Abstract
The paramyxovirus hemagglutinin-neuraminidase (HN) is a multifunctional protein mediating hemagglutination (HA), neuraminidase (NA), and fusion promotion activities. It has been a matter of debate whether HN contains combined or separate sites for HA and NA activities. To clear the issue, we determined the presence of the second binding site on human parainfluenza virus (hPIV) type 1, 2, and 3 and Sendai virus (SeV) HN proteins. Results of virus elution from erythrocytes at an elevated temperature and HA inhibition by NA inhibitor BCX-2798 suggest that all hPIVs bind to the receptor only through the NA catalytic site, while SeV HN has an additional receptor binding site. Comparison of SeV and hPIV1 HN sequences revealed two amino acid differences at residues 521 and 523 in the region close to the second binding site identified in Newcastle disease virus HN. We mutated hPIV1 HN at position 523 from Asn to the residue of SeV HN, Asp, and rescued a recombinant SeV that carries the mutated hPIV1 HN by a reverse genetics system. The hPIV1 HN with Asp at position 523 hemagglutinated in the presence of BCX-2798, suggesting that the amino acid difference at position 523 is critical for the formation of a second binding site. Creation of the second binding site on hPIV1 HN, however, did not significantly affect the growth or fusion activity of the recombinant virus. Our study indicates that the presence and requirement of a second binding site vary among paramyxoviruses.
The Paramyxoviridae family of enveloped, negative-stranded RNA viruses is comprised of many human pathogens such as human parainfluenza virus (hPIVs) types 1 to 4, as well as animal pathogens, including Sendai virus (SeV), simian virus 5 (SV5), and Newcastle disease virus (NDV). Paramyxovirus infection is initiated by two surface glycoproteins, hemagglutinin-neuraminidase (HN) and fusion (F) protein. HN is a multifunctional molecule which is responsible for virus attachment to a sialic acid-containing receptor and has a neuraminidase (NA) activity that removes sialic acids from progeny virus particles to prevent viral self-agglutination (27). The HN is also required for membrane fusion induced by F protein (18). Most paramyxovirus F proteins require HN protein from the homologous virus to induce membrane fusion, suggesting that in addition to its attachment function, HN protein interacts with a virus-type-specific F protein to bring about efficient membrane fusion (5, 10, 15, 16, 36). Some mutations in the HN protein eliminated its fusion promotion activity without affecting attachment or neuraminidase functions (8, 35, 39). Studies using coimmunoprecipitation assays indicated HN-F complex formation, suggesting that specific physical interaction is required for efficient membrane fusion (20, 21, 37). Both globular head and stalk regions of the HN are involved in the physical interaction with the F protein (5, 6, 10, 22, 40, 41). A current model for the fusion process mediated by HN and F proposes that HN and F proteins form a complex prior to HN attachment to the receptor (21). Sialic acid binding to the receptor/NA active site of HN induces a conformational change on the hydrophobic surface of the protein, which releases F from the complex and triggers the conformational change on F to merge viral and target cell membranes (20, 21, 39). Structural analysis of the crystal structures of NDV HN, with and without receptor analog, supports the idea that receptor binding to the NA catalytic site triggers conformational change of HN (9, 39). Therefore, sialic acid binding to the receptor/NA active site is essential for initiation of the virus infection process.
For years, it was a matter of debate whether the HN molecule possesses combined or separate active sites for hemagglutination (HA) and NA activities. Some evidence using SV5 or hPIV3 has indicated a single site for both activities (23, 30, 34). Other studies with an SeV temperature-sensitive mutant and monoclonal antibody inhibition of activities suggest two separate sites for HA and NA activities (31, 32). A crystal structure of NDV HN with thiosialoside provided direct evidence of the presence of a second sialic acid binding site (47). The second binding site was made up of hydrophobic residues from both monomers and involves interaction with sialic acid and not galactose. The side chain of Arg516 was involved in interaction with thiosialoside, suggesting that Arg516 may be a key residue involved in the formation of the second binding site. The presence of the second binding site in HN on virions was confirmed by a series of experiments using mutant viruses (7). Mutation of NDV HN at Arg516 resulted in the loss of the second site. The fusion promotion activity of HN was substantially reduced by the mutation. Furthermore, the mutation decreased the growth rate of the virus, suggesting that the second binding site on NDV HN facilitates virus infection and growth by enhancing the fusion promotion activity of HN (7).
Although the structural and biochemical data confirmed that NDV HN contains two receptor binding sites, it is not clear whether other viruses in the genera Respirovirus and Rubulavirus have HNs that can bind to receptors through two separate sites. It was reported that NA inhibitor 4-guanidino-Neu5Ac2en (zanamivir) inhibited not only NA activity but also the receptor interaction of hPIV3 HN (29). In contrast, NA inhibitor BCX-2798 (4-azido-5-isobutyrylamino-2,3-didehydro-2,3,4,5-tetradeoxy-d-glycero-d-galacto-2-nonulopyranosic acid) blocked only NA activity but not receptor binding of NDV HN (7). These previous studies suggest that the presence of the second receptor binding site may vary among the viruses of this group. Here, we present evidence of the second binding site on SeV HN, which does not exist on human parainfluenza virus type 1, 2, and 3 HN proteins. Our results indicate that the presence of separate receptor binding sites is not common among respiro-, rubula-, and avulaviruses. Furthermore, we found that a single mutation at residue 523 from Asn to Asp on hPIV1 HN created the second binding site. However, characterization of recombinant SeV carrying hPIV1 HN mutated at position 523 showed that the second binding site profited little in its fusion promotion or virus growth in tissue culture, suggesting that the requirement for a separate binding site for efficient virus growth differs between the viruses.
