Abstract
Here we present the genomic sequence of horsepox virus (HSPV) isolate MNR-76, an orthopoxvirus (OPV) isolated in 1976 from diseased Mongolian horses. The 212-kbp genome contained 7.5-kbp inverted terminal repeats and lacked extensive terminal tandem repetition. HSPV contained 236 open reading frames (ORFs) with similarity to those in other OPVs, with those in the central 100-kbp region most conserved relative to other OPVs. Phylogenetic analysis of the conserved region indicated that HSPV is closely related to sequenced isolates of vaccinia virus (VACV) and rabbitpox virus, clearly grouping together these VACV-like viruses. Fifty-four HSPV ORFs likely represented fragments of 25 orthologous OPV genes, including in the central region the only known fragmented form of an OPV ribonucleotide reductase large subunit gene. In terminal genomic regions, HSPV lacked full-length homologues of genes variably fragmented in other VACV-like viruses but was unique in fragmentation of the homologue of VACV strain Copenhagen B6R, a gene intact in other known VACV-like viruses. Notably, HSPV contained in terminal genomic regions 17 kbp of OPV-like sequence absent in known VACV-like viruses, including fragments of genes intact in other OPVs and approximately 1.4 kb of sequence present only in cowpox virus (CPXV). HSPV also contained seven full-length genes fragmented or missing in other VACV-like viruses, including intact homologues of the CPXV strain GRI-90 D2L/I4R CrmB and D13L CD30-like tumor necrosis factor receptors, D3L/I3R and C1L ankyrin repeat proteins, B19R kelch-like protein, D7L BTB/POZ domain protein, and B22R variola virus B22R-like protein. These results indicated that HSPV contains unique genomic features likely contributing to a unique virulence/host range phenotype. They also indicated that while closely related to known VACV-like viruses, HSPV contains additional, potentially ancestral sequences absent in other VACV-like viruses.
The genus Orthopoxvirus includes members of the family Poxviridae historically relevant to human health—variola virus (VARV), the etiologic agent of smallpox, and vaccinia virus (VACV), the vaccine virus used to eradicate smallpox (32). Other orthopoxviruses (OPVs), similar to VACV, are zoonotic and significant for human health, including monkeypox virus (MPXV) and cowpox virus (CPXV) (33). Still others, similar to VARV, remain restricted to specific, albeit nonhuman, hosts, including camelpox virus (CMLV) in camels and ectromelia virus (ECTV) in mice. Recent developments have heightened interest in OPV virulence and host range, including the threats of deliberate VARV reintroduction, virulence associated with preemptive smallpox vaccination and use of VACV-based recombinant vaccines, and the introduction of MPXV into the United States (16, 28, 69, 83). Isolation of OPV from infected animals and humans during limited disease outbreaks or from animals in the wild suggests that additional OPVs circulating in nature could represent an emerging disease threat (24, 25, 27, 32, 46, 49, 50, 90).
Given their importance, OPVs have been extensively studied as models of poxviral molecular biology, genomics, genetics, and virus-host interaction (19, 33, 59). Research has revealed that OPVs contain approximately 170 to 230 genes, with those in central genomic regions generally involved in poxviral intracytoplasmic replication and those in terminal genomic regions involved or potentially involved in virus-host interactions, including manipulation of host immune or cellular apoptotic responses (4, 19, 59, 60, 82, 87).
Comparative analysis of completely sequenced OPV genomes, including most known OPV species and several strains of VARV, VACV and the closely related rabbitpox virus (RPXV), MPXV, CMLV, and CPXV has begun to reveal the degree of variability within the genus Orthopoxvirus, verifying that terminal genomic regions are the most variable and thus likely to contribute to the virulence and host range characteristics of different OPVs (2, 9, 21, 22, 36, 39, 51, 52, 54, 58, 78, 80, 81). The precise roles and contributions of many variable genes and gene complements in OPV virulence and host range, however, remain to be fully characterized. It is likely that complete genomic data from uncharacterized OPV isolates will aid in OPV gene identification and functional characterization, while also providing information regarding the pathogenic potential of the virus.
Horsepox virus (HSPV) is an OPV causing horsepox, classically known as a poxviral disease of horses. Although common before the 20th century, horsepox is rare today to the point of being considered extinct (14, 44). Multiple clinical forms of horsepox have been described, including a benign, localized form involving lesions in the muzzle and buccal cavity known previously as contagious pustular stomatitis and a generalized, highly contagious form known as equine papular stomatitis (44, 94). Horsepox has also been associated with an exudative dermatitis of the pasterns described as “grease” or grease heel, a clinical syndrome also associated with other infectious and environmental agents (14, 33, 94). Horsepox is differentiated clinically from two other poxviral diseases of horses, equine molluscum contagiosum and Uasin Gishu disease. Equine molluscum contagiosum is a mild, self-limiting cutaneous disease similar to the human disease and is associated with a virus similar to molluscum contagiosum virus (88, 94). Uasin Gishu disease has been described in nonindigenous horses of eastern Africa and is associated with a poorly characterized OPV; however, generalized skin lesions are proliferative and papillomatous and the disease may be chronic in nature (33, 88, 94). HSPV is yet to be characterized molecularly, with no DNA sequence information available. Given the interest in understanding the genetic basis of viral host range and virulence and the relationships between OPVs, we have sequenced and analyzed the genome of a pathogenic field isolate of HSPV.
MATERIALS AND METHODS
Viral DNA isolation, cloning, sequencing, and sequence analysis.
