotides 677 to 766. pBSpuroA3 plasmid was amplified in each test as a positive control. (A) Specific amplification of TR DNA immunoprecipitated by antibodies against candidate genes. (B) Quantification of the immunoprecipitation based on quantification of the amplified specific bands. RD, relative density.
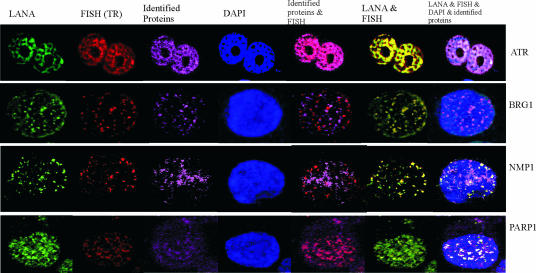
Colocalization of LANA, PARP-1, ATR, NPM1, and BRG1 with TR DNA.
Cellular and/or viral molecules that interact in cells typically would also colocalize in vitro. Colocalization in KSHV-positive BC-3 cells was detected by immunofluorescence in situ hybridization analysis. Candidate proteins (PARP-1, ATR, NPM1, and BRG1) were stained with specific antibodies, and the viral DNA was probed with TR-specific DNA fragments. Confocal microscopy showed that these proteins colocalized with the KSHV genome. Signals for the TR DNA, LANA protein, and candidate proteins (PARP-1, ATR, BRG1, and NPM1) were clearly colocalized, suggesting an association of these proteins in a complex in KSHV-infected cells (Fig. 5). These candidate proteins were all seen to colocalize with LANA in punctuate nuclear signals with the exclusion of the nucleolus. Blue DAPI staining showed nuclear chromatin stain. Brick-red merged panels showed the colocalization of the candidate proteins with the KSHV genome signals, the orange merged signal showed the colocalization of LANA and KSHV genome, as expected, and the white merged signals indicated colocalization of LANA, candidate protein ATR, BRG1, NPM1, or PARP-1, DAPI, and the KSHV genome (Fig. 5).
FIG. 5.
Colocalization of candidate proteins with TR and LANA. Shown is the colocalization between candidate proteins PARP-1, ATR, NPM1, and BRG1 and the TR as well as LANA in BC-3 cells. BC-3 cells were fixed and probed with the specific rabbit antibodies for LANA or human anti-LANA serum combining antibody against candidate protein ATR, BRG1, NPM1, or PARP-1. Slides were visualized by appropriate immunoglobulin-Alexa Fluor-conjugated secondary antibodies. Then cells were postfixed, treated with RNase A, and denatured. The slides were subjected to in situ hybridization with biotin-labeled Z6 cosmid probe and detected with a streptavidin-conjugated Alex Flour 594. The nuclei were counterstained with DAPI. FISH, fluorescence in situ hybridization.
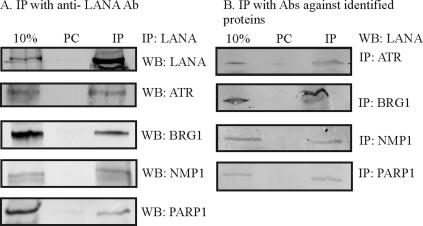
LANA associates with the candidate proteins ATR, NPM1, BRG1, and PARP-1 in KSHV-positive cells.
To further corroborate the above-mentioned observations, we performed immunoprecipitation analysis for each of the above-mentioned proteins or LANA and performed Western blotting for specific antigens to determine whether they associated in a complex in KSHV-positive cells. The results indicated that LANA associated with ATR, BRG1, NPM1, and PARP-1 in the KSHV-infected BC-3 cell line, using a specific antibody for LANA and Western blotting for each of the four candidate proteins (Fig. 6A). Importantly, in the reverse immunoprecipitation using specific antibodies against candidate proteins, Western blot analysis showed that LANA was detected in the immune complexes (Fig. 6B). Thus, ATR, BRG1, NPM1, and PARP-1 can associate with LANA and the TR DNA element and may also be important for TR function and KSHV persistence in the cells.
FIG. 6.
