Abstract
Earlier, our laboratory reported that purified glutathione S-transferase-virion host shutoff (GST-vhs) protein exhibited endoribonucleolytic activity in in vitro assays using as substrates in vitro-transcribed regions of IEX-1 mRNA. Here, we report that studies of the cleavage patterns of synthetic RNA oligonucleotides defined the activity of GST-vhs as being similar to that of RNase A. Thus, GST-vhs cleaved the RNA at the 3′ end of single-stranded cytidine and uridine residues. Since the GST-mvhs nuclease-defective mutant protein failed to cleave the synthetic RNAs, the results unambiguously attribute the activity to vhs.
Virion host shutoff (vhs) protein is the product of the UL41 open reading frame of herpes simplex virus 1. The protein, made late in infection, is packaged into virions and, in newly infected cells, shuts off host protein synthesis by degrading preexisting and newly transcribed mRNAs (8, 10, 15; reviewed in reference 14). Several lines of evidence accumulated in the past decade indicated that vhs acts as an mRNA-specific RNase. Specifically, it has been reported that (i) vhs degrades mRNA in the absence of other viral proteins, as shown by the inhibition of reporter gene expression in mammalian cells transiently cotransfected with a vhs expression vector (7, 13); (ii) extracts of detergent-solubilized virions have vhs-dependent nuclease activity which can be specifically blocked by anti-vhs antisera (18); (iii) vhs induces endonucleolytic cleavage of a number of RNA substrates when it is expressed as the only viral protein in the rabbit reticulocyte lysate in an in vitro translation system (2, 3); and (iv) vhs does not discriminate between cellular and viral mRNAs, but it exhibits a strong preference for mRNAs, as opposed to rRNA and tRNA, in all the systems tested (9, 12, 18).
However, the unambiguous proof of whether vhs is itself a nuclease in absence of any other cellular or viral proteins has been just recently reported by our laboratory (17). For the first time, a glutathione S-transferase (GST)-tagged full-length vhs has been purified in a soluble and biologically active form, inasmuch as it exhibited RNase activity in an in vitro assay. Moreover, the pattern of degradation of the RNA substrates into fragments of discrete sizes unequivocally defined vhs as an endoribonuclease. The substrates selected to test the in vitro activity of the purified GST-vhs protein were those representing the IEX-1 mRNA, based on earlier reports showing a distinctive vhs-dependent degradation pattern of this cellular mRNA in cells infected with herpes simplex virus 1 (4, 16). The in vitro-generated IEX-1 substrates incubated with the purified GST-vhs protein were degraded in a manner consistent with the analyses of the fate of this mRNA in the infected cells. The results pointed to a region immediately upstream or inside of the AU-rich elements as the preferential cleavage site (17). However, an attempt to identify specific sequence or secondary structure recognized by vhs by analyzing the sequences surrounding the potential cleavage sites in IEX-1 substrates was unsuccessful.
The objective of the experiments described in this report was to identify the sequences recognized by vhs as cleavage sites. We report that purified vhs resembles the bovine pancreatic RNase A inasmuch as it preferentially cleaves the RNA substrate at level of single-stranded (ss) cytidine (C) and uridine (U) residues. While it is unclear why HSV has evolved an enzyme with properties similar to those of RNase A, our results may contribute to a more comprehensive analysis defining the basis of the vhs selectivity for mRNAs.
The experimental approach selected was the comparison of the cleavage patterns generated following digestion of specific synthetic RNA oligonucleotides by the GST-vhs fusion protein with those produced by well-characterized ribonucleases. RNA oligonucleotides, varying in length between 21 and 48 nucleotides (nt), were all synthesized by Integrated DNA Technologies (Coralville, IA) and labeled at the 5′ end by incubation with T4 polynucleotide kinase in the presence of [γ-32P]ATP according to the protocol supplied with the mirVana probe and marker kit (Ambion, Austin, TX). The probes were then resolved on a denaturing polyacrylamide gel to separate the full-length fragment from the shorter products. The full-length probes were recovered by incubation at 37°C in probe elution buffer (Ambion), followed by ethanol precipitation to remove the salt and sodium dodecyl sulfate contained in the buffer. The probes were then resuspended in nuclease-free H2O and counted by using a Beckman (Fullerton, CA) LS8000 scintillation counter. Radiolabeled RNA markers were prepared from Decade marker RNA substrate (Ambion) as specified by the manufacturer.