The HSPV strain MNR-76 was isolated from sick horses in Bayan-somon of Khentei aimak, Mongolia, in 1976. MNR-76 causes severe disease in horses of the Mongolian breed, including pyrexia, pustular stomatitis with occasional lesions on udders and ears, and especially severe disease in foals and mares, in which death was noted (S. M. Mamadaliyev, personal communication). Viruses were passaged twice in sheep kidney cells, from which viral genomic DNA was extracted as previously described (93). Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509I endonuclease, cloned into the dephosphorylated EcoRI site of pUC19 plasmids, and grown in Escherichia coli DH10B cells (Gibco BRL, Gaithersburg, Md.). Double-stranded DNA templates were purified and sequenced from both ends with M13 forward and reverse primers using dideoxy chain terminator sequencing chemistries and the Applied Biosystems PRISM 3700 automated DNA sequencer (Applied Biosystems, Foster City, CA). Chromatogram traces were base called with Phred (30), which also produced a quality file containing a predicted probability of error at each base position. The sequences were assembled with Phrap (29) and CAP3 (43) using quality files and default settings to produce a consensus sequence with some subsequent manual editing using the Consed sequence editor (37). Gap closure was achieved by primer walking of gap-spanning clones and sequencing of PCR products. Final DNA consensus sequences represented on average sevenfold redundancy at each base position, contained no obvious polymorphisms, and demonstrated a Consed estimated error rate of less than 0.01 error per 10 kb.
Sequence analysis was conducted essentially as previously described (1). Briefly, DNA composition, structure, repeats, and restriction enzyme patterns were analyzed and open reading frame (ORF) maps created using EMBOSS (70), GCG v.10 (Accelrys, Inc., San Diego, CA), and MacVector (Accelrys, Inc) software packages. ORFs longer than 30 amino acids with a methionine start codon were evaluated for coding potential using the GLIMMER (71) computer program, and those greater than 60 amino acids were subjected to similarity searches against nonredundant protein databases and redundant viral protein databases using BLAST (8) and against viral nucleotide databases using TFASTA and TFASTX (65, 66). Here, 236 ORFs were annotated and numbered from left to right, with alphabetic subordering given to indicate multiple potential fragments of larger OPV ORFs. Given the predicted nature of all HSPV genes and gene products, ORF names were used throughout the text to indicate both the predicted gene and its putative protein product. Genomic, subgenomic, and protein alignments and comparisons were done using DIALIGN v2.2.1 (57) using anchors as generated by CHAOS (17), Multi-LAGAN (18), CLUSTAL W (89), BLAST, FASTA (64), SEAVIEW (34), and DOTTER (84) programs. Phylogenetic analyses were conducted on whole-genome sequences and genomic subregions, including a central region used previously for OPV phylogenetic analysis (positions 26800 to 170171) (22, 51), using PHYLIP (31); PHYLO_WIN (34), TREE-PUZZLE (73), and PHYML (40) programs, with evolutionary models selected using MrModeltest 2.2 (62) and additional analyses conducted on alignments in which poorly aligned regions were removed with Gblocks (20).
Nucleotide sequence accession number.
The HSPV MNR-76 genome sequence has been deposited in GenBank under accession no. DQ792504.
RESULTS AND DISCUSSION
Organization of the HSPV genome.
HSPV MNR-76 genome sequences were assembled into a contiguous sequence of 212,633 bp. The leftmost nucleotide was arbitrarily designated base 1. Similar to other OPVs, the HSPV genome contained 69% A+T nucleotide composition and a central coding region bounded by two identical inverted terminal repeat (ITR) regions.
HSPV ITRs were 7,527 bp and contained elements similar to repetitive and nonrepetitive sequences characterized in other OPVs, including a portion of the terminal hairpin loop-like sequence (positions 1 to 15 from each terminus) and nonrepetitive region 1 (NR1) (positions 21 to 101 from each terminus) and concatemer resolution (position 21 to 40 from each terminus) sequences identical to those present in VACV strain Copenhagen (CPN) (11, 36, 55). Notably, HSPV lacked extensive tandem repetition of terminally located sequences, containing only single copies of the 69-bp (positions 102 to 170, 100% identical to CPN) and 54-bp (positions 518 to 571, 96% identical to CPN) motifs repeated 8.5 to 42 times in VACV strains and RPXV (9, 10, 36, 51). Incomplete copies of 69-bp (positions 171 to 188), 54-bp (positions 572 to 601), and VACV 125-bp repeat-like (positions 494 to 517) motifs flanked complete 69-bp and 54-bp motifs, which were also separated from each other by an NR2-like sequence (positions 189 to 493, 92% identity to CPN positions 2867 to 3171). The HSPV ITR contained eight ORFs initiating and terminating in the ITR, with HSPV001/HSPV207 encompassing the 54-bp and 125-bp motif region (Table 1 ) . These data indicate that while similar to VACV in regions of the ITR, HSPV organizationally resembles other OPVs such as VARV, MPXV, and ECTV which contain fewer or single complete tandem repeat units in their termini (21, 53, 81).
TABLE 1.