Physical interactions between candidate proteins and LANA. (A) The interaction between candidate proteins PARP-1, ATR, NPM1, BRG1, and LANA in BC-3 cells were confirmed by coimmunoprecipitation assay by pulling down the above-mentioned proteins along with LANA using LANA-specific antibody. A total of 8 × 107 BJAB and BC-3 cells were lysed in RIPA buffer. After preclearing, the lysate was incubated with specific antibody against LANA and resolved with 8% SDS-PAGE. Western blotting for the indicated proteins was done by stripping and reprobing the same membrane. (B) Reverse immunoprecipitation with specific antibodies against proteins indicated was performed for BC-3 cells and probed with LANA polyclonal antibody. Ab, antibody; IP, immunoprecipitate; PC, preclear; WB, Western blot.
DISCUSSION
In this study, we have successfully combined stringent protein purification with DNA affinity column and sensitive MS analysis to identify a set of proteins binding to KSHV TR DNA. Here, we present the identification of 123 proteins that were bound to the TR DNA element of KSHV from KSHV-negative and -positive B cell lines. The identity of binding proteins was performed by analysis of multiple polypeptides for each protein and confirmed by LCQ sequencing.
The TR sequence of the KSHV genome contains at least two cis elements for LANA binding per copy of the TR element (2, 20). While two or more copies of TR are required for long-term maintenance, a single TR confers LANA-dependent origin activity on plasmid DNA (17). The TR can function like an autonomous replicating element, with LANA bound to the cognate sequences within the TR supporting viral replication (15). The fact that LANA can suppress transcription when bound to the TR seems to challenge a model by which the TR can directly contribute to KSHV DNA replication (14). Deletion mapping revealed a 71-bp-long minimal replicator containing two distinctive sequence elements: LANA binding sites (LBS1/2) and an adjacent 29- to 32-bp-long GC-rich sequence, which is referred to as the replication element (21). LANA interacts with many cellular factors, including RING3, pRb, p53, HP1, CREB binding protein, mSin3, MeCP2, DEK, histone H1, ORCs, and glycogen synthase kinase 3β (4, 12, 13, 24, 28, 29, 40, 41, 50, 52). The majority of these factors are involved in transcription, remodeling chromatin structure, and replication. Moreover, in support of our previous work, we noted an association of histone H1 isoforms C and D, especially in the BC-3 elution, suggesting a requirement for TR proteins which include LANA. However, no association of additional histone, including H2A and H2B, H3, and H4, was identified in this comprehensive screen. These proteins may be required for the maintenance of viral episomes as well as regulation of the transcription program during latency. Consistent with this idea, LANA is known to be an essential factor for maintaining the viral genome (18). However, it has been demonstrated that LANA does not have any enzymatic activity like helicase or polymerase. Therefore, enzymes which contain these activities and core components of the replication machinery are most likely recruited to the TR, contributing to the initiation and replication of the viral episome during latency.
Notably, the majority of the proteins identified are KSHV specific and were found to be associated with the TR from the BC-3, but not the BJAB, KSHV-negative nuclear extracts. This finding suggests that the binding between these proteins and TR DNA is KSHV dependent. Thus, the binding of these proteins to the TR element may be mediated by KSHV-encoded proteins, mainly LANA, as LANA is predominantly expressed during latent infection and binds to the TR. The identified proteins were grouped into several categories according to their nuclear functions. Among these are structural/nuclear matrix protein components, such as heterogeneous nuclear ribonuclear proteins and protein-associated splicing factor and chromatin regulatory and structural proteins, which include histones as well as proteins involved in other DNA-modulating activities, such as the chromatin-remodeling protein DEAD/DEAH proteins, and proteins whose nuclear functions are not yet fully known. These proteins are most likely in direct association with TR DNA or associated with other viral or cellular proteins bound to the TR in a macromolecular complex. Some of the identified proteins, for example the histones, have also been shown to be involved in DNA metabolic processes and in association with DNA (34). Some proteins identified are involved in the regulation of cell proliferation, for example, in proliferating cell nuclear proteins P120 and Ki-67 (11, 16). However, some have not been strictly classified in terms of their functions. The results presented here suggest that the TR DNA element is important for a number of biological processes likely to be associated with KSHV persistence in the infected cell.