MATERIALS AND METHODS
Cells and viruses.
LLC-MK2, HeLa T4+, and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. hPIV1 strain C35, hPIV2 strain Greer, hPIV3 strain C243, and SeV strain Enders were obtained from the American Type Culture Collection and grown in LLC-MK2 cells in DMEM supplemented with 0.15% bovine serum albumin (BSA) and 2 μg/ml of acetylated trypsin. The recombinant SeV (rSeVhHN) whose HN gene was replaced with that of hPIV1 (1) was also grown in DMEM supplemented with BSA and trypsin as described above. Vaccinia virus strains WR and vTF7.3 (11) were grown in HeLa T4+ cells.
Paramyxovirus neuraminidase inhibitor.
BCX-2798 was developed by BioCryst Pharmaceuticals, Inc. (Birmingham, Ala.). The drug was designed on the basis of the three-dimensional (3D) structure of the catalytic site of NDV HN. BCX-2798 is the derivative of Neu5Ac2en in which the O4 hydroxyl group has been replaced by an azido group and the methyl group of the acetamido moiety at C-5 has been replaced by an isopropyl group (1). The compound was provided as a lyophilized powder and stored at 4°C.
Recovery of a recombinant SeV.
To rescue rSeVhHN523D, we first mutated the hPIV1 HN gene. The hPIV1 HN gene was subcloned into pTF1 from the full-genome cDNA of rSeVhHN (1) using NotI and AscI. Mutation of amino acid 523N to 523D was done using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The mutated gene was inserted back into the full-genome rSeVhHN cDNA. The recombinant SeV (rSeVhHN523D) which expresses the mutated hPIV1 HN (Asp at position 523) was rescued using the reverse genetic system according to the procedure described previously (4, 17). The rescued recombinant virus was plaque purified and amplified in LLC-MK2 cells. To confirm the sequence of the HN gene of rSeVhHN523D, RNA was extracted from purified virus and amplified by reverse transcription-PCR using the StrataScript two-tube reverse transcription-PCR system (Stratagene) and appropriate primers. Mutation was verified by sequencing the PCR product.
Virus elution from RBC.
Viruses grown in LLC-MK2 cells were purified by ultracentrifugation through a 20 to 50% sucrose gradient (33). Each virus (128 HA units) was serially diluted in phosphate-buffered saline (PBS; pH 7.4) in a 96-well round-bottomed plate and incubated with 0.5% human or chicken red blood cells (RBC) for 1 or 2 h at 4°C. The plates were then shifted to 34°C, and HA titers were determined.
Hemagglutination inhibition (HI) test.
BCX-2798 was serially diluted in PBS and incubated with 8 HA units of purified viruses for 1 h at room temperature. Human or chicken RBC (0.5%, vol/vol) were added, and the HA titer was determined after incubation for 1 h at 4°C.
Neuraminidase and NI assays.
NA activities of the viruses were determined by a colorimetric method that detects N-acetylneuraminic acid released from N-acetylneuramin-lactose. Various amounts of purified viruses were incubated with the substrate in 0.2 M phosphate buffer (pH 7.4) for 1 h at 37°C, and released sialic acids were quantitated. The neuraminidase inhibition (NI) was determined as follows. BCX-2798 was serially diluted and incubated with a standard dose of virus for 1 h at room temperature. Then 50 μg of sialyllactose from human milk (Sigma) was added to the mixture and incubated for another 30 min at 37°C. NA activity was determined by the method of Aminoff (2).
Virus growth kinetics in LLC-MK2 cells.
Cells in six-well plates were infected with rSeVhHN or rSeVhHN523D at a multiplicity of infection (MOI) of 0.001 for 1 h at room temperature. After being washed with PBS, the cells were cultured at 34°C in DMEM supplemented with 0.15% BSA and 2 μg/ml of trypsin. At the indicated time points, 200-μl aliquots of medium were taken and replaced with equal volumes of fresh medium. The amount of infectious virus in each sample of medium was determined by titration in plaque assay with LLC-MK2 cells.
Content mixing assay for fusion.
Fusion activity of recombinant SeV was determined by the reporter gene assay described previously (3, 25) with some modifications. Briefly, one population of HeLa T4+ cells in a six-well plate was infected with indicated viruses (2.5 PFU/cell) and reinfected with vTF7.3 encoding T7 RNA polymerase (10 PFU/cell). After overnight incubation at 34°C, the monolayer of cells was treated with 50 U/ml of recombinant neuraminidase (New England BioLabs) for 30 min at 37°C and then with acetylated trypsin (5 μg/ml) to activate the F protein. A second population of HeLa T4+ cells was infected with vaccinia virus WR at an MOI of 10 and transfected with 1 μg/well of plasmid pRL-null, which includes the Renilla luciferase gene under the control of the T7 promoter (Promega). After overnight incubation, cells were collected and added to the recombinant SeV-infected cells. Cell fusion was monitored after 6 h of incubation at 34°C, using the Renilla Luciferase Assay System (Promega).
RESULTS
SeV, but not hPIV1, hPIV2, or hPIV3, has a second receptor binding site on HN.