The GST-vhs, GST alone, and GST-mvhs (the GST-vhs mutant protein bearing substitutions of three amino acid residues, E192, D194, and D195, which are essential for the nuclease activity) recombinant proteins were expressed and purified as reported elsewhere (17). RNase A, RNase T1, and RNase V1 were purchased from Ambion. Their preferred cleavage sites are as follows: at the 3′ end of ss C and U residues (RNase A) and at the 3′ end of ss guanidine (G) residues (RNase T1) and base-paired nucleotides (RNase V1). The cleavage assay was carried out under the experimental conditions described in the previous report (17) with a variation only regarding the inactivation of the nuclease activity and the subsequent precipitation of the cleavage products which, in this series of experiments, were carried out in the presence of the precipitation/inactivation buffer (Ambion) supplied with the purchased ribonucleases.
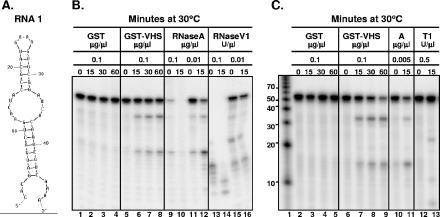
The first oligonucleotide (RNA 1) tested contained 48 nt (5′-CACGAUGACUGAACUACC GCAUGAAAGUGCGGAUCACAGUCGUCAAAA-3′). The predicted secondary structure obtained with the aid of the mfold program (version 3.2; available at the website http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi) (11, 19) is shown in Fig. 1A. An equal amount (0.1 μg/μl) of GST-vhs fusion protein or GST alone as well as different dilutions of RNase A, RNase V1, and RNase T1 were incubated at 30°C with the RNA 1 in a total volume of 50 μl as previously reported (17). Samples were withdrawn at various times and analyzed by electrophoresis in a 15% polyacrylamide denaturing gel, followed by autoradiography. The results, shown in Fig. 1B and C, were as follows. The RNA substrate mixed with GST protein alone remained stable through the 60-min incubation time (Fig. 1B, lanes 1 to 4; Fig. 1C, lanes 2 to 5), whereas that reacted with the GST-vhs fusion protein was cleaved into two major fragments (Fig. 1B, lanes 5 to 8; Fig. 1C, lanes 6 to 9). On the basis of the migration pattern of the RNA markers (Fig. 1C, lane 1), the lengths of the two fragments, which appeared as early as 15 min after incubation (Fig. 1B, lane 6; Fig. 1C, lane 7), were estimated to be 10 to 20 nt and 30 to 40 nt, respectively. Based on the loop structures (Fig. 1A), the incubation of RNA 1 with RNase A was expected to generate four major cleavage products of 14, 15, 34, and 35 nt, respectively, plus two very small fragments of 1 and 3 nt, whereas RNase T1 was predicted to produce a unique fragment of 23 nt. Three different dilutions of RNase A were tested (Fig. 1B, lanes 9 to 12; Fig. 1C, lanes 10 and 11) to identify the one showing the optimal level of activity in our experimental conditions. As shown in panel C, lane 11, the incubation of RNA 1 with 0.005 μg/μl of RNase A for 15 min generated two major bands whose size was close to the predicted values, i.e., 34 to 35 nt and 14 to 15 nt, respectively. It is conceivable that the percentage of the gel used (i.e., 15%) was not entirely appropriate to obtain an accurate resolution of the bands that differ by only one nucleotide (34 versus 35; 14 versus 15). In our experimental conditions, RNase T1 generated not only the expected fragment of 23 nt, due to the cleavage at the 3′ end of the only ss G residue present on the top loop (Fig. 1A), but also two unpredicted smaller fragments (Fig. 1C, lane 13). Their lengths (∼12 nt and less than 10 nt) pointed to the two G residues present in positions 4 and 11, respectively, as potential cleavage sites. Even though we cannot provide definitive proof, we hypothesize that the cleavage of the only ss G residue at position 23 changed the secondary structure of RNA 1, allowing the other two G residues to become exposed to RNA T1. Finally, two different dilutions of RNase V1 were tested. RNase V1 is reported to cleave base-paired nucleotides and, based on the secondary structure of RNA 1, the generation of several fragments ranging from 4 to 22 nt was expected. The cleavage pattern of RNase V1 is shown in Fig. 1B, lanes 13 to 16. Taken together, these data clearly indicate that the pattern of degradation of RNA 1 substrate generated upon incubation with our GST-vhs protein highly resembles that of RNase A.