DNA damage can be experimentally induced by irradiation, UV exposure, and chemical treatment (19, 45). However, this damage may also arise naturally as a consequence of DNA replication (31). Many of the repair and recombination proteins are reported to be redistributed to double-stranded breaks after irradiation or chemical treatment and colocalize with cellular DNA replication sites during S phase (32). It is believed that the presence of stalled replication forks or stretches of single-stranded DNA is responsible for recruiting these factors (27). In the absence of such repair functions, genomic integrity degrades. In this study, multiple proteins associated with DNA damage repair, such as ATR and poly(ADP-ribose) polymerase (PARP-1), have been identified. PARP-1 is a nuclear enzyme which is activated in response to genotoxic insults by binding to damaged DNA and attaching polymers of ADP-ribose to nuclear proteins at the expense of its substrate NAD+ (35). PARP-1 is important in DNA damage signaling and has been reported to bind KSHV TR DNA (38). Our result confirmed the interaction between PARP-1 and TR DNA and further demonstrated that in spite of binding directly to the TR, PARP-1 also associates with LANA independently of the TR, suggesting a role of PARP-1 in the context of LANA function. ATM and rad3-related (ATR) proteins are members of the phosphoinositide 3-kinase related kinase family (7). ATR plays a crucial role in the normal cell-cycle and early development and is also responsible for the DNA damage response that halts the progression of the cell cycle, in particular, in response to a variety of DNA damage signals (8). Importantly, and likely to be related to its association with the KSHV TR element, ATR is speculated to modulate the progression of DNA replication by regulating S-phase kinases at unfired origins from its vantage point at active or stalled replication forks and sites of damage (5). Thus, it may play a role in ensuring the continued firing of the initiation of replication at the TR during latent infection.
Two main groups of chromatin remodeling complexes exist in mammalian cells: (i) those requiring ATP hydrolysis to alter histone-DNA contacts, such as SWI/SNF protein (49), and (ii) those that covalently modify histone proteins by a variety of posttranslational modifications (phosphorylation, acetylation, and methylation), such as histone acetyltransferases, methyltransferases, and histone deacetylase complexes (17). We have detected members of both types of complexes as TR binding proteins from our study. BRG1 is an ATPase associated with SWI/SNF protein (33). NPM1 (B23 protein) is a multifunctional nucleolar protein whose molecular chaperone activity is proposed to play a role in ribosome assembly (10). The identification of these proteins suggests a role for these proteins in KSHV replication and transcription. There is evidence that interactions with transcription factors may target SWI/SNF complexes to specific promoters to regulate transcription. It is also possible that KSHV uses a similar mechanism of targeted regulation by recruiting SWI/SNF complexes to weak viral promoters in early infection or during reactivation to enhance the transcription of a selected set of promoters and also the TR element controlling transcription of some viral genes which include K1 (25, 26).
We believe that the large number of binding proteins reflects the multiple roles of the KSHV TR element in viral DNA replication and gene expression. Additionally, not all of the binding proteins are likely to interact directly with the TR DNA. Therefore, some of the interactions may be dependent on interaction with intermediate viral binding partners like LANA. It remains to be determined which TR binding proteins physically interact with TR independently of viral proteins. Many of the identified proteins associate with numerous other proteins, including other TR binding proteins. It is possible that the association of TR with one or two proteins may lead to the recruitment of numerous other proteins or complexes required for specific functions.
We hypothesize that KSHV recruits some of these cellular proteins to the TR element which contains a latent replication ori to participate directly in KSHV replication. Alternatively, they may be targeted to damaged viral DNA that arises during replication. The determination of the functions of these proteins in the context of KSHV infection is likely to be complex. A number of the TR binding proteins function in the major cellular recombination repair pathways; thus, it will be of interest to determine whether these proteins have a necessary role in KSHV replication, are recruited to sites of DNA damage or stalled replication forks, or may increase the efficiency of KSHV replication initiation at the TR. Further studies are needed to determine the specific role of the proteins identified in viral infection and long-term genome persistence.