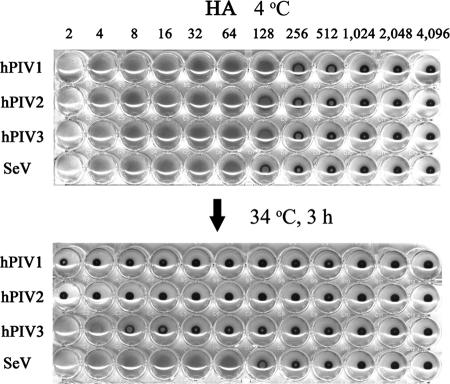
We previously reported that, in addition to a large binding pocket which mediates both attachment and NA activity, NDV HN contains a second sialic acid binding site (7, 47). To determine whether the presence of the second receptor binding site is a common feature among viruses in the genera Respirovirus and Rubulavirus, we characterized the receptor binding activities of SeV, hPIV1, hPIV2, and hPIV3 at different temperatures. Purified viruses (128 HA) were serially diluted (1:2 ratio) in PBS and incubated with an equal amount of RBC at 4°C. We used human RBC because hPIV3 does not hemagglutinate chicken RBC. The 96-well plate was then shifted to 34°C and incubated for 3 h. If HN binds to the sialic acid-containing receptor only through the NA catalytic site, and if the NA can cleave the linkage, the association will be released by its enzymatic activity at the elevated temperature (7). Among the four viruses that we tested, hPIV1, hPIV2, and hPIV3 were eluted from the RBC at 34°C, while SeV remained bound to RBC even after 3 h of incubation at 34°C (Fig. 1). This result is the same with NDV, which remained attached to the RBC even at the elevated temperature, while mutant NDVs, whose HNs were mutated at residue 516, a key residue for forming the second binding site, eluted from RBC at an elevated temperature (7).
FIG. 1.
Virus adsorption and elution from RBC. Attachment activity of hPIV types 1, 2, and 3 and SeV was analyzed by standard HA assay. Purified viruses (128 HA units) were serially diluted in PBS, and an equal volume of 0.5% human RBC was added to the wells. After incubation for 2 h at 4°C, the plate was then shifted to 34°C and incubated for another 3 h.
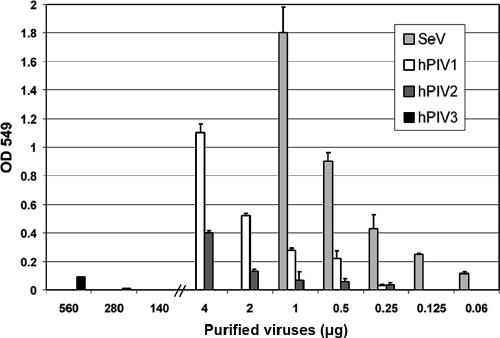
It is possible that different levels of NA activity reflect the elution results. HN with low NA activity may stay attached longer than viruses with highly active NA. We therefore compared the NA activities of SeV, hPIV1, hPIV2, and hPIV3 by colorimetric NA assay using N-acetylneuramin-lactose as a substrate. Various amounts of purified viruses were incubated with sialyllactose in phosphate buffer (pH 7.4), and the released N-acetylneuraminic acids were quantitated. The results indicate that the NA activity of SeV HN was much higher than those of hPIVs (Fig. 2). With 1 μg of virus, SeV HN showed 6 and 25 times higher activity than those of hPIV1 and hPIV2, respectively. The NA activity of hPIV3 using N-acetylneuramin-lactose substrate was very weak compared with those of other viruses tested. The HN contents of SeV, hPIV1, hPIV2, and hPIV3 were 21, 19, 18, and 28% of total viral proteins, respectively, as determined by densitometries of a sodium dodecyl sulfate (SDS)-polyacrylamide gel (data not shown). Although SeV HN has much higher NA activity, the virus remained attached to RBC after 3 h of incubation at 34°C, while hPIVs were eluted from the RBC under the same condition (Fig. 1). These results suggest that hPIV1, hPIV2, and hPIV3 HNs bind to RBC only through the NA active site, which was cleaved at an elevated temperature because of the NA activity. In contrast to hPIVs, SeV remained attached to the RBC, possibly due to the binding through an additional binding site on its HN as observed for NDV HN (7).
FIG. 2.
Comparison of the NA activities of parainfluenza viruses. Various amounts of purified viruses were used to analyze viral NA activity by the colorimetric method. All values are averages ± standard deviations (error bars) from three independent experiments. OD 549, optical density at 549 nm.
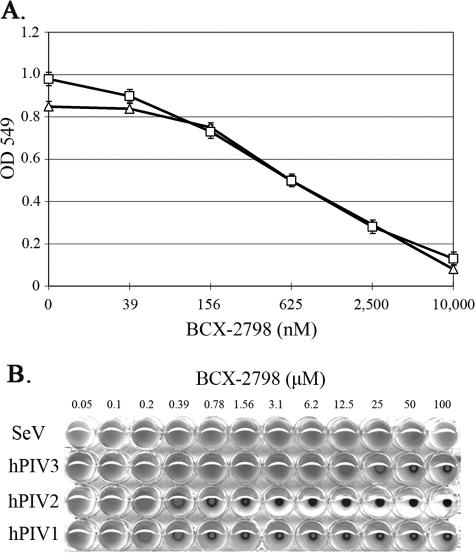
To further confirm the presence or absence of the second binding site on HNs, we determined the HA activity of the viruses in the presence of the NA inhibitor BCX-2798 (1). BCX-2798 was designed based on the 3D structure of the catalytic site of NDV HN and is highly effective in inhibiting NA activities in vitro (1). We first tested the NI activity of BCX-2798 on SeV HN and compared it with that on hPIV1 HN (Fig. 3A). The 50% inhibitory concentration value of BCX-2798 for SeV HN NA activity was almost equivalent with that of hPIV1 HN, indicating that BCX-2798 blocks the SeV NA as efficiently as that of hPIV1. We next determined whether BCX-2798 inhibited the HA of SeV, hPIV1, hPIV2, or hPIV3. As shown in Fig. 3B, BCX-2798 blocked the HA of all hPIVs. Inhibitory activity against hPIV3 HN was less efficient than that against hPIV1 or hPIV2, which correlated with the NI activity of BCX-2798 against these viruses, suggesting that the binding affinity of BCX-2798 at the NA active site of hPIV3 HN is lower than that at the NA site of hPIV1 or hPIV2 (1). Importantly, BCX-2798 did not inhibit the HA activity of SeV even at 100 μM (Fig. 3B), although the NI activities of BCX-2798 against hPIV1 and SeV HNs were almost equivalent (Fig. 3A). Together with the data from the RBC elution assay (Fig. 1), these results indicate that SeV, but not hPIV, HN has the second receptor binding site.