FIG. 1.
Comparison of the cleavage pattern of RNA 1 produced by GST-vhs fusion protein with those generated by RNases A, T1, and V1. (A) Secondary structure of the RNA 1 oligonucleotide as predicted by the mfold program. (B) Either GST (0.1 μg/μl; lanes 1 to 4) or GST-vhs fusion protein (0.1 μg/μl; lanes 5 to 8) as well as two different concentrations of RNase A (0.1 μg/μl, lanes 9 and 10; 0.01 μg/μl, lanes 11 and 12) and RNase V1 (0.1 U/μl, lanes 13 and 14; 0.01 U/μl, lanes 15 and 16) were incubated with 5′-end-labeled RNA 1 substrate. Aliquots were removed at the indicated times (in minutes; top of gel) and analyzed as described in the text. (C) Either GST (0.1 μg/μl; lanes 2 to 5) or GST-vhs fusion protein (0.1 μg/μl; lanes 6 to 9) as well as RNase A (0.005 μg/μl; lanes 10 and 11) and RNase T1 (0.5 U/μl; lanes 12 and 13) were incubated with 5′-end-labeled RNA 1 substrate for the time intervals (in minutes; top of gel) shown and analyzed as for panel B. Lane 1, 5′-end-labeled Decade RNA markers. The lengths of the fragments (in nucleotides) are reported on the side.
To verify this observation, seven additional RNA oligonucleotides with only one or two possible cleavage sites were designed and tested under the same experimental conditions. Furthermore, GST-mvhs, which bears substitutions of three critical amino acid residues (E192, D194, D195) and which was shown earlier to be nuclease defective (17), was tested with the new substrates to verify the specificity of the RNase activity of our purified GST-vhs fusion protein. The results, shown in Fig. 2, 3, and 4, were as follows.
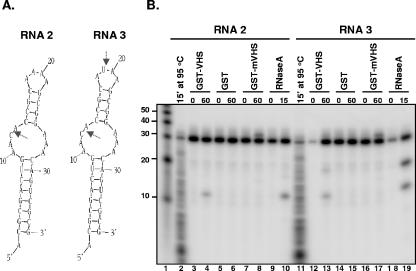
FIG. 2.
GST-vhs fusion protein, but not GST-mvhs mutant, cleaves RNAs 2 and 3 similarly to RNase A. (A) Secondary structure of either RNA 2 (left) or RNA 3 (right) oligonucleotides as predicted by the mfold program; the arrows mark the sites of RNase A expected cleavage. (B) GST-vhs fusion protein (lanes 3, 4, 12, and 13), GST alone (lanes 5, 6, 14, and 15), or GST-mvhs mutant protein bearing alanine substitutions for E192, D194, and D195 residues (lanes 7, 8, 16, and 17) (each at 0.1 μg/μl) as well as 0.01 μg/μl RNase A (lanes 9, 10, 18, and 19) were incubated with either 5′-end-labeled RNA 2 substrate (lanes 3 to 10) or 5′-end-labeled RNA 3 substrate (lanes 12 to 19) for the time intervals shown (in minutes) and analyzed as described in the text. Lane 1, 5′-end-labeled Decade RNA marker. The lengths of the fragments (in nucleotides) are reported on the side. RNA 2 (lane 2) and RNA 3 (lane 11) were incubated for 15 min (15′) at 95°C in the presence of Ambion alkaline hydrolysis buffer (50 mM sodium carbonate [pH 9.2], 1 mM EDTA) in the attempt to generate a ladder of hydrolyzed RNA fragments.
