Abstract
Baculoviruses, a family of large, rod-shaped viruses that mainly infect lepidopteran insects, have been widely used to transduce various cells for exogenous gene expression. Nonetheless, how a virus controls its transcription program in cells is poorly understood. With a custom-made baculovirus DNA microarray, we investigated the recombinant Autographa californica multiple nucleopolyhedrosis virus (AcMNPV) gene expression program in lepidopteran Sf21 cells over the time course of infection. Our analysis of transcription kinetics in the cells uncovered sequential viral gene expression patterns possibly regulated by different mechanisms during different phases of infection. To gain further insight into the regulatory network, we investigated the transcription program of a mutant virus deficient in an early transactivator (pe38) and uncovered several pe38-dependent and pe38-independent genes. This study of baculovirus dynamic transcription programs in different virus genetic backgrounds provides new molecular insights into how gene expression in viruses is regulated.
Members of the Baculoviridae family have enveloped, double-stranded, circular DNA genomes of about 90 to 160 kb. For more than 20 years, baculoviruses have been widely used to obtain expression of a large variety of functional recombinant proteins in lepidopteran cells (14, 47, 52), and because they have a relatively narrow range of hosts, they have also been used for a long time as bioinsecticides (5, 22, 40, 48, 50). One of the best-characterized baculoviruses is Autographa californica multiple nucleopolyhedrosis virus (AcMNPV), whose genome encodes at least 154 open reading frames (ORFs) in which may be found a potential coding sequence (1). Gene expression of AcMNPV may be mainly regulated at the transcription level, and the virus replication cycle may be closely regulated by both viral factors and the cellular milieu, sequentially expressing early, late, and very late genes (12, 35).
The early transcripts of AcMNPV are synthesized by host RNA polymerase II, while its late transcripts are mediated by virus-encoded RNA polymerase (15, 21). To utilize the host cell transcriptional machinery, baculoviruses have evolved promoters that closely resemble cellular RNA polymerase II-responsive promoters containing the TATA element (3, 4, 10, 16, 45). Other cis-acting regulatory elements, such as the initiator element CAGT motif (3, 44) and downstream activating region elements (24, 45), have also been found in promoters of several early AcMNPV genes. Some unconventional promoter motifs, e.g., CGTGC (33, 42, 53), are highly responsive to transactivators expressed by the baculovirus. In DNA viruses, the potent viral transactivators are synthesized in an early phase to stimulate early viral gene expression, while the host cell viral DNA concentration is still low. Four baculovirus transactivators have been found to stimulate early gene transcription, i.e., IE0, IE1, IE2, and PE38 (13). The pe38-deficient baculovirus has a reduced capacity for genome replication, budded-virus (BV) production, and virulence in Heliothis virescens larvae (39). The roles of the other early transactivators in viral infection, however, still need to be specified.
Late-phase infection is divided into two stages, late and very late, which extend from around 6 to 76 h postinfection (hpi). In late-stage infection (6 to 12 hpi), studies have identified the genes involved in replication of the viral genome and production of BVs, both known as late expression factors (lef genes) (30, 36, 46). Also from these late RNA transcripts, a TAAG sequence motif has been identified and found to constitute a primary element for late and very late promoter activity (35). In the very late stage of infection (18 to 76 hpi), also known as the occlusion phase, proteins associated with occlusion, lysis of the cell, and release of mature occluded viruses are expressed (35). At this stage, other gene products, some not essential for BV production, are also hyperexpressed. One such product, polyhedrin (polh), for example, functions as a major matrix protein for mature occluded viruses.
Although many temporally expressed viral genes have been identified, very little is known about the kinetics of genome-wide expression and regulation during the virus replication cycle in their host lepidopteran cells. To better understand how transcription regulatory mechanisms operate in cells, the temporal dynamic transcription program that occurs during the baculovirus infection cycle can be investigated. To do this, we took advantage of the recent advances in genome sequences of several baculovirus species (1, 18) and developed a whole-genome microarray analysis system. By using AcMNPV to infect Sf21 cells, we examined the viral transcription cascade in natural host cells. Our analysis of the array profile suggests that expression kinetics correlate with the viral infection, replication, and pathological effect in infected cells. An early-stage baculovirus transactivator pe38 deletion mutant virus strain was used to study its interface with the regulatory network. A comprehensive view of the baculovirus temporal transcription program in different virus genetic backgrounds would makes it possible to decipher the molecular mechanisms underlying the regulation of gene expression during infection cycles, which could help in making better use of this virus in biomedical research.
MATERIALS AND METHODS
Cell culture and viruses.
Spodoptera frugiperda Sf21 cells were cultured at 27°C in Grace's medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone), 100 U/ml penicillin, and 100 μg/ml streptomycin.
Several recombinant baculoviruses were used for temporal gene expression profiling. Bac-PH-EGFP, in which the coding region of the polh gene was replaced with a sequence encoding enhanced green fluorescent protein, was used to infect Sf21 cells. The recombinant baculovirus's titer was determined on and it was propagated in Sf21 cells as previously described (28). Wild-type AcMNPV (strain E2) and pe38 deletion virus Δpe38-E9/E9, generously donated by David Theilmann (39), were applied in our differential gene expression profile analysis.
Baculovirus microarray preparation.
To prepare PCR-amplified probes to be spotted onto microarray slides, 156 pairs of primers (shown at http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SI.pdf) were designed with PROBEWIZ (41). PCR amplification was performed to produce 155 nonredundant cDNA probes representing the potential coding ORFs (1) in the genome of AcMNPV strain C6 (NC_001623) as shown at the NCBI website (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=NC_001623). Genomic DNA of wild-type AcMNPV was extracted with the Viral RNA Mini kit (Viogen). It was then used as the PCR template. PCR products were purified with the QIAquick Mini kit (QIAGEN), dried in a SpeedVac, and resuspended in a spotting solution containing 50% dimethyl sulfoxide. The concentration of each amplicon was adjusted to 0.2 to 0.5 mg/ml, and then it was spotted onto aminosilane-coated UltraGAPS glass slides (Corning) by a homemade robotic microarray spotter.
In order to obtain replicates from each microarray hybridization experiment, each cDNA probe was printed four times on the glass slide. Additionally, a series of control DNA probes in the Lucidea Universal ScoreCard (Amersham Bioscience), which were derived from sequences of yeast intergenic regions, were also spotted onto different regions of the baculovirus microarray slide. These were used to generate a calibration curve for universal reference and normalization (see below).
RNA extraction, spiked-in targets, and fluorescent dye labeling.