aaaaaaaaaa
Acknowledgments
This work was supported by grants from the Leukemia and Lymphoma Society of America and by public health service grants NCI CA072510 and CA091792 and NIDCR DE01436 (to E.S.R.). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
We also thank Chao-Xing Yuan and the Proteomics Core Facility at the University of Pennsylvania School of Medicine for their technical support.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borah, S., S. C. Verma, and E. S. Robertson. 2004. ORF73 of herpesvirus saimiri, a viral homolog of Kaposi's sarcoma-associated herpesvirus, modulates the two cellular tumor suppressor proteins p53 and pRb. J. Virol. 78:10336-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Cimprich, K. A., T. B. Shin, C. T. Keith, and S. L. Schreiber. 1996. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA 93:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8:841-853. [DOI] [PubMed] [Google Scholar]
- 11.Fonagy, A., C. Swiderski, A. Wilson, W. Bolton, N. Kenyon, and J. W. Freeman. 1993. Cell cycle regulated expression of nucleolar antigen P120 in normal and transformed human fibroblasts. J. Cell. Physiol. 154:16-27. [DOI] [PubMed] [Google Scholar]
- 12.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 13.Fujimuro, M., and S. D. Hayward. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3β. J. Virol. 77:8019-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 15.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes, J., L. Li, C. Schlueter, M. Duchrow, C. Wohlenberg, C. Gerlach, I. Stahmer, S. Kloth, E. Brandt, and H. D. Flad. 1991. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am. J. Pathol. 138:867-873. [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory, P. D., K. Wagner, and W. Horz. 2001. Histone acetylation and chromatin remodeling. Exp. Cell Res. 265:195-202. [DOI] [PubMed] [Google Scholar]
- 18.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 20.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, J., and R. Renne. 2005. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J. Virol. 79:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadonaga, J. T., and R. Tjian. 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 83:5889-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagunoff, M., and D. Ganem. 1997. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology 236:147-154. [DOI] [PubMed] [Google Scholar]
- 26.Lagunoff, M., D. M. Lukac, and D. Ganem. 2001. Immunoreceptor tyrosine-based activation motif-dependent signaling by Kaposi's sarcoma-associated herpesvirus K1 protein: effects on lytic viral replication. J. Virol. 75:5891-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert, S., and A. M. Carr. 2005. Checkpoint responses to replication fork barriers. Biochimie 87:591-602. [DOI] [PubMed] [Google Scholar]
- 28.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 29.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 30.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 32.Lisby, M., and R. Rothstein. 2005. Localization of checkpoint and repair proteins in eukaryotes. Biochimie 87:579-589. [DOI] [PubMed] [Google Scholar]
- 33.Lusser, A., and J. T. Kadonaga. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25:1192-1200. [DOI] [PubMed] [Google Scholar]
- 34.Marzluff, W. F., and R. J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14:692-699. [DOI] [PubMed] [Google Scholar]
- 35.Masutani, M., H. Nakagama, and T. Sugimura. 2005. Poly(ADP-ribosyl)ation in relation to cancer and autoimmune disease. Cell. Mol. Life Sci. 62:769-783. [DOI] [PubMed] [Google Scholar]
- 36.Moore, P. S., and Y. Chang. 2003. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 57:609-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, P. S., S. J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohsaki, E., K. Ueda, S. Sakakibara, E. Do, K. Yada, and K. Yamanishi. 2004. Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 78:9936-9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 40.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 42.Renne, R., M. Lagunoff, W. Zhong, and D. Ganem. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakakibara, S., K. Ueda, K. Nishimura, E. Do, E. Ohsaki, T. Okuno, and K. Yamanishi. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 78:7299-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 46.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Si, H., and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stedman, W., Z. Deng, F. Lu, and P. M. Lieberman. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566-12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 50.Verma, S. C., T. Choudhuri, R. Kaul, and E. S. Robertson 2006. Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J. Virol. 80:2243-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma, S. C., and E. S. Robertson. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155-163. [DOI] [PubMed] [Google Scholar]
- 52.Viejo-Borbolla, A., M. Ottinger, E. Bruning, A. Burger, R. Konig, E. Kati, J. A. Sheldon, and T. F. Schulz. 2005. Brd2/RING3 interacts with a chromatin binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618-13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, J., C. Chau, Z. Deng, W. Stedman, and P. M. Lieberman. 2005. Epigenetic control of replication origins. Cell Cycle 4:889-892. [DOI] [PubMed] [Google Scholar]