FIG. 3.
Inhibition of NA and HA activities by BCX-2798. (A) NI activities of BCX-2798 against SeV (triangles) and hPIV1 (squares) HN. Purified SeV (0.6 μg) or hPIV1 (5 μg) was preincubated with various concentrations of BCX-2798, and NA activities of the mixture were determined using a colorimetric NA assay. All values are the averages ± standard deviations (error bars) from three independent experiments. (B) HA inhibition of SeV and hPIVs by BCX-2798. Eight HA units of purified viruses were incubated with serially diluted BCX-2798 for 1 h at room temperature. An equal volume of 0.5% human RBC was added to the mixtures and incubated for 1 h at 4°C.
Identification of the residue responsible for formation of the second binding site on type 1 parainfluenza viruses.
The 3D structure of NDV HN protein in complex with thiosialoside revealed the precise location of the second sialic acid binding site on NDV HN, which indicated that the side chain of Arg516 located at β5S4 was involved in direct interaction with the receptor (47). Characterization of mutated NDV HNs supported the finding that Arg516 plays an important role in the formation of the second receptor binding site (7). Sequence analysis of SeV and hPIV1 HNs showed that these two HNs are highly homologous, sharing 72% identity (12). However, our results suggest that SeV HN possesses the second binding site, while hPIV1 HN does not (Fig. 1 and 3). The sequence comparison of SeV and hPIV1 HNs indicates that both SeV and hPIV1 HNs have Arg at position 520, which corresponds to Arg516 in NDV HN. However, there are two amino acid differences between SeV and hPIV1 HN close to Arg520: SeV HN has Ile and Asp at positions 521 and 523, respectively, while hPIV1 HN has Leu521 and Asn523 (Fig. 4).
FIG. 4.
3D structure of NDV HN showing the location of the second binding site and partial HN amino acid alignment. Sequence comparison between SeV (strain Enders), hPIV1 (strain C35), and NDV (strain Kansas) HNs around the second receptor binding site is shown. Dashes in the hPIV1 HN sequence indicate amino acids identical to those of SeV HN.
We first attempted to rescue mutant SeVs containing HN with Leu at position 521 (rSeV521L) or Asn at position 523 (rSeV523N) by reverse genetics (17). rSeV521L was successfully rescued. Characterization of SeV521L suggests that the amino acid difference at residue 521 has little effect on the formation of the second binding site. rSeV521L bound to RBC at 4°C did not elute after incubation at 34°C for 3 h. Furthermore, BCX-2798 did not block the HA activity of rSeV521L (data not shown). rSeV523N was not rescued despite many attempts. These results may suggest that the amino acid difference at residue 523, but not residue 521, plays a critical role in the formation of a second binding site, which is required for the growth of SeV. The effect of the Ile and Leu difference at 521 may be minimal because of their similarity, while the difference at 523 between acidic Asp and neutral Asn could be critical to the formation of the second binding site. To determine whether the Asp-Asn difference at position 523 is responsible for the lack of the second binding site on hPIV1 HN, we mutated hPIV1 HN at 523 from Asn to Asp and rescued the recombinant SeV that carries the mutated hPIV1 HN (rSeVhHN523D) by a reverse genetics system. The production of rSeVhHN whose HN was replaced with that of wild-type (wt) hPIV1 HN was previously reported (1). Recombinant SeV whose HN gene was replaced with hPIV1 HN is viable because hPIV1 HN functions well with SeV F to promote fusion (5). We first characterized the protein contents of the rescued virus and compared them with that of rSeVhHN. SDS-polyacrylamide gel electrophoresis analysis of the purified viruses confirmed that the rescued rSeVhHN523D, like rSeVhHN, contains HN which migrates at the same position with hPIV1 HN (Fig. 5A). The virion HN content was not significantly different between rSeVhHN and rSeVhHN523D, showing that these recombinant SeVs carry similar levels of hPIV1 HN. We next compared the NA activities of the recombinant viruses. The NA activity of rSeVhHN523D was almost equivalent to that of rSeVhHN (Fig. 5B), which is consistent with our previous observation that mutation in the second binding site of NDV HN does not affect the NA activity (7). The effect of the mutation on attachment activity of rSeVhHN523D was analyzed by virus adsorption to and elution from RBC. The HA titer of SeV, hPIV1, rSeVhHN, and rSeVhHN523D was adjusted to 128 HA units. The presence of the second binding site was determined by shifting the same HA plate to 34°C (Fig. 6A). Both hPIV1 and rSeVhHN were completely eluted from chicken RBC after incubation for 1 h at 34°C, while SeV remained bound to RBC even at a higher temperature. Unlike rSeVhHN, rSeVhHN523D remained attached to the RBC after 1 h of incubation at 34°C although attachment activity of hPIV1 HN was not completely restored by mutation of Asn523 to Asp.