HSPV ORFs in terminal genomic regions compared to best-matching ORFs annotated in VACVs, RPXV, and CPXVsa
| HSPV ORF | Position (lengthb) | VACVc
|
RPXVd
|
CPXVe
|
Putative function/similarityh | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPN
|
WR
|
Tian
|
MVA
|
mO
|
ORF | Length | GRI
|
BRI
|
||||||||||||||||||||||||||||||||
| ORF | Length | % Idf | ORF | Length | ORFg | Length | ORF | Length | ORF | Length | ORF | Length | % Idf | ORF | Length | |||||||||||||||||||||||||
| Left-terminal genomic region
|
||||||||||||||||||||||||||||||||||||||||
| HSPV001 | 688-473 (72) | 001 | 64 | |||||||||||||||||||||||||||||||||||||
| HSPV002 | 1547-804 (248) | C23L | 244 | 86 | 001 | 244 | C20L | 244 | 001L | 136 | L10L | 258 | 001 | 258 | D1L | 255 | 87 | 003 | 246 | Chemokine binding protein | ||||||||||||||||||||
| HSPV003 | 2723-1676 (349) | C22L | 122 | 91 | 002 | 61 | 002L | 176 | L09L | 34 | 63 | D2L | 351 | 95 | 005 | 355 | TNFR II-like protein, CrmB | |||||||||||||||||||||||
| 004 | 122 | L08L | 122 | 122 | ||||||||||||||||||||||||||||||||||||
| HSPV004 | 4570-2810 (587) | C21L | 113 | 100 | 005 | 48 | L07L | 48 | 113 | D3L | 586 | 95 | 006 | 619 | Ankyrin repeat protein | |||||||||||||||||||||||||
| C20L | 103 | 69 | 006 | 64 | L06L | 128 | 109 | |||||||||||||||||||||||||||||||||
| C19L | 259 | 90 | 007 | 109 | L04L | 109 | 77 | |||||||||||||||||||||||||||||||||
| 008 | 112 | 149 | ||||||||||||||||||||||||||||||||||||||
| HSPV005a | 5051-4779 (91) | L03L | 93 | D4L | 672 | 97 | 008 | 672 | Ankyrin repeat protein | |||||||||||||||||||||||||||||||
| HSPV005b | 5530-5081 (150) | C18L | 150 | 98 | L03L | 93 | 163 | D4L | 672 | 92 | 008 | 672 | ||||||||||||||||||||||||||||
| HSPV005c | 6797-5607 (397) | C17L | 386 | 92 | 003L | 102 | L02L | 416 | 385 | D4L | 672 | 94 | 008 | 672 | ||||||||||||||||||||||||||
| 004L | 233 | |||||||||||||||||||||||||||||||||||||||
| HSPV006 | 7419-6961 (153) | C16L | 181 | 97 | 189R | 188 | L01L | 147 | 002 | 184 | D5L | 153 | 97 | 009 | 153 | |||||||||||||||||||||||||
| HSPV007 | 8041-7589 (151) | D6L | 219 | 76 | 010 | 215 | ||||||||||||||||||||||||||||||||||
| HSPV008 | 9327-8509 (273) | D7L | 273 | 89 | BTB/POZ domain protein | |||||||||||||||||||||||||||||||||||
| HSPV009 | 10133-9681 (151) | D12L | 202 | 96 | 014 | 202 | Chemokine binding domain protein | |||||||||||||||||||||||||||||||||
| HSPV010 | 10529-10197 (111) | D13L | 111 | 99 | 015 | 110 | TNFR, CD30-like protein | |||||||||||||||||||||||||||||||||
| HSPV011a | 11413-10625 (263) | D14L | 764 | 95 | 016 | 764 | Ankyrin repeat protein | |||||||||||||||||||||||||||||||||
| HSPV011b | 12197-11334 (288) | D14L | 764 | 95 | 016 | 764 | ||||||||||||||||||||||||||||||||||
| HSPV011c | 12637-12191 (149) | D14L | 764 | 97 | 016 | 764 | ||||||||||||||||||||||||||||||||||
| HSPV012 | 14485-13175 (437) | C1L | 437 | 97 | 017 | 435 | Ankyrin repeat protein | |||||||||||||||||||||||||||||||||
| HSPV013 | 15127-14852 (92) | C2L | 178 | 98 | 018 | 171 | ||||||||||||||||||||||||||||||||||
| HSPV014a | 15498-15220 (93) | C3L | 833 | 95 | 019 | 796 | Ankyrin repeat protein | |||||||||||||||||||||||||||||||||
| HSPV014b | 16216-15473 (248) | C3L | 833 | 94 | 019 | 796 | ||||||||||||||||||||||||||||||||||
| HSPV014c | 16975-16367 (203) | C3L | 833 | 94 | 019 | 796 | ||||||||||||||||||||||||||||||||||
| HSPV014d | 17605-17111 (165) | C3L | 833 | 89 | 019 | 796 | ||||||||||||||||||||||||||||||||||
| HSPV015a | 17913-17695 (73) | C4L | 170 | 91 | 020 | 170 | VACV C7L-like protein | |||||||||||||||||||||||||||||||||
| HSPV015b | 18205-17960 (82) | C4L | 170 | 94 | 020 | 170 | ||||||||||||||||||||||||||||||||||
| HSPV016 | 18365-18784 (140) | C11R | 142 | 95 | 009 | 140 | C18R | 140 | 005R | 140 | 005R | 140 | 006 | 138 | C5R | 138 | 88 | 021 | 139 | Growth factor | ||||||||||||||||||||
| HSPV017 | 19934-18942 (331) | C10L | 331 | 96 | 010 | 331 | C17L | 331 | 006L | 326 | 007L | 331 | 007 | 331 | C6L | 331 | 96 | 022 | 331 | |||||||||||||||||||||