Cells infected by a specified recombinant baculovirus were harvested at various times postinfection and washed with culture medium. Total RNA of each sample was then extracted with the RNeasy Mini kit (QIAGEN) without further purification. To reduce the possibility of RNA degradation during preparation before fluorescent dye labeling, the integrity of all RNA samples was examined with a Bioanalyzer 2100 (Agilent) with an RNA Nano Labchip (Agilent).
The samples were then labeled with fluorescent dye; 20 μg of total RNA harvested from infected cells was reverse transcribed to Cy3- or Cy5-labeled cDNA with the CyScribe First Strand labeling kit (Amersham Bioscience) in the presence of Cy3-dUTP or Cy5-dUTP (Amersham Bioscience). We used oligo(dT)18-20 as the primer for reverse transcription.
It was important to generate a calibration curve for normalization of gene expression data across experiments. To do this, the mixture of spiked-in poly(A) RNA control targets of the Lucidea Universal ScoreCard (Amersham Bioscience) was also included in the labeling reaction mixtures, according to the manufacturer's instructions.
Microarray hybridization.
Cy3- or Cy5-labeled cDNA was hybridized to a baculovirus array (at 16 h and 42°C in 25% formamide-5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.1% sodium dodecyl sulfate [SDS]-0.1 mg/ml salmon sperm DNA). For posthybridization processing, the array was washed twice (5 min each time, 42°C) in buffer I (2× SSC, 0.1% SDS), followed by a wash (10 min, room temperature) in buffer II (0.1× SSC, 0.1× SDS). Finally, the array was washed four times (1 min each time) in buffer III (0.1× SSC). With a GenePix 4000B laser scanner (Axon), the microarray image was then retrieved and processed with GenePix Pro software (Axon).
Real-time quantitative RCR.
Templates for real-time quantitative PCR analysis were made from samples (1 μl) of a 10-fold dilution of cDNA. Superscript II (Invitrogen) was used for this cDNA synthesis based on total RNA (2 μg) samples harvested at 12 specific time points throughout the course of the viral infection. Real-time PCRs were carried out with the LightCycler DNA master SYBR green I kit (Roche) in the LightCycler (Roche) machine. All but one of the real-time PCR primers for the genes tested were the same as those used to amplify viral probes for microarray production. The one exception was the orf-68 gene, for which 5′-AATCTATCCGATGGCAAATAC was used as the forward primer and 5′-CGGACGGGTCTGTTCA was used as the reverse primer.
Data analysis.
To normalize microarray data, fluorescence signal intensities of the external RNA spiked-in controls (see above) with predetermined quantities of input mRNA were used to fit a Bayesian isotonic regression model. This regression curve was regarded as the plot of the expected intensity versus the relative expression level and was used to transform each probe's signal intensity to obtain its normalized expression level. We note that this approach uses a Bayesian model to take into account the smoothness and the monotonicity of the regression function and falls into the framework outlined in our previous work (8). This has recently been detailed in work emphasizing its application in microarray data analysis (7). For each gene at a specific postinfection time, the mean of the four normalized expression levels was used as the basis for further data analysis.
We analyzed the gene expression kinetics of Bac-PH-EGFP in Sf21 cells throughout the infection time course (0 to 72 hpi). For mathematical convenience, we transformed unequal intervals into a time scale (0 to 1) with equal intervals between adjacent time points. Calibrated signal intensities were used to fit a piecewise polynomial regression model with the Markov chain Monte Carlo algorithms.
This gene-specific regression function represents the expression pattern of a gene during the time course of infection. Generally speaking, this function begins with a value of 0 and at some point increases until it reaches its maximum. Two key features of each gene-specific regression function were used to characterize the expression kinetics of that gene during the time course of infection. One was the time to onset (Ton), i.e., the point where the expression level of a gene first exceeds the background noise. The time to maximum (Tmax) was estimated on the basis of the time when the expression of a gene was at its highest. To group genes according to distinct kinetic features, a two-dimensional clustering of Ton and Tmax was performed with the algorithm described by Hall and Heckman (17).
Dynamically regulated gene expression patterns were also clustered by the K-means algorithm. The expression levels shown were normalized by a constant so that the sum of the 12 squares of the expression levels (at 12 time points) for each gene equals 1. The average Ton for each gene cluster was expressed as the mean ± the standard deviation.
We used GeneSpring software (Agilent) to identify possible common upstream regulatory sequences in a given set of clustered genes. The common upstream regulatory sequence denoted sequences ranging from 15 to 190 bases upstream of an ORF, and the entire span of the sequences was about 5 to 6 oligonucleotides in length. Only the exact match of a consensus sequence was retrieved. A cutoff value of 0.1 for the probability of a false-positive result was used.
RESULTS
Customized baculovirus microarray analysis system.
Strategically, it is important to select appropriate probes to design the array. Proper selection ensures the specificity and sensitivity of a customized microarray. To help generate gene-specific probes for our microarray, we performed systematic genomic sequence analysis of AcMNPV. For each gene, a probe sequence was selected. We created a total of 155 unique DNA probes. These represent those selected ORFs that potentially encode AcMNPV proteins as predicted by Ayres and colleagues (1). Each probe was PCR amplified with specific primer pairs (shown at http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SI.pdf) and genomic DNA of wild-type baculovirus strain C6 as the template. All probes were spotted in quadruplicate onto a microarray slide (Fig. 1A).
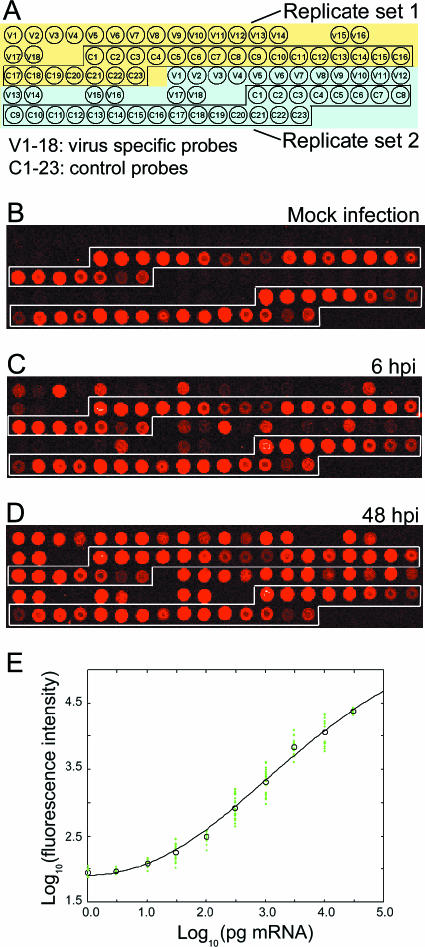
FIG. 1.