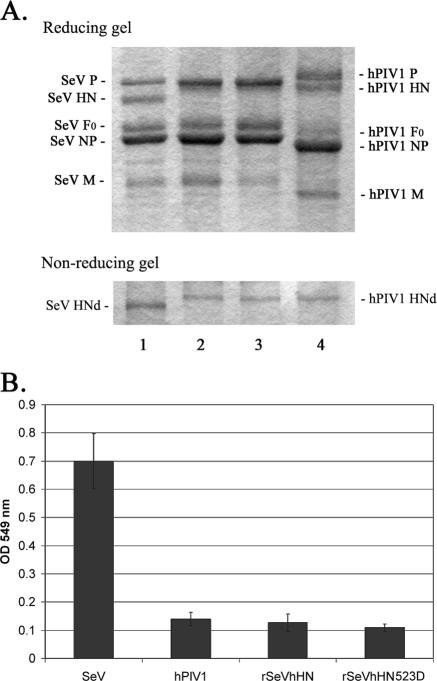
FIG. 5.
Characterization of rSeVhHN523D. (A) SDS-polyacrylamide gel electrophoresis analysis of purified viruses. Three micrograms of purified SeV (lane 1), rSeVhHN (lane 2), rSeVhHN523D (lane 3), or hPIV1 (lane 4) was fractionated in SDS-10% polyacrylamide gels in reducing and nonreducing conditions. HNd, HN dimer. (B) NA activities of SeV, hPIV1, rSeVhHN, and rSeVhHN523D. Purified viruses (0.4 μg) were used to analyze the NA activity by the colorimetric method. All values are the averages ± standard deviations from three independent experiments. OD 549 nm, optical density at 549 nm.
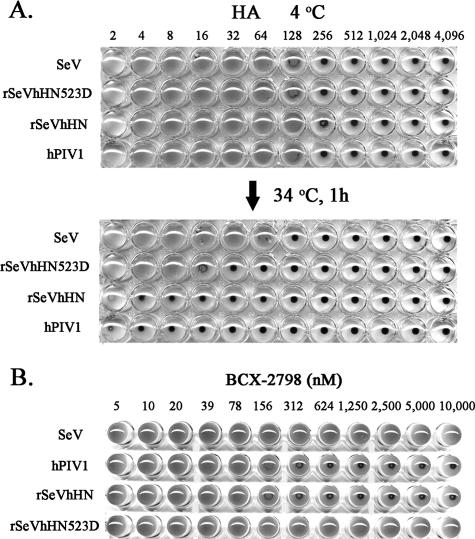
FIG. 6.
Effect of Asn-to-Asp mutation at residue 523 of hPIV1HN on virus attachment activity. (A) An equal amount of purified virus (128 HA units) was serially diluted (1:2 ratio) in PBS and incubated with 0.5% chicken RBC in a 96-well plate for 1 h at 4°C. The plate was then shifted to 34°C and incubated for another 1 h. (B) HI activity of BCX-2798. Eight HA units of purified viruses was incubated with serially diluted BCX-2798 for 1 h at room temperature. An equal volume of 0.5% chicken RBC was added to the mixtures and incubated for another 1 h at 4°C.
The presence of the second binding site on rSeVhHN523D was also determined by the HI test in the presence of BCX-2798. As shown in Fig. 6B, HA by hPIV1 or rSeVhHN was blocked by 312 nM of BCX-2798, while rSeVhHN523D, like SeV, bound to RBC even in the presence of 10 mM of the NA inhibitor. Together with the elution assay described above, these results suggest that the amino acid difference at residue 523 plays a critical role in the formation of a second binding site on hPIV1 HN.
The fusion promotion activity of hPIV1 HN is unaffected by the second binding site.
Our previous study of NDV HN suggested that the loss of the second binding site by a single mutation at residue 516 substantially reduced the fusion promotion activity of NDV (7). Therefore, we next determined the effect of the second receptor binding site on hPIV1 HN in its fusion promotion activity. The fusion activity was compared by syncytium formation in HeLa T4+ cells infected with the viruses (5). Both rSeVhHN and rSeVhHN523D induced similar levels of syncytium formation (Fig. 7A). Infection with rSeVhHN at an MOI of 1 fused 19% of the cells in the culture. Similarly, infection with rSeVhHN523D resulted in syncytium formation in 21% of the cells. To more accurately quantitate promotion of membrane fusion, the activity of rSeVhHN and rSeVhHN523D was determined by a content mixing reporter gene assay. HeLa T4+ cells were infected with rSeVhHN or rSeVhHN523D together with vTF7.3, which expresses T7 RNA polymerase. The cells were then overlaid with HeLa T4+ cells transfected with a plasmid that carries the Renilla luciferase gene under the control of the T7 promoter. After 6 h of incubation, cells were lysed and the lysates were analyzed for luciferase activity (Fig. 7B). rSeVhHN523D induced fusion at slightly higher levels than rSeVhHN did in this assay, consistent with the results of syncytium formation observed earlier (Fig. 7A). These results suggest that the presence of the second binding site does not increase significantly the fusion promotion activity of hPIV1 HN.
FIG. 7.
Fusion activity of rSeVhHN and rSeVhHN523D. (A) Syncytium formation of HeLa T4+ cells infected with rSeVhHN or rSeVhHN523D at an MOI of 1. Infected cells were cultured for 24 h and then treated with acetylated trypsin (5 μg/ml) for 30 min and incubated with DMEM containing 10% fetal calf serum for another 4 h. (B) Membrane fusion activity of recombinant SeVs in infected HeLa T4+ cells was determined by content mixing assay. All values are the averages ± standard deviations from three independent experiments.