| HSPV018 | 20448-20765 (106) | 011 | 181 | C16R | 44 | 007R | 91 | 008R | 239 | 008 | 242 | C7R | 242 | 88 | 023 | 242 | ECTV p28-like host range protein | |||||||||||||||||||||||
| HSPV019 | 21707-21330 (126) | 013 | 126 | C15L | 68 | 008L | 120 | 009L | 124 | 009 | 126 | C8L | 124 | 78 | 024 | 126 | Secreted IL-18 binding protein | |||||||||||||||||||||||
| HSPV020a | 22038-21769 (90) | 014 | 237 | 009L | 90 | 010L | 90 | 409 | C9L | 668 | 96 | 025 | 668 | Ankyrin repeat host range protein | ||||||||||||||||||||||||||
| HSPV020b | 22479-22054 (142) | 014 | 237 | C14L | 142 | 010L | 142 | 011L | 142 | 409 | C9L | 668 | 95 | 025 | 668 | |||||||||||||||||||||||||
| HSPV020c | 22999-22571 (143) | 015 | 137 | C13L | 115 | 011L | 135 | 012L | 137 | 409 | C9L | 668 | 97 | 025 | 668 | |||||||||||||||||||||||||
| HSPV020d | 23508-23278 (77) | 016 | 77 | C12L | 77 | 012L | 90 | 013L | 77 | 77 | C9L | 668 | 91 | 025 | 668 | |||||||||||||||||||||||||
| HSPV020e | 23745-23533 (71) | 017 | 71 | C11L | 76 | 013L | 71 | 014L | 71 | 71 | C9L | 668 | 95 | 025 | 668 | |||||||||||||||||||||||||
| HSPV021 | 24072-23884 (63) | 018 | 60 | C10L | 59 | 59 | 59 | 010 | 59 | C10L | 62 | 95 | 026 | 68 | ||||||||||||||||||||||||||
| HSPV022 | 26149-24248 (634) | C9L | 634 | 99 | 019 | 634 | C9L | 634 | 014L | 109 | 016L | 634 | 011 | 634 | C11L | 614 | 69 | 027 | 632 | Ankyrin repeat protein | ||||||||||||||||||||
| 015L | 96 | |||||||||||||||||||||||||||||||||||||||
| 016L | 297 | |||||||||||||||||||||||||||||||||||||||
| HSPV023 | 26725-26195 (177) | C8L | 184 | 89 | 020 | 177 | C8L | 177 | 017L | 177 | 019L | 177 | 012 | 177 | C12L | 182 | 88 | 028 | 186 | |||||||||||||||||||||
| HSPV024 | 27249-26800 (150) | C7L | 150 | 97 | 021 | 150 | C7L | 150 | 018L | 150 | 020L | 150 | 013 | 150 | C13L | 150 | 97 | 029 | 150 | Host range protein | ||||||||||||||||||||
| HSPV025 | 27949-27485 (155) | C6L | 151 | 89 | 022 | 151 | C6L | 151 | 019L | 157 | 021L | 151 | 014 | 151 | C14L | 156 | 96 | 030 | 155 | |||||||||||||||||||||
| HSPV026a | 28280-28089 (64) | C5L | 204 | 98 | 023 | 204 | C5L | 204 | 022L | 204 | 015 | 204 | C15L | 205 | 98 | 031 | 69 | |||||||||||||||||||||||
| HSPV026b | 28690-28328 (121) | C5L | 204 | 98 | 023 | 204 | C5L | 204 | 022L | 204 | 015 | 204 | C15L | 205 | 96 | 032 | 125 | BTB/POZ domain protein | ||||||||||||||||||||||
| HSPV027a | 29175-28765 (137) | C4L | 316 | 97 | 024 | 316 | C4L | 255 | 023L | 316 | 016 | 316 | C16L | 316 | 97 | 033 | 316 | |||||||||||||||||||||||
| HSPV027b | 29689-29489 (67) | C4L | 316 | 98 | 024 | 316 | C4L | 255 | 023L | 316 | 016 | 316 | C16L | 315 | 93 | 033 | 316 | |||||||||||||||||||||||
| HSPV028 | 30547-29759 (263) | C3L | 263 | 96 | 025 | 263 | C3L | 263 | 025L | 263 | 017 | 263 | C17L | 259 | 93 | 034 | 263 | Complement binding protein | ||||||||||||||||||||||
| HSPV029 | 32153-30618 (512) | C2L | 512 | 97 | 026 | 512 | C2L | 512 | 026L | 512 | 018 | 512 | C18L | 512 | 97 | 035 | 512 | Kelch-like protein | ||||||||||||||||||||||
| HSPV030 | 32891-32223 (223) | C1L | 224 | 97 | 027 | 229 | C1L | 224 | 027L | 224 | 019 | 224 | C19L | 231 | 96 | 036 | 231 | |||||||||||||||||||||||
| HSPV031 | 33231-32881 (117) | N1L | 117 | 100 | 028 | 117 | NIL | 117 | 020L | 113 | 028L | 117 | 020 | 117 | Q1L | 117 | 94 | 037 | 117 | TLR/IL-1R/TNFR signaling inhibitor | ||||||||||||||||||||
| HSPV032 | 33895-33371 (175) | N2L | 175 | 97 | 029 | 175 | N2.1L* | 175 | 021L | 170 | 029L | 175 | 021 | 176 | Q2L | 175 | 98 | 038 | 177 | Alpha-amanitin-sensitive protein | ||||||||||||||||||||
| HSPV033a | 34155-33940 (72) | M1L | 472 | 94 | 030 | 472 | M1L | 472 | 030L | 472 | 022 | 472 | P1L | 474 | 93 | 039 | 473 | Ankyrin repeat protein | ||||||||||||||||||||||
| HSPV033b | 34798-34148 (217) | M1L | 472 | 97 | 030 | 472 | M1L | 472 | 030L | 472 | 022 | 472 | P1L | 474 | 97 | 039 | 473 | |||||||||||||||||||||||
| HSPV033c | 35347-34871 (159) | M1L | 472 | 96 | 030 | 472 | M1L | 472 | 030L | 472 | 022 | 472 | P1L | 474 | 93 | 039 | 473 | |||||||||||||||||||||||