AcMNPV microarray design and calibration method. (A) Illustration of a small region in an array with two out of four replicates of virus-specific and external control probes (represented by V1 to -18 and C1 to -23, respectively). (B) Array image of a mock infection experiment revealing virtually no cross hybridization to virus-specific probes. (C and D) Array assays of Sf21 cells infected with Bac-PH-EGFP at an MOI of 10 at 6 and 48 hpi, respectively. (E) Nonlinear calibration curve showing the relationship between signal intensities obtained from external control probes and predetermined quantities of spiked-in control targets.
A set of external control probes that was used in data normalization was included in the array (Fig. 1A to D). For each of our array experiments, external spiked-in control targets with measured concentrations were added to the labeling reaction mixtures from the very beginning. Signal intensities of these control probes were used to develop a customized normalization method as described in Materials and Methods. A calibration curve for correction of the gene expression level was obtained for each array experiment to normalize all systematic differences between array hybridizations (Fig. 1E). These curves provided a reliable benchmark to calibrate all possible systematic errors of the platforms. They also allowed us to transform nonlinear fluorescent signal intensities into expression levels of viral transcripts over a wide dynamic range (at least 4 orders of magnitude).
Probe specificity was validated by hybridizations with cellular targets derived from mock-infected Sf21 cells. There was no evident hybridization signal detected by those baculovirus probes for mock-infected cells, compared to those for Bac-PH-EGFP-infected cells at 6 and 48 hpi (Fig. 1B to D). This difference suggests that all of the 155 predicted potentially coding ORFs were transcribed in Sf21 cells and that the selected probes on the custom-made microarray did not cross-hybridize with those control targets (external spiked-in yeast mRNAs) or nontarget genes (mRNAs of Sf21 cells).
Viral gene expression cascade in permissive cells.
To obtain a comprehensive view of the gene expression profile throughout the course of infection, microarray analysis was performed. This was done with RNA extracted from cells at 12 postinfection time points. Bac-PH-EGFP recombinant baculovirus (28) was employed to infect the fully permissive Sf21 cells at a multiplicity of infection (MOI) of 10. Total RNAs from cell lysates were harvested at 12 different time points between 0 and 72 hpi. Array analysis was done with each of these samples. The normalized hybridization signals for each gene at different time points were then obtained (shown at http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SII.pdf). A temporal ordering of expression profiles was created from a two-dimensional heat map (Fig. 2A) to illustrate a genome-wide view of specific gene expression patterns observed at each of the times during the course of infection. If the global baculovirus gene expression pattern in Sf21 cells can be viewed as a sequential array of cellular events, the microarray results revealed a highly ordered, orchestrated transcription program process. We detected approximately 20% of the transcripts in the viral genome from 3 to 6 hpi (Fig. 2A, group I), 50 to 60% from 6 to 9 hpi (Fig. 2A, group II), and the remaining 20 to 30% from 12 to 15 hpi (Fig. 2A, group III).
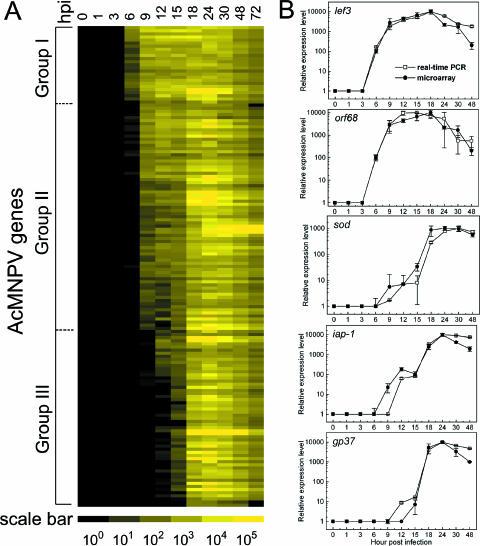
FIG. 2.
Time course microarray analysis of recombinant AcMNPV gene expression patterns in Sf21 cells. (A) Sf21 cells infected with Bac-PH-EGFP at an MOI of 10 were harvested at the indicated time points for microarray analysis. The patterns of viral gene expression during the course of infection are depicted as a two-dimensional heat map. Black blocks represent zero expression, and yellow blocks symbolize different expression levels corresponding to the scale bar at the bottom. (B) Quantitative real-time PCR confirmation of microarray results for five randomly selected genes. For data comparison, results derived by both methods were adjusted to 10,000 (lef-3, orf-68, iap1, and gp37) or 1,000 (sod) arbitrary units. All PCR data are averages of duplicate tests, and microarray data are averages of four replicates.
The customized array system is a good representation of the global transcription program of AcMNPV. To validate the temporal expression patterns of the microarray results, two genes in both group I and group II and one gene in group III were randomly selected for real-time quantitative PCR assay with cells harvested in different infection phases. The gene expression patterns over the infection course determined by these two quantitative methods were very similar (Fig. 2B).
Viral gene expression kinetics.