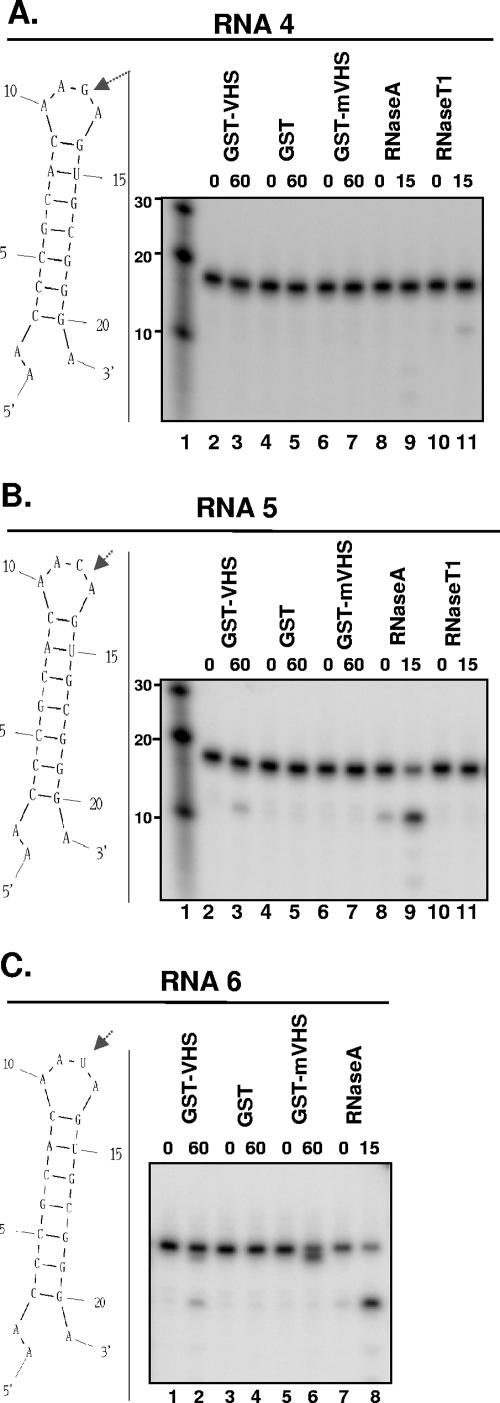
FIG. 3.
GST-vhs fusion protein cleaves RNAs 4, 5, and 6 similarly to RNase A. (A) Left, secondary structure of RNA 4 oligonucleotide as predicted by the mfold program; the arrow marks the site of RNase T1 expected cleavage. Right, 0.1 μg/μl of GST-vhs fusion protein (lanes 2 and 3), GST alone (lanes 4 and 5), or GST-mvhs mutant protein (lanes 6 and 7) as well as 0.01 μg/μl RNase A (lanes 8 and 9) and 0.01 U/μl of RNase (T1) were incubated with 5′-end-labeled RNA 4 for the time intervals shown (in minutes; top of gel) and analyzed as described in the text. Lane 1, 5′-end-labeled Decade RNA marker. The lengths of the fragments (in nucleotides) are reported on the side. (B) Left, secondary structure of RNA 5 oligonucleotide as predicted by the mfold program; the arrow marks the site of RNase A expected cleavage. Right, 0.1 μg/μl of GST-vhs fusion protein (lanes 2 and 3), GST alone (lanes 4 and 5), or GST-mvhs mutant protein (lanes 6 and 7) as well as 0.01 μg/μl RNase A (lanes 8 and 9) and 0.01 U/μl of RNase (T1) were incubated 5′-end-labeled RNA 5 for the time intervals shown (in minutes; top of gel) and analyzed as for panel A. Lane 1, 5′-end-labeled Decade RNA marker. The lengths of the fragments (in nucleotides) are reported on the side. (C) Left, secondary structure of RNA 6 oligonucleotide as predicted by the mfold program; the arrow marks the site of RNase A expected cleavage. Right, 0.1 μg/μl of GST-vhs fusion protein (lanes 1 and 2), GST alone (lanes 3 and 4), or GST-mvhs mutant protein (lanes 5 and 6) as well as 0.01 μg/μl RNase A (lanes 7 and 8) were incubated with 5′-end-labeled RNA 6 substrate for the time intervals shown (in minutes; top of gel), and the samples were analyzed as for panels A and B.