| HSPV034 | 35987-35328 (220) | M2L | 220 | 99 | 031 | 220 | M2L | 196 | 031L | 220 | 023 | 220 | P2L | 163 | 99 | 040 | 220 | |||||||||||||||||||||||
| HSPV035 | 36976-36125 (284) | K1L | 284 | 97 | 032 | 284 | K1L | 189 | 022L | 98 | 032L | 284 | 024 | 284 | M1L | 284 | 95 | 041 | 284 | Ankyrin repeat host range protein | ||||||||||||||||||||
| HSPV036 | 38328-37210 (373) | K2L | 369 | 97 | 033 | 369 | K2L | 369 | 023L | 369 | 035L | 369 | 025 | 369 | M2L | 378 | 97 | 042 | 373 | Serpin SPI-3 | ||||||||||||||||||||
| HSPV037 | 38644-38381 (88) | K3L | 88 | 96 | 034 | 88 | K3L | 88 | 024L | 88 | 037L | 88 | 026 | 88 | M3L | 88 | 97 | 043 | 88 | eIF2α-like PKR inhibitor | ||||||||||||||||||||
| HSPV038 | 39979-38708 (424) | K4L | 424 | 99 | 035 | 424 | K5L* | 424 | 025L | 424 | 038L | 424 | 027 | 424 | M4L | 424 | 97 | 044 | 424 | Phospholipase D-like protein | ||||||||||||||||||||
| HSPV039 | 40845-40159 (229) | K5L | 136 | 95 | 037 | 134 | K6L | 134 | 026L | 170 | 040L | 134 | 121 | M5L | 276 | 94 | 045 | 276 | Monoglyceride lipase | |||||||||||||||||||||
| K6L | 81 | 96 | 038 | 81 | 041L | 84 | 84 | |||||||||||||||||||||||||||||||||
| HSPV040 | 40984-41430 (149) | K7R | 149 | 100 | 039 | 140 | K8R | 140 | 028R | 149 | 042R | 140 | 028 | 149 | M6R | 161 | 96 | 046 | 149 | |||||||||||||||||||||
| HSPV041 | 42262-41501 (254) | F1L | 226 | 88 | 040 | 226 | F1L | 226 | 029L | 222 | 044L | 226 | 029 | 227 | G1L | 238 | 85 | 048 | 251 | Apoptosis inhibitor | ||||||||||||||||||||
| HSPV042 | 42717-42277 (147) | F2L | 147 | 98 | 041 | 147 | F2L | 147 | 030L | 147 | 045L | 147 | 030 | 147 | G2L | 147 | 97 | 049 | 147 | dUTPase | ||||||||||||||||||||
| HSPV043 | 44183-42744 (480) | F3L | 480 | 99 | 042 | 480 | F3L | 480 | 031L | 476 | 046L | 480 | 031 | 480 | G3L | 485 | 98 | 050 | 480 | Kelch-like protein | ||||||||||||||||||||
| HSPV044 | 45153-44197 (319) | F4L | 319 | 99 | 043 | 319 | F4L | 319 | 032L | 319 | 049L | 319 | 032 | 319 | G4L | 319 | 99 | 051 | 333 | Ribonucleotide reductase small subunit | ||||||||||||||||||||
| HSPV045 | 46150-45188 (321) | F5L | 321 | 96 | 044 | 322 | F5L | 322 | 033L | 97 | 050L | 321 | 033 | 321 | G5L | 323 | 95 | 052 | 323 | |||||||||||||||||||||
| 034L | 218 | |||||||||||||||||||||||||||||||||||||||
| HSPV046 | 46404-46183 (74) | F6L | 74 | 100 | 045 | 74 | F6L | 74 | 035L | 74 | 051L | 74 | 034 | 74 | G6L | 74 | 100 | 053 | 71 | |||||||||||||||||||||
| HSPV047 | 46674-46423 (84) | F7L | 92 | 90 | 046 | 80 | F7L | 82 | 036L | 80 | 052L | 80 | 035 | 84 | G7L | 80 | 96 | 054 | 81 | |||||||||||||||||||||
| HSPV048 | 47035-46841 (65) | F8L | 65 | 93 | 047 | 65 | F8L | 50 | 037L | 65 | 053L | 65 | 036 | 65 | G8L | 65 | 98 | 055 | 65 | |||||||||||||||||||||
| HSPV049 | 47733-47098 (212) | F9L | 212 | 99 | 048 | 212 | F9L | 212 | 038L | 212 | 054L | 212 | 037 | 212 | G9L | 212 | 99 | 056 | 212 | |||||||||||||||||||||
| HSPV050 | 49039-47723 (439) | F10L | 439 | 99 | 049 | 439 | F10L | 439 | 039L | 439 | 056L | 439 | 038 | 439 | G10L | 439 | 99 | 057 | 439 | Ser/Thr protein kinase | ||||||||||||||||||||
| HSPV051 | 50126-49065 (354) | F11L | 354 | 99 | 050 | 348 | F11L | 354 | 040L | 84 | 057L | 354 | 039 | 354 | G11L | 354 | 99 | 059 | 354 | RhoA-interacting protein | ||||||||||||||||||||
| 041L | 100 | |||||||||||||||||||||||||||||||||||||||
| HSPV052 | 52091-50187 (635) | F12L | 635 | 99 | 051 | 635 | F12.1L* | 635 | 042L | 635 | 059L | 635 | 040 | 635 | G12L | 634 | 97 | 060 | 634 | IEV protein | ||||||||||||||||||||
| HSPV053 | 53243-52128 (372) | F13L | 372 | 99 | 052 | 372 | F13L | 372 | 043L | 372 | 060L | 372 | 041 | 372 | G13L | 372 | 99 | 061 | 372 | Palmitylated EEV envelope lipase | ||||||||||||||||||||
| HSPV054 | 53482-53264 (73) | F14L | 73 | 98 | 053 | 73 | F14L | 73 | 044L | 73 | 061L | 73 | 042 | 73 | G14L | 73 | 97 | 062 | 73 | |||||||||||||||||||||
| HSPV055 | 54230-53757 (158) | F15L | 158 | 98 | 054 | 147 | F15L* | 158 | 045L | 158 | 063L | 158 | 043 | 158 | G15L | 158 | 99 | 064 | 158 | |||||||||||||||||||||
| HSPV056 | 54932-54240 (231) | F16L | 231 | 98 | 055 | 231 | F16L | 231 | 046L | 231 | 064L | 231 | 044 | 231 | G16L | 231 | 97 | 065 | 231 | |||||||||||||||||||||