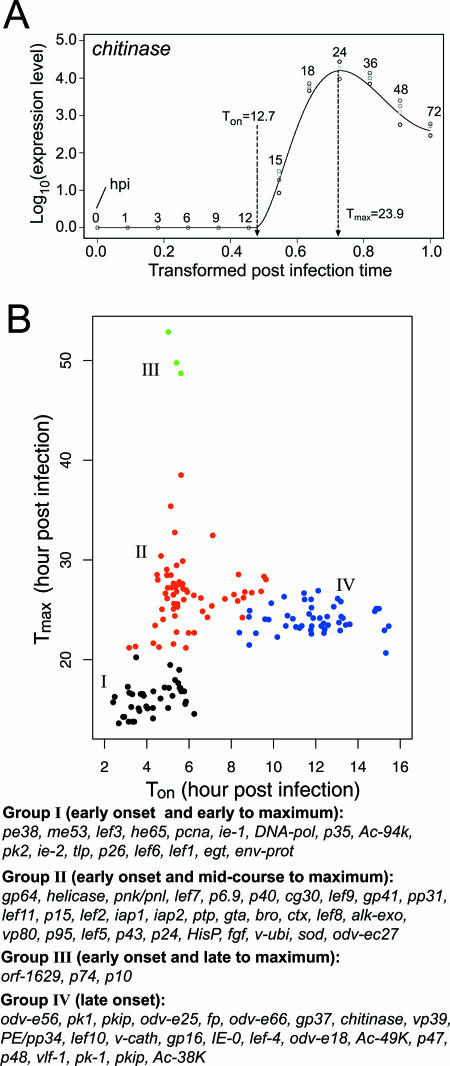
To allow us to further clarify the events within the expression cascade that are essential for AcMNPV infection, we introduce two parameters, Ton and Tmax, representing the postinfection time of onset of gene expression and the time to the maximal expression level, respectively (shown at http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SII.pdf). For example, in chitinase (orf-126) gene expression kinetics (Fig. 3A), Ton is 12.7 hpi and Tmax is 23.9 hpi. While Ton points to the time when a viral transcript is produced in a given host cell type, Tmax reflects the time for the ultimate steady-state accumulation of a viral transcript during the infection process. The genome-wide Ton and Tmax were calculated and scatter plotted in two dimensions (Fig. 3B). Ton varied from 2 to 16 hpi, while Tmax had a much wider range of 13 to 53 hpi. However, most transcripts were centered between 13 and 30 hpi. By 30 hpi, more than 95% (149 out of 156) of the AcMNPV genes had reached their highest levels of expression. However, the coefficients of correlation between Ton and Tmax (r = 0.20) for all of the viral genes were weak, meaning that there was no constant lag between the onset time of transcription and the time to the maximal expression level. This indicates that the onset of transcription and time to the maximal accumulation of a transcript in host cells were not necessarily related, a finding similar to that for human herpesvirus 8 (43). Collectively, these data suggest that Ton and Tmax are the two key kinetic parameters that show the dynamic gene expression pattern in the infection process.
FIG. 3.
Gene expression kinetics and classification. (A) Ton (time to onset of gene expression) and Tmax (time to maximum expression level) were demonstrated by using chitinase (orf-126) as an example. (B) A classification methodology for AcMNPV genes based on the criteria of Ton and Tmax kinetics. Selected known genes in each classified group are listed at the bottom.
On the basis of the above-mentioned transcript kinetic parameters, we clustered (by two-dimensional clustering analysis; for details, see Materials and Methods) the global transcriptional patterns into four groups, I (early onset, early to maximum), II (early onset, mid-course to maximum), III (early onset, late to maximum), and IV (late onset) (Fig. 3B and http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SII.pdf). Three genes on top of the others in group I (early-onset genes) were previously characterized as the immediate-early genes or early genes that are expressed in the earlier phase after infection. These three genes were the primary early-transcribed transactivator pe38 (26), the major early-transcribed DNA synthesis regulator me53 (23), and the early gene lef-3 (31), with Tons of 2.4, 2.5, and 2.9 hpi, respectively. Other members of groups I and II have also been reported as early genes. They include the immediate-early transactivator ie-1 gene, the major BV envelope glycoprotein gp64 gene, and the virus DNA polymerase DNA pol gene. In these early-transcribed genes (Ton = 2.5 to 9.6 hpi), transcripts in group I reached Tmax in the early phase of infection (13.6 to 19.5 hpi). Genes in group II or group III arrived at Tmax relatively late (range, 21.2 to 38.5 and 48.7 to 52.9 hpi, respectively). In contrast, genes in group IV appeared to initiate their transcription and reach their maximal expression in the late phase of infection (Ton, >8.4 hpi; Tmax, 20.7 to 26.9 hpi).
The late stage of the infection life cycle initiates at the onset of viral genome replication, which is about 5 to 6 hpi. During this stage, the genes for late structural proteins in groups II and IV, including vp39, vp80, and p24, together with the newly synthesized viral DNA, were assembled to form BV for cell-to-cell transmission of the virus within a single host or in cell culture. As a final point in the very late phase (Tmax = 49 to 53 hpi), genes in group III encoding envelope- or capsid-related structural proteins, such as p10, orf-1629, and p74, were heavily transcribed for virus packaging. Therefore, the expression cascade of viral transcripts in the late and very late phases appeared to be reflected in our transcription kinetic model.
Gene expression pattern and cytogenetic distribution.
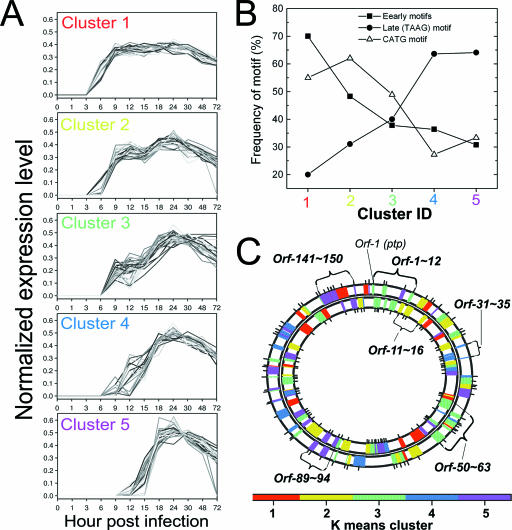
Genes participating in the same biological process may be transcriptionally coregulated (2, 27, 38, 54). Thus, baculovirus genes having the same temporal expression pattern during the time course of infection may imply sets of functionally correlated genes or suggest a common regulatory sequence motif in the genome. On the basis of K means clustering analysis, the 155 viral gene expression patterns in Sf21 cells were grouped into five clusters (Fig. 4A). These clusters were rearranged in order of increasing average Tons, ranging from top to bottom as 3.3 ± 0.5, 5.0 ± 0.8, 5.5 ± 0.7, 8.8 ± 1.1, and 12.5 ± 1.4 for clusters 1, 2, 3, 4, and 5, respectively. The time course expression profile cluster seemed to be dependent on Ton, because the amplitude of variability of Ton within the same cluster was much smaller than that among different clusters. Also, many genes in the same cluster appeared to be functionally correlated, although the factors behind this are not known. For example, many genes classified in cluster 1 were functioning as viral transcriptional activators, DNA replication factors, and apoptotic suppressors, whereas many genes grouped in clusters 4 and 5 were viral capsid-associated proteins or occlusion-derived virus envelope proteins (shown at http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SII.pdf).
FIG. 4.
Cluster analysis of gene expression patterns and colocalization in baculovirus genomic map. (A) Five gene clusters obtained by K means clustering analysis were arranged in an increasing order of the average Tons. (B) Correlation between the frequency of promoter sequence motifs and gene clusters. (C) Genome map view of the five gene clusters color coded in the AcMNPV genome. Gene clusters that colocalized to nearby loci are indicated. A colocalized cluster is defined as a region that contains at least five consecutive genes from the same gene cluster where no more than one interruption occurs by a gene from other gene clusters in either the plus or the minus strand. ID, identification.