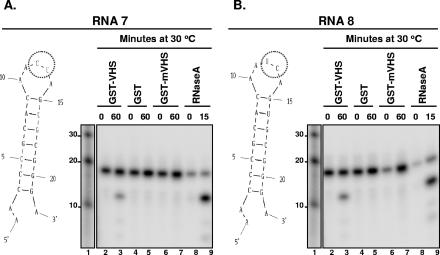
FIG. 4.
GST-vhs fusion protein cleaves RNAs 7 and 8 similarly to RNase A. (A and B) Left, secondary structure of either RNA 7 (A) or RNA 8 (B) oligonucleotides as predicted by the mfold program; the circle marks the sites of RNase A predicted cleavage. Right, 0.1 μg/μl of GST-vhs fusion protein (lanes 2 and 3), GST alone (lanes 4 and 5), or GST-mvhs mutant protein (lanes 6 and 7) as well as 0.01 μg/μl RNase A (lanes 8 and 9) were incubated with either 5′-end-labeled RNA 7 substrate (A) or 5′-end-labeled RNA 8 substrate (B) for the time intervals shown (in minutes). Lane 1, 5′-end-labeled Decade RNA marker. The lengths of the fragments (in nucleotides) are reported on the side.
(i) RNA 2 and RNA 3 oligonucleotides (36 nt) have the same sequence except for the adenine residue in position 19 of the second loop of RNA 2 that is replaced by a U residue in RNA 3. Based on the predicted secondary structure (Fig. 2A), RNase A should cleave RNA 2 substrate only at level of the unique ss C residue present in position 12 in the first loop, whereas it should generate two distinct fragments, of 12 and 19 nt, respectively, upon digestion of RNA 3. This was indeed the case (Fig. 2B, lanes 9, 10, 18, and 19). As observed with RNA 1 substrate, RNA 2 and RNA 3 substrates incubated with GST-vhs protein were cleaved in a manner comparable to that of RNase A (Fig. 1B, lanes 3, 4, 12, and 13). The low level of the 19-nt fragment generated by incubation of GST-vhs with RNA 3 (Fig. 1B, lane 13) compared to RNase A (Fig. 1B, lane 19) suggests that vhs may have a preference for ss C residues over ss U residues. As expected, no cleavage products were detected when the RNA substrates were incubated with either GST alone (Fig. 1B, lanes 5, 6, 14, and 15) or GST-mvhs mutant protein (Fig. 1B, lanes 7, 8, 16, and 17), confirming that the RNase activity was specifically driven by vhs and not by a contaminating bacterial nuclease that copurified with GST or the chimeric protein. The slower migrating band observed with the samples incubated with the GST-mvhs mutant protein (Fig. 1B, lanes 8 and 17) could be a result of a tight association of the inactive form of vhs protein with the RNA substrates. This band disappeared in a parallel experiment in which the cleavage products were subjected to phenol-chloroform extraction prior to analysis with the gel (data not shown).