| HSPV057 | 54995-55297 (101) | F17R | 101 | 98 | 056 | 101 | F17R | 101 | 047R | 101 | 065R | 101 | 045 | 101 | G17R | 101 | 99 | 066 | 101 | DNA binding virion core protein | ||||||||||||||||||||
| HSPV058 | 56736-55300 (479) | E1L | 479 | 99 | 057 | 479 | E1L | 479 | 048L | 479 | 067L | 479 | 046 | 479 | F1L | 479 | 98 | 067 | 479 | Poly(A) polymerase large subunit | ||||||||||||||||||||
| HSPV059 | 58946-56736 (737) | E2L | 737 | 99 | 058 | 737 | E2L | 737 | 049L | 737 | 068L | 737 | 047 | 737 | F2L | 737 | 97 | 068 | 737 | |||||||||||||||||||||
| HSPV060 | 59619-59050 (190) | E3L | 190 | 97 | 059 | 190 | E3L* | 190 | 050L | 190 | 069L | 190 | 048 | 190 | F3L | 190 | 97 | 069 | 190 | dsRNA binding PKR inhibitor | ||||||||||||||||||||
| HSPV061 | 60453-59677 (259) | E4L | 259 | 99 | 060 | 259 | E5L | 259 | 051L | 259 | 070L | 259 | 049 | 259 | F4L | 259 | 98 | 070 | 261 | RNA polymerase subunit RPO30 | ||||||||||||||||||||
| HSPV062 | 60530-61522 (331) | E5R | 331 | 97 | 061 | 341 | E6R | 257 | 052R | 331 | 071R | 341 | 050 | 341 | F5R | 331 | 94 | 071 | 319 | |||||||||||||||||||||
| Right-terminal genomic region
|
||||||||||||||||||||||||||||||||||||||||
| HSPV146a | 146239-146030 (70) | A25L | 65 | 91 | 145 | 65 | A27L | 233 | 136L | 65 | 185L | 210 | 233 | A26L | 1,279 | 67 | 158 | 1,284 | ATI protein | |||||||||||||||||||||
| HSPV146b | 146819-146211 (203) | A26L | 322 | 93 | 146 | 154 | A27L | 233 | 185L | 210 | 233 | A26L | 1,279 | 94 | 158 | 1,284 | ||||||||||||||||||||||||
| HSPV146c | 147691-147167 (175) | 147 | 227 | A29L | 230 | 187L | 227 | 227 | A26L | 1,279 | 94 | 158 | 1,284 | |||||||||||||||||||||||||||
| HSPV146d | 149828-147654 (725) | 148 | 725 | A31L | 725 | 189L | 725 | 725 | A26L | 1,279 | 97 | 158 | 1,284 | |||||||||||||||||||||||||||
| HSPV147 | 151378-149876 (501) | A26L | 322 | 98 | 149 | 500 | A33L | 168 | 137L | 230 | 191L | 502 | 134 | 500 | A27L | 518 | 94 | 159 | 192 | IMV ATI-like protein P4c | ||||||||||||||||||||
| A34L | 71 | 161 | 260 | |||||||||||||||||||||||||||||||||||||
| A35L | 229 | |||||||||||||||||||||||||||||||||||||||
| HSPV148 | 151761-151432 (110) | A27L | 110 | 99 | 150 | 110 | A36L | 110 | 138L | 110 | 192L | 110 | 135 | 110 | A28L | 110 | 99 | 162 | 110 | IMV membrane protein | ||||||||||||||||||||
| HSPV149 | 152202-151765 (146) | A28L | 146 | 99 | 151 | 146 | A37L* | 146 | 139L | 146 | 193L | 146 | 136 | 146 | A29L | 146 | 99 | 163 | 146 | IMV membrane protein | ||||||||||||||||||||
| HSPV150 | 153120-152206 (305) | A29L | 305 | 99 | 152 | 305 | A38L | 305 | 140L | 305 | 195L | 305 | 137 | 305 | A30L | 305 | 98 | 164 | 305 | RNA polymerase subunit RPO35 | ||||||||||||||||||||
| HSPV151 | 153316-153086 (77) | A30L | 77 | 100 | 153 | 77 | A39L | 77 | 141L | 77 | 196L | 77 | 138 | 77 | A31L | 77 | 100 | 165 | 76 | Virion core protein | ||||||||||||||||||||
| HSPV152 | 153476-153874 (133) | A31R | 124 | 90 | 154 | 124 | A40R | 141 | 142R | 125 | 197R | 127 | 139 | 124 | A32R | 145 | 90 | 166 | 140 | |||||||||||||||||||||
| HSPV153 | 154656-153847 (270) | A32L | 300 | 99 | 155 | 270 | A41L | 327 | 143L | 269 | 199L | 270 | 140 | 300 | A33L | 300 | 99 | 167 | 311 | ATPase, DNA packaging | ||||||||||||||||||||
| HSPV154 | 154774-155328 (185) | A33R | 185 | 100 | 156 | 185 | A42R* | 185 | 144R | 185 | 200R | 185 | 141 | 185 | A34R | 185 | 98 | 168 | 187 | EEV envelope protein | ||||||||||||||||||||
| HSPV155 | 155355-155858 (168) | A34R | 168 | 99 | 157 | 168 | A44R | 168 | 145R | 168 | 201R | 168 | 142 | 168 | A35R | 168 | 97 | 169 | 168 | EEV envelope protein | ||||||||||||||||||||
| HSPV156 | 155905-156432 (176) | A35R | 176 | 98 | 158 | 176 | A45R | 176 | 146R | 176 | 202R | 176 | 143 | 177 | A36R | 176 | 98 | 171 | 176 | |||||||||||||||||||||
| HSPV157 | 156502-157170 (223) | A36R | 221 | 99 | 159 | 221 | A46R | 221 | 147R | 208 | 204R | 221 | 144 | 224 | A37R | 223 | 98 | 172 | 224 | IEV protein | ||||||||||||||||||||