Of great interest, we found a relationship between the abundance of a specific promoter motifs and a cluster of expression profiles. Earlier work by Ayres and colleagues attempted to correlate promoter motifs in upstream ORF sequences of AcMNPV with temporal expression properties (1). To date, there have been reports of several upstream sequence motifs of AcMNPV genes, including the early promoter TATA box, the initiator CAGT, and upstream regulatory element CGTGC, or the TAAG motif upstream of most of the late genes (12, 35). For AcMNPV genes, early and CATG motifs were found in the 160 nucleotides upstream of an ORF, and the TAAG late motif has been discovered within 80 nucleotides upstream of an ATG initiation codon (1). By calculating the abundance of different promoter motifs in a cluster of genes, we noted that the abundance of the early promoter motifs dropped dramatically, from 70% (cluster 1) to 31% (cluster 4). In contrast, the frequency of the late promoter motif ATTG increased sharply from 20% (cluster 1) to 64% (cluster 4) (Fig. 4B). The frequency of those previously identified promoter motifs correlates well with different clusters of genes that exhibit unique temporal expression patterns, suggesting that the viral cis-acting regulatory elements may orchestrate a complex, hierarchical gene expression cascade during the course of infection.
Besides the previously described promoter motifs, there are at least 23 genes in different expression clusters whose regulatory sequence motifs are still unidentified. It is also possible that there are other novel regulatory motifs or machineries coexisting with the identified motifs in the same gene cluster and that they also regulate the transcription pattern. In support of this idea, we found that the expression patterns of some genes appeared to be uncorrelated with their upstream sequence motifs. For example, some early-expressed genes, including orf-81, orf-66, tlp, and orf-43, possessed only late promoter motifs (http://www.nhri.org.tw/nhri_org/mm/jljuang/supplementary/AcMNPV-Sf21/Table_SII.pdf), whereas some late-expressed genes, such as orf-92 and orf-107, possessed only early motifs. These data, together with other recent evidence (37), indicate that the finding of an early or late promoter motif does not necessarily predict gene expression patterns because there might be other motifs or trans-acting factors involved in the regulation of transcription.
In addition to trying to discover if there is a correlation between the expression pattern and upstream sequence motifs, we also wanted to explore if there are any chromosomally adjacent genes that are transcriptionally coregulated. Marking the AcMNPV cytogenetic loci of genes with the color of their specific expression clusters, we found most genes to be randomly interspersed throughout the genome, with no obvious correlation with their specific patterns of expression (Fig. 4C). Nevertheless, some genes displaying synchronized expression patterns were clustered in a particular region of the genome map. There were about six such regional clusters (Fig. 4C), including the orf-1-to-orf-12 cluster, at which 9 out of 12 genes were grouped in cluster 3, and the orf-141-to-orf-150 cluster, where 9 out of 10 genes were members of cluster 5.
Physically adjacent genes could possibly share similar patterns of expression because they may be subject to the same regulatory control when transcribed. Exploring this possibility, we attempted to identify a potential common upstream regulatory sequence that might correlate with the regional clusters in the AcMNPV genome. Of the regional clusters we examined, only one was found to have a putative coregulatory motif sequence. A novel GTGCG sequence motif, located between upstream positions 15 and 190, was found in seven (orf-5, ctx, orf-1629, ptp, orf-4, lef-2, and orf-603) out of nine genes (P = 0.05) in the orf-1-to-12 cluster (data not shown), but this motif was far less common in the upstream sequences of other ORFs of the AcMNPV genome. The biological significance of this common sequence for the regulation of gene expression is not clear. The clustered genes that had very short conserved upstream sequence motifs may offer a spatial or positional advantage, making possible the efficient production of functionally related gene products that facilitate host infection.
Transcription network revealed by mutation of a very early-transcribed transactivator gene.
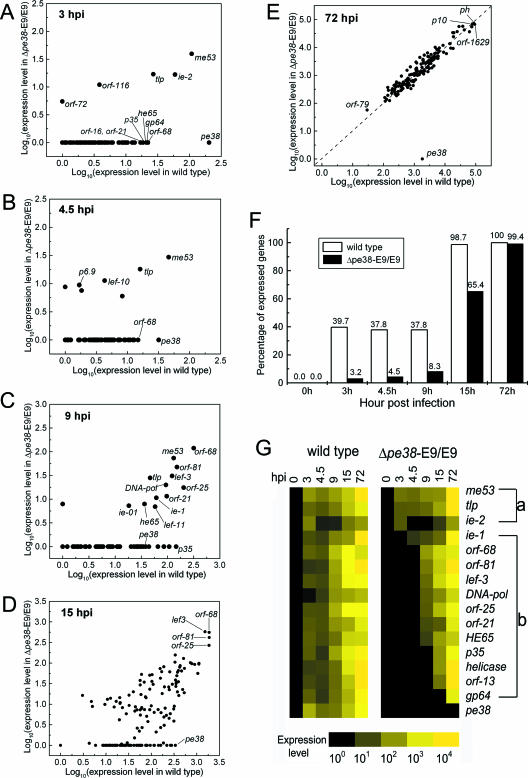
The transcription network in a baculovirus infection was closely coordinated by a sequential expression of early, late, and very late genes (Fig. 2A). Genes may have been interdependent along the time course, with later genes relying on other genes previously expressed. By microarray analysis, we studied the effect of a mutant baculovirus, Δpe38-E9/E9 (39), whose earliest transcribed baculovirus gene, pe38 (Ton = 2.4 hpi), was deleted, on the transcription network. A parallel global gene expression analysis of wild-type AcMNPV versus the Δpe38-E9/E9 mutant virus was conducted at six different time points (0 to 72 hpi) over the infection period. In theory, a scatter plot analysis of expression levels for wild-type and mutant viruses would reveal that pe38-independent genes fall along the diagonal and differentially expressed genes deviate from the diagonal. At an early stage (3 hpi), only five viral transcripts, me53, ie-2, orf-116, orf-72, and tlp, were clearly expressed in cells infected with the Δpe38-E9/E9 mutant virus. These were also coincidently expressed in the same manner as the wild-type virus at later stages (Fig. 5A and G, group a), suggesting that the expression of these five genes was essentially pe38 independent. The above possibility of independent transcription of me53 was supported by a previous study using transient expression of a reporter construct containing the me53 promoter to demonstrate that the transcription of me53 may be independent of other trans-acting viral factors (23). In contrast, 12 other genes (Fig. 5G, group b), including pe38, orf-68, gp64, he65, p35, and orf-16, were expressed only in cells infected with the wild-type virus and not in cells infected with the Δpe38-E9/E9 virus at 3 hpi. This suggests that expression of pe38 is vital to the regulation of many early genes in AcMNPV.