Growth kinetics of rSeVhHN and rSeVhHN523D in culture cells.
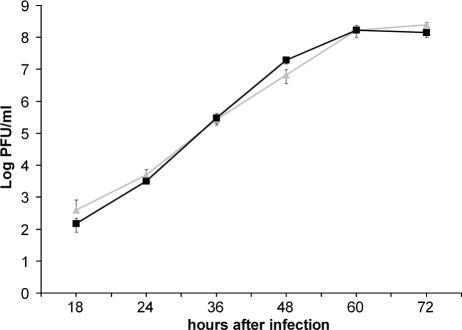
To determine whether the presence of the additional binding site has an advantage for the growth of the recombinant virus, we infected LLC-MK2 cells with rSeVhHN or rSeVhHN523D at an MOI of 0.001 and cultured the cells in the medium containing trypsin at 1 μg/ml for 72 h at 34°C. Culture medium was harvested at various time points after infection, and the amount of infectious virus was quantified by plaque titration (Fig. 8). No difference in the growth abilities of rSeVhHN and rSeVhHN523D was observed. We also compared the plaque formation by the recombinant SeVs in LLC-MK2 cells. Both rSeVhHN and rSeVhHN523D developed ∼2-mm plaques in 6 days after infection, and no significant difference of the plaque size was observed (data not shown). Together with the results of growth kinetics and syncytium formation, these results show that the presence of a separate binding site on hPIV1 HN is unlikely to provide a clear advantage in the process of rSeVhHN infection.
FIG. 8.
Growth kinetics of rSeVhHN (gray triangles) and rSeVhHN523D (black squares) in LLC-MK2 cells. Virus titers were measured at the times indicated on the x axis by plaque assay. The data are means of three experiments with standard errors.
DISCUSSION
Attachment of paramyxovirus to a sialic acid-containing receptor is mediated by HN protein, which also carries NA activity to cleave the terminal sialic acid from the glycoconjugate. For a long time, it had been unclear whether a single site mediates both receptor binding and NA activity or whether HN has an additional receptor binding site in addition to the NA catalytic site. The 3D structures of the globular head of NDV, hPIV3, and SV5 HN proteins revealed the presence of a large binding pocket which mediates both receptor binding and NA activities (9, 19, 46). NDV HN has been shown to have a separate sialic acid binding site in addition to the NA active site (7, 47). We showed in this study that, like NDV, SeV has a separate binding site, while human parainfluenza virus types 1, 2, and 3 do not have the second site on HN, indicating that the presence of the separate receptor binding site varies among paramyxoviruses. Purified hPIVs bound to RBC at 4°C were eluted at an elevated temperature, suggesting that virus HN bound to sialic acid-containing receptors only through the NA catalytic site. However, SeV was not eluted from the RBC at the elevated temperature even though the HN has higher NA activity than those of hPIVs (Fig. 1 and 2). Preincubation of purified hPIVs with NA inhibitor BCX-2798 blocked the HA activities of the viruses, indicating that the NA catalytic site is also responsible for the HA activity of these viruses (Fig. 3B). Consistent with our data, previous reports showed that zanamivir, a potent inhibitor of influenza virus neuraminidase, blocks the receptor binding of hPIV3 (23, 30). In sharp contrast, the HA activity of SeV was not inhibited by BCX-2798 (Fig. 3). Because BCX-2798 has similar 50% inhibitory concentrations to block NA activities of SeV and hPIV1, it is highly likely that SeV HN has an additional receptor binding site. Our finding of a separate receptor binding site on SeV HN is consistent with a previous report of anti-SeV HN monoclonal antibodies that inhibit NA, but not HA (33). It may also explain the expanded spectrum of SeV receptor specificity. Using different types of purified gangliosides, we previously showed that hPIV1 and hPIV3 recognize only limited types of neolacto-series gangliosides as receptors, whereas SeV can bind to various types of neolacto- and ganglio-series gangliosides containing a terminal sialic acid (38). Our finding that SeV, but not hPIV1 and hPIV3, contains a separate binding site on HN is consistent with its extended spectrum of receptor recognition.
Evidence of the second receptor binding site was provided by the structural study of NDV HN cocrystallized with thiosialoside (47). These structural data identified the location of the second site on NDV HN, which indicated that Arg at 516 plays a key role in the formation of the binding site. Like NDV, SeV HN has a separate receptor binding site, while closely related hPIV1 HN, which shares 72% identity with SeV HN, does not have the second site (Fig. 1 and 3). Taking advantage of the sequence homology between SeV and hPIV1 HNs and structural data for the second site on NDV HN, we revealed that residue 523 on type 1 parainfluenza virus HN plays a critical role in the formation of the second receptor binding site. Rescued rSeVhHN523D hemagglutinated in the presence of a high concentration of BCX-2798, while the rSeVhHN, which projected wt hPIV1 HN from the virion, failed to hemagglutinate (Fig. 6B). Elution of the rSeVhHN523D from RBC at an elevated temperature was limited, while rSeVhHN or hPIV1 was completely eluted by incubation at 34°C for 1 h (Fig. 6A). Asn at position 523 is well conserved among hPIV1 isolates (14). These data suggest that the absence of the second binding site on hPIV1 HN is due to the amino acid difference at position 523, which is located at the similar position of the second site of NDV HN (7, 47).