| HSPV158 | 157237-158025 (263) | A37R | 263 | 99 | 160 | 263 | A47R | 268 | 148R | 263 | 205R | 263 | 145 | 263 | A38R | 268 | 95 | 173 | 263 | |||||||||||||||||||||
| HSPV159 | 158139-158324 (62) | 64 | 161 | 62 | 64 | 57 | 207R | 62 | 56 | A39R | 64 | 95 | 174 | 63 | ||||||||||||||||||||||||||
| HSPV160 | 159157-158327 (277) | A38L | 277 | 95 | 162 | 277 | Unknown | 277 | 149L | 277 | 208L | 277 | 146 | 277 | A40L | 277 | 97 | 175 | 277 | CD47-like membrane glycoprotein | ||||||||||||||||||||
| HSPV161 | 159173-160150 (326) | A39R | 403 | 93 | 163 | 295 | A49R | 228 | 150R | 83 | 209R | 403 | 147 | 401 | A41R | 402 | 94 | 176 | 409 | Semaphorin-like protein | ||||||||||||||||||||
| 164 | 142 | A50R | 142 | 151R | 210 | |||||||||||||||||||||||||||||||||||
| HSPV162 | 160402-160896 (165) | A40R | 168 | 97 | 165 | 159 | A51R | 159 | 152R | 168 | 211R | 159 | 148 | 159 | A42R | 166 | 95 | 177 | 160 | C-type lectin-like membrane protein | ||||||||||||||||||||
| HSPV163 | 161656-161000 (219) | A41L | 219 | 97 | 166 | 219 | A52L | 219 | 153L | 219 | 212L | 219 | 149 | 219 | A43L | 219 | 97 | 178 | 218 | Secreted immunomodulatory protein | ||||||||||||||||||||
| HSPV164 | 161827-162225 (133) | A42R | 133 | 97 | 167 | 133 | A53R | 133 | 154R | 128 | 213R | 133 | 150 | 133 | A44R | 133 | 100 | 179 | 133 | Profilin-like protein | ||||||||||||||||||||
| HSPV165 | 162266-162850 (195) | A43R | 194 | 93 | 168 | 194 | A54R | 194 | 155R | 190 | 214R | 194 | 151 | 194 | A45R | 196 | 95 | 180 | 194 | |||||||||||||||||||||
| HSPV166 | 164226-163189 (346) | A44L | 346 | 98 | 170 | 346 | A55L | 346 | 157L | 346 | 216L | 346 | 153 | 346 | A47L | 346 | 98 | 182 | 346 | Hydroxysteroid dehydrogenase | ||||||||||||||||||||
| HSPV167 | 164273-164647 (125) | A45R | 125 | 99 | 171 | 125 | A56R | 125 | 158R | 121 | 217R | 125 | 154 | 125 | A48R | 125 | 98 | 183 | 125 | Superoxide dismutase-like protein | ||||||||||||||||||||
| HSPV168 | 164640-165359 (240) | A46R | 214 | 100 | 172 | 240 | A57R | 210 | 159R | 240 | 218R | 240 | 155 | 240 | A49R | 240 | 97 | 184 | 242 | TLR/IL-1R signaling inhibitor | ||||||||||||||||||||
| HSPV169 | 166183-165452 (244) | A47L | 244 | 99 | 173 | 252 | A58L | 252 | 160L | 238 | 220L | 252 | 156 | 244 | A50L | 244 | 97 | 185 | 244 | |||||||||||||||||||||
| HSPV170 | 166282-166893 (204) | A48R | 204 | 100 | 174 | 227 | A59R | 204 | 161R | 204 | 221R | 204 | 157 | 204 | A51R | 227 | 99 | 186 | 227 | Thymidylate kinase | ||||||||||||||||||||
| HSPV171 | 166944-167429 (162) | A49R | 162 | 96 | 175 | 162 | A60R | 162 | 162R | 162 | 222R | 162 | 158 | 162 | A52R | 162 | 95 | 187 | 162 | |||||||||||||||||||||
| HSPV172 | 167464-169119 (552) | A50R | 552 | 98 | 176 | 552 | A61R | 552 | 163R | 552 | 223R | 552 | 159 | 552 | A53R | 552 | 97 | 188 | 554 | DNA ligase | ||||||||||||||||||||
| HSPV173a | 169175-169414 (80) | A51R | 334 | 88 | 177 | 334 | A62R* | 334 | 164R | 310 | 226R | 73 | 84 | A54R | 334 | 92 | 189 | 334 | ||||||||||||||||||||||
| HSPV173b | 169359-170168 (270) | A51R | 334 | 96 | 177 | 334 | A62R | 334 | 164R | 310 | 227R | 266 | 126 | A54R | 334 | 96 | 189 | 334 | ||||||||||||||||||||||
| 120 | ||||||||||||||||||||||||||||||||||||||||
| HSPV174 | 170241-170810 (190) | A52R | 190 | 98 | 178 | 190 | A63R* | 190 | 228R | 190 | 160 | 190 | A55R | 190 | 96 | 190 | 190 | TLR/IL-1R signaling inhibitor | ||||||||||||||||||||||
| HSPV175 | 171134-171439 (102) | A53R | 108 | 91 | 179 | 103 | * | 137 | 229R | 186 | 161 | 102 | A56R | 186 | 88 | 191 | 186 | TNFR, CrmC | ||||||||||||||||||||||
| AORFT | 81 | 100 | * | 103 | ||||||||||||||||||||||||||||||||||||
| HSPV176 | 171969-173660 (564) | A55R | 564 | 99 | 180 | 564 | A65R* | 564 | 232R | 564 | 162 | 564 | A57R | 564 | 97 | 193 | 563 | Kelch-like protein | ||||||||||||||||||||||
| HSPV177 | 173713-174654 (314) | A56R | 315 | 98 | 181 | 314 | A66R | 315 | 165R | 315 | 233R | 310 | 163 | 206 | A58R | 314 | 94 | 194 | 297 | EEV hemagglutinin | ||||||||||||||||||||
| HSPV178a | 174675-174860 (62) | A57R | 151 | 93 | 182 | 151 | A67R | 151 | 234R | 37 | 164 | 151 | A59R | 197 | 96 | 195 | 197 | Guanylate kinase | ||||||||||||||||||||||