FIG. 5.
Differential gene expression profiling of a pe38 deletion mutant virus in strain Sf21 cells. (A to E) Scatter plot analysis for the dynamic differential gene expression delay of the Δpe38-E9/E9 mutant. Both wild-type and mutant viruses at an MOI of 10 were used to infect Sf21 cells. The gene expression levels shown here have been subjected to normalization. (F) Statistics of the delayed gene expression patterns at various times postinfection. An expressed gene is defined as one whose normalized expression level at the indicated time postinfection was greater than zero. (G) Temporal expression profiles of selective AcMNPV genes that were expressed in the wild-type virus in an earlier phase (<4.5 hpi) and their counterparts in the Δpe38-E9/E9 mutant virus. Genes in groups a and b are pe38 independent and pe38 dependent, respectively.
Although many genes were not expressed in Δpe38-E9/E9-infected cells at early stages of infection, their expressions were deferred only at the beginning. Later, they gradually returned to normal levels at later stages of infection (Fig. 5B to F). Differences in gene expression patterns between these two viruses became less noticeable at 72 hpi (Fig. 5E). Expression of early genes (e.g., ie-1, lef-3, DNA-pol, he65, lef-11, and several novel ORFs) was not found until 4.5 to 9 hpi. Some (e.g., p143, p35, and orf-13) were only detectable at 9 to 15 hpi (Fig. 5G). Most affected by the mutation of pe38 was the expression of gp64, which was transcribed only from 15 to 72 hpi in Sf21 cells (Fig. 5G). These findings suggest that the very early-transcribed pe38 gene product may be critical at the beginning of infection but less important in the later stages in Sf21 cells. Alternatively, one might speculate that there are redundant viral or cellular factors, compensating for the deficiency of pe38 in regulation of the transcription network.
DISCUSSION
In this study, we used a customized AcMNPV microarray system to collect global baculovirus gene expression data to characterize the expression program for potential coding ORFs selected by Ayres et al. (1), including previously known and uncharacterized ones. Our microarray analysis suggests that all 155 of the selected ORFs were expressed in permissive cells. It also provides us with comprehensive time-related information on the transcription that occurs and dynamic and quantitative information on the expression of individual genes at specific time points over the course of infection. In other words, microarray analysis has been proved in this study to be an important tool for the initial identification and characterization of potential ORFs. By the same approach, we should be able to identify the rest of the potential AcMNPV ORFs predicted from the genome sequence.
We did find, however, a low correlation between raw array data and the real quantitative results of virus transcripts during infection (data not shown). Therefore, direct interpretation of raw array data should be done cautiously. Appropriate transformations are needed for the interpretation of data, particularly with regard to early-expressed genes whose expression levels are relatively low at the onset of expression. We found that an objective benchmark is needed in order to calibrate experimental and biological deviations when trying to characterize gene expression kinetics at different time points over the infection period from a set of array data that are inherently noisy. This need has not been demonstrated by previous related DNA virus microarray studies (6, 43, 49). We used external control probes plus spiked-in targets to create a calibration system and used an innovative statistical method (Fig. 1) to systematically process and normalize large amounts of gene expression data. As shown in Fig. 2B, the results of the array analysis, after data transformation, were very similar to the results obtained by the generally accepted real-time PCR method. The calibration and normalization schemes established in this study can be applied to other, related microarray studies for analysis of viral gene expression profiles.
This study also proposes a new strategy for statistically investigating the kinetics of global baculovirus transcription. The kinetics of a specific viral gene was highlighted from two independent indexes, Ton and Tmax. These were individually generated through modeling procedures with array data obtained at different time points over the infection period. On the basis of the results obtained for previously characterized genes, the expression kinetics provided clues to possible viral strategies, which may be involved in the activation or inactivation of downstream genes in target cells. For example, several members of group I in Fig. 3B were early transactivators, including ie-1, ie-2, and pe38, and the DNA synthesis regulator gene me53. The products of these genes are critically needed at the very beginning of the regulatory events, when the baculovirus aggressively utilizes host RNA polymerase for viral gene transcription. Later, they quickly become less important, when viral RNA polymerase starts to take over the transcription machinery. Therefore, it is reasonable to assume that genes in group I are transcribed early and quickly reach a maximum expression level. In contrast, several genes in group II are needed throughout the whole period until the later stages of infection, ensuring proper DNA replication. Four such genes are gp64, which encodes an envelope fusion protein essential for BV production; the helicase p143 gene (9, 34); lef-2 (25); and lef-11 (32), which are required for DNA replication. Thus, the groups of expression kinetics seem to be functionally associated with the baculovirus replication program in permissive cells.
The study of baculovirus transcription kinetics can characterize the global transcription program of known genes, as well as provide clues to potential functions of previously uncharacterized ORFs in the network. For example, a putative ORF, orf-68, located upstream of an early gene, lef-3, was shown to have Tons and Tmaxs similar to those of the three most immediate-early transcripts (i.e., pe38, me53, and lef-3), suggesting that orf-68 may play a parallel role in the early infection cycle.
Another approach used to portray the transcription program, unsupervised clustering analysis, is typically used for extracting particular subsets of expression patterns from microarray data. By this approach, we discovered two major findings regarding the regulation of baculovirus transcription programs. First, transcription onset timing was strongly influenced by the conserved promoter sequence motifs. Second, the onset time of expression might predict the subsequent viral gene expression pattern. Furthermore, because functionally correlated or coregulated genes in an operon of a bacterial genome may be located in nearby loci of the physical genome (29), we thought it might be interesting to investigate whether a similar gene organization exists in the AcMNPV genome. We tagged these gene clusters into the cytogenetic map and analyzed the relationship between the gene clusters in the viral genome and the temporal expression patterns. Most of the gene clusters were randomly dispersed in the baculovirus genome, although a few gene groups were located in adjacent regions. To our surprise, two putative late-operon-like regions were found in the orf-89-to-orf-94 and orf-141-to-orf-150 regions, where most (n = 5 and n = 7 in orf-89 to orf-94 and orf-141 to orf-150, respectively) gene products function as structural proteins or display a late expression pattern. To further confirm the existence of two late-operon-like regions in AcMNPV, we conducted a comparative genomic analysis of these sequences in three phylogenetically closely related baculoviruses: AcMNPV, Bombyx mori nucleopolyhedrosis virus, and Orgyia pseudotsugata multicapsid nucleopolyhedrosis virus. Both regions were found to be evolutionarily conserved (data not shown), supporting the argument that they had grouped together in common regulatory clusters. Although similar operon-like structures have been reported in the human herpesvirus 8 (Kaposi's sarcoma virus) genome (11, 51), it is still unclear whether an operon-like strategy is indeed used by baculoviruses to coregulate the transcription of genes clustered in the neighboring viral genomic loci. Therefore, it is important to gather information that may help answer the question of whether the observed gene organization in the AcMNPV genome actually affects viral gene expression. In addition to two gene cassettes, we discovered a common GTGCG sequence motif in the upstream coding sequences of many genes in the orf-1-to-orf-12 region. Still further studies are needed to determine whether such a motif may also coregulate transcription.