Interestingly, an increase of receptor binding avidity by zanamivir was reported in NDV HN (26). It was proposed that the second binding site on NDV HN was activated by occupation of the NA catalytic site with zanamivir. Consistent with the result, comparison of the NDV HN crystallized alone and in complex with a receptor analog revealed the conformational changes in restricted areas including residue 516 on strand β5S4 (9, 39). Because Arg at residue 516 is a key residue for the second binding site (7), it is possible that the second binding site is activated upon receptor binding to the NA catalytic site of HN (26, 47).
It is not clear how these two functions, receptor binding and neuraminidase, are regulated during the initiation of infection and why some viruses do not have the second binding site. A current model of paramyxovirus infection proposes that receptor binding induces a conformational change on the hydrophobic surface of the HN protein, which dissociates HN-F interaction and activates the F protein to initiate membrane fusion (20, 21, 39). Comparison of the structure of NDV HN with and without Neu5Ac2en showed significant differences in the catalytic site and the area that forms the hydrophobic surface of the protein (9, 39). Analysis of the mutated HN proteins revealed that the hydrophobic area is directly involved in the fusion promotion activity of the protein (39). Recent work by McGinnes and Morrison (21) showed evidence that HN and F proteins reside in a complex prior to HN protein attachment and receptor binding induces dissociation of the complex. These studies indicate that receptor binding to the NA catalytic site is essential for the initiation of virus infection. However, involvement of the second binding site in the process of virus infection is not clear.
What is the advantage of having the second binding site in the process of virus infection? NDV mutants that lack the second site can infect cells in vitro, indicating that the second site is optional (7). However, fusion promotion activity of wt NDV HN, which has the second site, was higher than that of mutant NDV HN that lacks the second site. In addition, the enhanced fusion promotion activity of wt HN correlates with faster growth kinetics of the virus (7). However, the creation of the second site on hPIV1 HN did not affect the fusion promotion and growth kinetics of the recombinant virus (Fig. 7 and 8). Like NDV, SeV HN, which contains the second site, has much higher NA activity than those of hPIVs (Fig. 2). One explanation why viruses that have high NA activity carry the second site could be that the additional binding site helps the virus stay in close proximity to the cellular membrane during the infection process. If highly active NA quickly cleaves the link between the cell receptor and the virus, the target membrane may not be in close contact with the virus long enough for the virus to complete membrane fusion. In other words, viruses with low NA activity may not require an additional binding site to keep the virus in close proximity to the target membrane because of the slow catalytic activity. In fact, fusion promotion activity of hPIV1 HN was not enhanced significantly by the created second binding site (Fig. 7). In support of this hypothesis, neuraminidase-deficient mumps virus variants were reported to be more fusogenic than wild-type viruses (43, 44). It was also suggested that differences in neuraminidase and receptor binding activities of hPIV3 HNs influence their fusion promotion activity (28).
A second sialic acid binding site has also been discovered on the N1 and N9 subtype NA of influenza A viruses (13, 45). The receptor binding activity of the second site has been successfully transferred to the N2 subtype of NA by site-directed mutagenesis, suggesting that a few residues are involved in the formation of the second site (24). The X-ray structure of a complex of N9 NA with sialic acid revealed the location of a second binding site on the surface of the enzyme (42). Interestingly, the residues that interact with a sialic acid at the second site are mostly conserved in avian strains, but not in human strains (42). This is similar to our finding that human parainfluenza virus types 1 to 3 lack the second binding site on HN, while SeV (a mouse pathogen) or NDV (an avian virus) possesses the second site. The role of NA in the process of influenza virus infection is apparently different from that of HN of paramyxovirus. A unique feature of HN protein is that paramyxoviruses utilize the NA catalytic site as the receptor binding site to initiate virus infection. Therefore, there must be a balance between receptor binding avidity and neuraminidase activities for efficient virus infection and spread. Viruses which infect different hosts may require a precise adjustment of each of the activities of the multifunctional HN protein for optimized infection and transmission within and among the hosts.
Acknowledgments
We thank Dong Kim for technical assistance and BioCryst Pharmaceuticals, Inc. (Birmingham, Ala.), for providing BCX-2798.
This work was supported by Public Health Service grant AI055940 from the National Institutes of Health.