(ii) To determine whether GST-vhs protein preferentially cleaves ss C residues over ss U residues, five additional oligonucleotides, RNA 4 to RNA 8, were designed. These are shorter (21 to 22 nt) and have a simpler secondary structure than RNA 1 to RNA 3, containing two loops (Fig. 3A to C, left; Fig. 4A and B, left). The five substrates have the same nucleotide sequence except for the residue in position 12. This residue is a G in RNA 4, a C in RNA 5, a U in RNA 6, or the dinucleotide CU or UC in RNA 7 or RNA 8, respectively. Based on the presence of only an ss G residue, RNA 4 was cleaved as expected only by RNase T1 (Fig. 3A, lanes 10 and 11), albeit with a very low efficiency, but not by either RNase A (lanes 8 and 9) or GST-vhs (lanes 2 and 3). In contrast, RNA 5 and 6, as expected, were cleaved by either RNase A (Fig. 3B, lanes 8 and 9; Fig. 3C, lanes 7 and 8) or GST-vhs (Fig. 3B, lanes 2 and 3; Fig. 3C, lanes 1 and 2), generating a major fragment of 12 nt. Similar results were obtained with RNA 7 (Fig. 4A) and RNA 8 (Fig. 4B), indicating that GST-vhs cleaves after C and U residues with equal efficiency. These results suggest that other features of the RNA substrates, such as secondary structures of RNA 2 and 3 that are slightly more complex than those of the last oligonucleotides tested (RNA 4 to 8), can affect the vhs cleavage preference.
In conclusion, the results presented here show that our purified GST-vhs fusion protein is an endoribonuclease similar to RNase A with respect to the cleavage specificity at the level of C and U residues in single-stranded stretches of RNA. The evidence that the GST-mvhs is inactive indicates that the activity exhibited by the GST-vhs protein is not due to contamination. We should note that an alignment of the primary protein sequence of vhs with those of known RNases A did not show regions of homology. It is noteworthy that the substrate specificity of vhs we reported here differs from the previously described tendency of vhs-dependent endoribonuclease activity to cleave between purine residues (2, 3, 18). Instead of the purified active and mutated chimeric GST-vhs proteins used in the present and previous studies (17), Zelus and colleagues (18) and Elgadi and colleagues (2, 3) used as a source of vhs either crude virion extracts or the products of in vitro translation systems. The uncertainty belaboring the earlier results may be deduced from the conclusion of the previous studies that “the available data argue that vhs-induced endoribonuclease activity displays a relatively relaxed sequence specificity, suggesting that other features, such as RNA secondary structure, may play a major role in defining the sites of preferential cleavage” (2). It is not very likely that vhs could have a different specificity from that of the chimeric protein, even though we cannot completely rule out the possibility that the large GST tag could impose oligomeric properties on its fusion protein partners, which could impact their specificities. However, vhs has been reported to aggregate with cellular or viral proteins that could define its selectivity for specific mRNAs. The list includes the cellular translation initiation factor eIF4H and the related factors eIF4B and eIF4A, and these interactions have been proposed as a mechanism for targeting vhs to the regions of translation initiation of mRNAs (1, 5, 6). Further studies to define the basis of the selectivity of vhs for mRNAs are currently in progress.
Acknowledgments
We thank R. Haselkorn for helpful discussions and Weiran Zhang for technical support.
These studies were aided by National Cancer Institute grants CA87661, CA83939, CA71933, CA78766, and CA88860.
REFERENCES
- 1.Doepker, R. C., W. Hsu, H. A. Saffran, and J. R. Smiley. 2004. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J. Virol. 78:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elgadi, M. M., and J. R. Smiley. 1999. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff. J. Virol. 73:9222-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esclatine, A., B. Taddeo, L. Evans, and B. Roizman. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. USA 101:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (vhs) protein of herpes simplex virus: genetic and biochemical evidence that vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, P., D. N. Everly, Jr., and G. S. Read. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones, F. E., C. A. Smibert, and J. R. Smiley. 1995. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J. Virol. 69:4863-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karr, B. M., and G. S. Read. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195-220. [DOI] [PubMed] [Google Scholar]
- 9.Krikorian, C. R., and G. S. Read. 1991. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65:112-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 12.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pak, A. S., D. N. Everly, K. Knight, and G. S. Read. 1995. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology 211:491-506. [DOI] [PubMed] [Google Scholar]
- 14.Smiley, J. R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taddeo, B., A. Esclatine, W. Zhang, and B. Roizman. 2003. The stress-inducible immediate-early responsive gene IEX-1 is activated in cells infected with herpes simplex virus 1, but several viral mechanisms, including 3′ degradation of its RNA, preclude expression of the gene. J. Virol. 77:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taddeo, B., W. Zhang, and B. Roizman. 2006. The UL41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA 103:2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]