To uncover the role of a viral gene in the regulatory pathway, a viral gene needs to be mutated or deleted and the functional consequence evaluated. We analyzed the differential transcription profile of a mutant virus defective in pe38 (Δpe38-E9/E9), an immediate-early viral gene presumed to be vital but probably not essential in viral DNA synthesis and BV production (39). As expected, global gene expression patterns were indeed greatly altered at the early and late stages (Fig. 5A to D) while the delayed transcripts were gradually returned to the same levels as those of the wild-type control in the very late stage (Fig. 5E), suggesting that although the pe38-encoded gene product plays a critical role in the regulation of early transcription units, it might not be as important for all regulatory cascades, especially later stages. Furthermore, the delayed temporal expression pattern we observed also might give clues helpful in understanding why the mutant virus has the phenotype of delayed BV production but still produces infectious virus particles similar to the wild-type virus (39). Most importantly, the sequential expression map in the pe38-deficient background revealed a regulatory hierarchy. Expression of the several early-transcribed genes at the top of the hierarchy (e.g., me53, tlp, and ie-2) was not altered by the mutation of pe38. An early transactivator, ie-1, was only affected in the early stage, whereas the expression levels of most of the early-transcribed genes were greatly affected by the mutation of pe38. Data from earlier studies also support this. For example, the expression of the helicase p143 (33) and gp64 (39) genes is regulated by pe38. To display the putative regulatory cascade mediated by pe38, we extracted 16 early-transcribed genes and compared the differences in temporal expression patterns between wild-type and mutant viruses. pe38 seemed crucial for the transactivation of five genes (lef-3, DNA-pol, he65, p143, and lef-11) (Fig. 5G). These five, in turn, seemed to affect the transcription of p35 and gp64. The regulatory cascade involving the pe38-deficient virus may well be complemented by another early transactivator, possibly ie-1, to modulate downstream gene expression.
This is the first study to use array analysis to elucidate the regulatory pathway controlled by a specific baculovirus early-expressed gene. Systematic mutation of other transactivator genes in the AcMNPV genome will allow further exploration of the transcription regulatory network. The same strategy can possibly be used to uncover novel roles of known genes or putative functions of uncharacterized ORFs in the genome. In the present study, for example, we identified the delayed expression of he65, a gene encoding a putative RNA ligase (19), in cells infected with the pe38 mutant virus. On the basis of this finding, it is tempting to investigate whether PE38's modulation of he65 expression is a counterreaction to the host defense. After all, he65 has been implicated in the repair of essential RNA molecules damaged by the host defense mechanism to block infection. Although this is speculation, further investigation of this possibility may forge a path into the heart of the regulatory network involving pe38. Moreover, this array-based strategy for the study of the transcription regulatory network may be particularly useful for those randomly generated mutant viruses that display specific phenotypes although the underlying genes or mechanisms need elucidation (20).
By introducing an array strategy to systematically characterize the previously established baculovirus mutants, this study points to a possibly very productive direction for constructing a baculovirus gene regulation network. The discovery and collection of these insights can allow us to build an integrated database that can greatly enhance the research of baculoviruses and their future applications.
Acknowledgments
We thank David A. Theilmann for providing the wild-type and pe-38 deletion mutant baculoviruses and Tom Kost and Ing-Ming Chiu for comments on the manuscript. We also thank Yin-Lan Liu and Shiang-Min Lin for efforts in baculovirus microarray chip production, microarray experiment, and real-time RT-PCR analysis. We are grateful to Wei-Chen Chen and Pramod Gupta for professional assistance in the programming and statistical analysis of microarray data and James Steed for English editing of the manuscript.
This work was supported by NHRI.
REFERENCES
- 1.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 2.Baliga, N. S., M. Pan, Y. A. Goo, E. C. Yi, D. R. Goodlett, K. Dimitrov, P. Shannon, R. Aebersold, W. V. Ng, and L. Hood. 2002. Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc. Natl. Acad. Sci. USA 99:14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blissard, G. W., P. H. Kogan, R. Wei, and G. F. Rohrmann. 1992. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology 190:783-793. [DOI] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and G. F. Rohrmann. 1991. Baculovirus gp64 gene expression: analysis of sequences modulating early transcription and transactivation by IE1. J. Virol. 65:5820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonning, B. C., and B. D. Hammock. 1996. Development of recombinant baculoviruses for insect control. Annu. Rev. Entomol. 41:191-210. [DOI] [PubMed] [Google Scholar]
- 6.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, I.-S., C. A. Hsiung, C.-C. Wen, and Y.-J. Wu. 2006. A note on the consistency in Bayesian shape-restricted regression with random Bernstein polynomials, p. 36-44. In C. H. Zhang, Z. Ying, and C. A. Hsiung (ed.), Random walks, sequential analysis and related topics: a festschrift in honor of Y. S. Chow. World Scientific, Singapore.
- 8.Chang, I.-S., C. A. Hsiung, Y.-J. Wu, and C.-C. Yang. 2005. Bayesian survival analysis using Bernstein polynomials. Scand. J. Stat. 32:447-466. [Google Scholar]
- 9.Chen, T., D. Sahri, and E. B. Carstens. 2004. Characterization of the interaction between P143 and LEF-3 from two different baculovirus species: Choristoneura fumiferana nucleopolyhedrovirus LEF-3 can complement Autographa californica nucleopolyhedrovirus LEF-3 in supporting DNA replication. J. Virol. 78:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson, J. A., and P. D. Friesen. 1991. Identification of upstream promoter elements mediating early transcription from the 35,000-molecular-weight protein gene of Autographa californica nuclear polyhedrosis virus. J. Virol. 65:4006-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-166. In L. K. Miller (ed.), The baculoviruses. Plenum, New York, N.Y.