REFERENCES
- 1.Alymova, I. V., G. Taylor, T. Takimoto, T. H. Lin, P. Chand, Y. S. Babu, C. Li, X. Xiong, and A. Portner. 2004. Efficacy of novel hemagglutinin-neuraminidase inhibitors BCX 2798 and BCX 2855 against human parainfluenza viruses in vitro and in vivo. Antimicrob. Agents Chemother. 48:1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aminoff, D. 1961. Methods for the quantitative estimation of N-acetylneuraminic acid and their application to hydrolysates of sialomucoids. Biochem. J. 81:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagai, S., and R. A. Lamb. 1995. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J. Virol. 69:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bousse, T., T. Matrosovich, A. Portner, A. Kato, Y. Nagai, and T. Takimoto. 2002. The long noncoding region of the human parainfluenza virus type 1 F gene contributes to the read-through transcription at the M-F gene junction. J. Virol. 76:8244-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bousse, T., T. Takimoto, W. L. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 204:506-514. [DOI] [PubMed] [Google Scholar]
- 6.Bousse, T., T. Takimoto, and A. Portner. 1995. A single amino acid changes enhances the fusion promotion activity of human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Virology 209:654-657. [DOI] [PubMed] [Google Scholar]
- 7.Bousse, T. L., G. Taylor, S. Krishnamurthy, A. Portner, S. K. Samal, and T. Takimoto. 2004. Biological significance of the second receptor binding site of Newcastle disease virus hemagglutinin-neuraminidase protein. J. Virol. 78:13351-13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 10.Deng, R., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman, W. L., D. S. Gill, R. A. Scroggs, and A. Portner. 1990. The hemagglutinin-neuraminidase glycoproteins of human parainfluenza virus type 1 and Sendai virus have high structure-function similarity with limited antigenic cross-reactivity. Virology 175:211-221. [DOI] [PubMed] [Google Scholar]
- 13.Hausmann, J., E. Kretzschmar, W. Garten, and H. D. Klenk. 1995. N1 neuraminidase of influenza virus A/FPV/Rostock/34 has haemadsorbing activity. J. Gen. Virol. 76:1719-1728. [DOI] [PubMed] [Google Scholar]
- 14.Henrickson, K. J., and L. L. Savatski. 1996. Two distinct human parainfluenza virus type 1 genotypes detected during the 1991 Milwaukee epidemic. J. Clin. Microbiol. 34:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66:4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, X. L., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 19.Lawrence, M. C., N. A. Borg, V. A. Streltsov, P. A. Pilling, V. C. Epa, J. N. Varghese, J. L. McKimm-Breschkin, and P. M. Colman. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335:1343-1357. [DOI] [PubMed] [Google Scholar]
- 20.McGinnes, L. W., K. Gravel, and T. G. Morrison. 2002. Newcastle disease virus HN protein alters the conformation of the F protein at cell surfaces. J. Virol. 76:12622-12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinnes, L. W., and T. G. Morrison. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 80:2894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melanson, V. R., and R. M. Iorio. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murrell, M., M. Porotto, T. Weber, O. Greengard, and A. Moscona. 2003. Mutations in human parainfluenza virus type 3 hemagglutinin-neuraminidase causing increased receptor binding activity and resistance to the transition state sialic acid analog 4-GU-DANA (zanamivir). J. Virol. 77:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuss, J. M., and G. M. Air. 1991. Transfer of the hemagglutinin activity of influenza virus neuraminidase subtype N9 into an N2 neuraminidase background. Virology 183:496-504. [DOI] [PubMed] [Google Scholar]
- 25.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porotto, M., M. Fornabaio, O. Greengard, M. T. Murrell, G. E. Kellogg, and A. Moscona. 2006. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J. Virol. 80:1204-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porotto, M., O. Greengard, N. Poltoratskaia, M. A. Horga, and A. Moscona. 2001. Human parainfluenza virus type 3 HN-receptor interaction: effect of 4-guanidino-Neu5Ac2en on a neuraminidase-deficient variant. J. Virol. 75:7481-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porotto, M., M. Murrell, O. Greengard, L. Doctor, and A. Moscona. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porotto, M., M. Murrell, O. Greengard, M. C. Lawrence, J. L. McKimm-Breschkin, and A. Moscona. 2004. Inhibition of parainfluenza virus type 3 and Newcastle disease virus hemagglutinin-neuraminidase receptor binding: effect of receptor avidity and steric hindrance at the inhibitor binding sites. J. Virol. 78:13911-13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portner, A. 1981. The HN glycoprotein of Sendai virus: analysis of site(s) involved in hemagglutinating and neuraminidase activities. Virology 115:375-384. [DOI] [PubMed] [Google Scholar]
- 32.Portner, A., R. A. Scroggs, P. S. Marx, and D. W. Kingsbury. 1975. A temperature-sensitive mutant of Sendai virus with an altered hemagglutinin-neuraminidase polypeptide: consequences for virus assembly and cytopathology. Virology 67:179-187. [DOI] [PubMed] [Google Scholar]
- 33.Portner, A., R. A. Scroggs, and D. W. Metzger. 1987. Distinct functions of antigenic sites of the HN glycoprotein of Sendai virus. Virology 158:61-68. [DOI] [PubMed] [Google Scholar]
- 34.Scheid, A., and P. W. Choppin. 1974. The hemagglutinating and neuraminidase protein of a paramyxovirus: interaction with neuraminic acid in affinity chromatography. Virology 62:125-133. [DOI] [PubMed] [Google Scholar]
- 35.Sergel, T., L. W. McGinnes, M. E. Peeples, and T. G. Morrison. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717-726. [DOI] [PubMed] [Google Scholar]
- 36.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, T., A. Portner, R. A. Scroggs, M. Uchikawa, N. Koyama, K. Matsuo, Y. Suzuki, and T. Takimoto. 2001. Receptor specificities of human respiroviruses. J. Virol. 75:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]
- 42.Varghese, J. N., P. M. Colman, A. van Donkelaar, T. J. Blick, A. Sahasrabudhe, and J. L. McKimm-Breschkin. 1997. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA 94:11808-11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waxham, M. N., and J. Aronowski. 1988. Identification of amino acids involved in the sialidase activity of the mumps virus hemagglutinin-neuraminidase protein. Virology 167:226-232. [DOI] [PubMed] [Google Scholar]
- 44.Waxham, M. N., and J. S. Wolinsky. 1986. A fusing mumps virus variant selected from a nonfusing parent with the neuraminidase inhibitor 2-deoxy-2,3-dehydro-N-acetylneuraminic acid. Virology 151:286-295. [DOI] [PubMed] [Google Scholar]
- 45.Webster, R. G., G. M. Air, D. W. Metzger, P. M. Colman, J. N. Varghese, A. T. Baker, and W. G. Laver. 1987. Antigenic structure and variation in an influenza virus N9 neuraminidase. J. Virol. 61:2910-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan, P., T. B. Thompson, B. A. Wurzburg, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803-815. [DOI] [PubMed] [Google Scholar]
- 47.Zaitsev, V., M. von Itzstein, D. Groves, M. Kiefel, T. Takimoto, A. Portner, and G. Taylor. 2004. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 78:3733-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]