- 13.Friesen, P. D., and L. K. Miller. 2001. Insect viruses, p. 602-612. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields' virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 14.Gillespie, L. S., K. K. Hillesland, and D. J. Knauer. 1991. Expression of biologically active human antithrombin III by recombinant baculovirus in Spodoptera frugiperda cells. J. Biol. Chem. 266:3995-4001. [PubMed] [Google Scholar]
- 15.Grula, M. A., P. L. Buller, and R. F. Weaver. 1981. α-Amanitin-resistant viral RNA synthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvae and Spodoptera frugiperda cells. J. Virol. 38:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarino, L. A., and M. Smith. 1992. Regulation of delayed-early gene transcription by dual TATA boxes. J. Virol. 66:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, P., and N. E. Heckman. 2002. Estimating and depicting the structure of a distribution of random functions. Biometrika 89:145-158. [Google Scholar]
- 18.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 19.Ho, C. K., and S. Shuman. 2002. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl. Acad. Sci. USA 99:12709-12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, Y., H. R. Lo, T. C. Lee, C. P. Wu, and Y. C. Chao. 2004. Enhancement of correct protein folding in vivo by a non-lytic baculovirus. Biochem. J. 382:695-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huh, N. E., and R. F. Weaver. 1990. Identifying the RNA polymerases that synthesize specific transcripts of the Autographa californica nuclear polyhedrosis virus. J. Gen. Virol. 71(Pt. 1):195-201. [DOI] [PubMed] [Google Scholar]
- 22.Inceoglu, A. B., S. G. Kamita, A. C. Hinton, Q. Huang, T. F. Severson, K. Kang, and B. D. Hammock. 2001. Recombinant baculoviruses for insect control. Pest Manag. Sci. 57:981-987. [DOI] [PubMed] [Google Scholar]
- 23.Knebel-Mörsdorf, D., A. Kremer, and F. Jahnel. 1993. Baculovirus gene ME53, which contains a putative zinc finger motif, is one of the major early-transcribed genes. J. Virol. 67:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogan, P. H., X. Chen, and G. W. Blissard. 1995. Overlapping TATA-dependent and TATA-independent early promoter activities in the baculovirus gp64 envelope fusion protein gene. J. Virol. 69:1452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krappa, R., and D. Knebel-Mörsdorf. 1991. Identification of the very early transcribed baculovirus gene PE-38. J. Virol. 65:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagreid, A., T. R. Hvidsten, H. Midelfart, J. Komorowski, and A. K. Sandvik. 2003. Predicting gene ontology biological process from temporal gene expression patterns. Genome Res. 13:965-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. F., C. C. Chen, T. A. Hsu, and J. L. Juang. 2000. A baculovirus superinfection system: efficient vehicle for gene transfer into Drosophila S2 cells. J. Virol. 74:11873-11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewin, B. 2000. The operon. Oxford University Press, New York, N.Y.
- 30.Li, L., S. H. Harwood, and G. F. Rohrmann. 1999. Identification of additional genes that influence baculovirus late gene expression. Virology 255:9-19. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., A. L. Passarelli, and L. K. Miller. 1993. Identification, sequence, and transcriptional mapping of lef-3, a baculovirus gene involved in late and very late gene expression. J. Virol. 67:5260-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 76:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, A., and E. B. Carstens. 1993. Immediate-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology 195:710-718. [DOI] [PubMed] [Google Scholar]
- 34.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 35.Lu, A., and L. K. Miller. 1997. Regulation of baculovirus late and very late gene expression, p. 193-211. In L. K. Miller (ed.), The baculoviruses. Plenum, New York, N.Y.
- 36.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mans, R. M., and D. Knebel-Morsdorf. 1998. In vitro transcription of pe38/polyhedrin hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J. Virol. 72:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald, M. J., and M. Rosbash. 2001. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107:567-578. [DOI] [PubMed] [Google Scholar]
- 39.Milks, M. L., J. O. Washburn, L. G. Willis, L. E. Volkman, and D. A. Theilmann. 2003. Deletion of pe38 attenuates AcMNPV genome replication, budded virus production, and virulence in heliothis virescens. Virology 310:224-234. [DOI] [PubMed] [Google Scholar]
- 40.Miller, L. K. 1995. Genetically engineered insect virus pesticides: present and future. J. Invertebr. Pathol. 65:211-216. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen, H. B., and S. Knudsen. 2002. Avoiding cross hybridization by choosing nonredundant targets on cDNA arrays. Bioinformatics 18:321-322. [DOI] [PubMed] [Google Scholar]
- 42.Ohresser, M., N. Morin, M. Cerutti, and C. Delsert. 1994. Temporal regulation of a complex and unconventional promoter by viral products. J. Virol. 68:2589-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pullen, S. S., and P. D. Friesen. 1995. The CAGT motif functions as an initiator element during early transcription of the baculovirus transregulator ie-1. J. Virol. 69:3575-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pullen, S. S., and P. D. Friesen. 1995. Early transcription of the ie-1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5′ noncoding leader region. J. Virol. 69:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, G. E., M. D. Summers, and M. J. Fraser. 1983. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 3:2156-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, L. M., M. Hirst, M. Lopez Ferber, A. T. Merryweather, P. J. Cayley, and R. D. Possee. 1991. Construction of an improved baculovirus insecticide containing an insect-specific toxin gene. Nature 352:85-88. [DOI] [PubMed] [Google Scholar]
- 49.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szewczyk, B., L. Hoyos-Carvajal, M. Paluszek, I. Skrzecz, and M. Lobo de Souza. 2005. Baculoviruses—re-emerging biopesticides. Biotechnol. Adv. 24(2):143-160. [DOI] [PubMed] [Google Scholar]
- 51.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 52.Taylor, A. L., A. Haze-Filderman, A. Blumenfeld, B. Shay, L. Dafni, E. Rosenfeld, Y. Leiser, E. Fermon, Y. Gruenbaum-Cohen, and D. Deutsch. 2006. High yield of biologically active recombinant human amelogenin using the baculovirus expression system. Protein Expr. Purif. 45:43-53. [DOI] [PubMed] [Google Scholar]
- 53.Tomalski, M. D., J. G. Wu, and L. K. Miller. 1988. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology 167:591-600. [PubMed] [Google Scholar]
- 54.Tureci, O., H. Bian, F. O. Nestle, L. Raddrizzani, J. A. Rosinski, A. Tassis, H. Hilton, M. Walstead, U. Sahin, and J. Hammer. 2003. Cascades of transcriptional induction during dendritic cell maturation revealed by genome-wide expression analysis. FASEB J. 17:836-847. [DOI] [PubMed] [Google Scholar]