| HSPV178b | 174975-175265 (97) | A57R | 151 | 100 | 182 | 151 | A67R | 151 | 166R | 97 | 235R | 151 | 164 | 151 | A59R | 197 | 95 | 195 | 197 | |||||||||||||||||||||
| HSPV179 | 175419-176318 (300) | B1R | 300 | 99 | 183 | 300 | B1R | 300 | 167R | 300 | 236R | 300 | 165 | 300 | B1R | 300 | 99 | 196 | 299 | Ser/Thr protein kinase, DNA replication | ||||||||||||||||||||
| HSPV180a | 176412-177068 (219) | B2R | 219 | 96 | 184 | 219 | B2R | 219 | 168R | 96 | 238R | 219 | 219 | B2R | 503 | 93 | 197 | 505 | ||||||||||||||||||||||
| 169R | 143 | |||||||||||||||||||||||||||||||||||||||
| HSPV180b | 177107-177478 (124) | B3R | 124 | 96 | 185 | 167 | B3R | 124 | 170R | 179 | 240R | 124 | 124 | B2R | 503 | 91 | 197 | 505 | Schlafen-like protein | |||||||||||||||||||||
| HSPV181 | 178138-179811 (558) | B4R | 558 | 98 | 186 | 558 | B4R | 558 | 171R | 177 | 242R | 558 | 166 | 558 | B3R | 558 | 95 | 198 | 558 | Ankyrin repeat protein | ||||||||||||||||||||
| 172R | 409 | |||||||||||||||||||||||||||||||||||||||
| HSPV182 | 179917-180867 (317) | B5R | 317 | 97 | 187 | 317 | B5R | 317 | 173R | 317 | 243R | 317 | 167 | 317 | B4R | 317 | 96 | 199 | 317 | EEV host range protein | ||||||||||||||||||||
| HSPV183a | 180953-181285 (111) | B6R | 173 | 99 | 188 | 173 | B6R | 173 | 174R | 173 | 244R | 173 | 168 | 173 | B5R | 183 | 93 | 200 | 179 | |||||||||||||||||||||
| HSPV183b | 181288-181482 (65) | B6R | 173 | 100 | 188 | 173 | B6R | 173 | 174R | 173 | 244R | 173 | 168 | 173 | B5R | 183 | 93 | 200 | 179 | |||||||||||||||||||||
| HSPV184 | 181523-182068 (182) | B7R | 182 | 100 | 189 | 182 | B7R | 182 | 175R | 177 | 246R | 182 | 169 | 182 | B6R | 182 | 97 | 201 | 181 | Chemokine binding domain | ||||||||||||||||||||
| HSPV185 | 182149-182964 (272) | B8R | 272 | 97 | 190 | 272 | B8R | 272 | 176R | 226 | 247R | 272 | 170 | 272 | B7R | 271 | 97 | 202 | 266 | IFN-γ receptor | ||||||||||||||||||||
| HSPV186 | 183054-183284 (77) | B9R | 77 | 100 | 191 | 77 | B9R | 77 | 177R | 72 | 248R | 77 | 61 | B8R | 221 | 97 | 203 | 225 | MYXV M-T4-like protein | |||||||||||||||||||||
| HSPV187 | 183384-183746 (121) | B10R | 166 | 99 | 192 | 166 | B10R | 166 | 178R | 158 | 249R | 166 | 172 | 166 | B9R | 501 | 94 | 204 | 501 | Kelch-like protein | ||||||||||||||||||||
| HSPV188 | 183821-184054 (78) | B11R | 88 | 96 | 193 | 72 | B11R | 76 | 179R | 74 | 250R | 72 | 173 | 72 | B10R | 105 | 96 | 205 | 90 | |||||||||||||||||||||
| HSPV189 | 184124-184972 (283) | B12R | 283 | 98 | 194 | 283 | B12R | 283 | 180R | 283 | 251R | 283 | 174 | 283 | B11R | 283 | 98 | 206 | 285 | Ser/Thr protein kinase | ||||||||||||||||||||
| HSPV190 | 185074-186108 (345) | B14R | 222 | 97 | 195 | 345 | B14R | 222 | 181R | 116 | 253R | 222 | 175 | 345 | B12R | 345 | 93 | 207 | 341 | Serpin, SPI-2 | ||||||||||||||||||||
| 182R | 222 | |||||||||||||||||||||||||||||||||||||||
| HSPV191 | 186186-186632 (149) | B15R | 149 | 97 | 196 | 149 | B15R | 149 | 183R | 143 | 254R | 149 | 176 | 149 | B13R | 149 | 98 | 208 | 149 | |||||||||||||||||||||
| HSPV192 | 186736-187713 (326) | B16R | 290 | 97 | 197 | 326 | B16R | 290 | 184R | 326 | 255R | 326 | 207 | B14R | 326 | 95 | 209 | 326 | IL-1 receptor | |||||||||||||||||||||
| 134 | ||||||||||||||||||||||||||||||||||||||||
| HSPV193 | 188784-187765 (340) | B17L | 340 | 97 | 198 | 340 | B17L | 340 | 185L | 340 | 257L | 340 | 177 | 340 | B15L | 340 | 96 | 210 | 340 | |||||||||||||||||||||
| HSPV194 | 188921-190642 (574) | B18R | 574 | 98 | 199 | 574 | B18R | 574 | 186R | 574 | 258R | 413 | 178 | 574 | B16R | 574 | 95 | 211 | 574 | Ankyrin repeat protein | ||||||||||||||||||||
| HSPV195 | 190711-191775 (355) | B19R | 353 | 98 | 200 | 351 | B19R | 353 | 187R | 234 | 179 | 351 | B17R | 351 | 91 | 212 | 366 | IFN-α/β binding protein | ||||||||||||||||||||||
| HSPV196 | 191850-194222 (791) | B20R | 127 | 96 | 202 | 53 | B20R* | 613 | 180 | 791 | B18R | 795 | 94 | 213 | 800 | Ankyrin repeat protein | ||||||||||||||||||||||||
| 203 | 309 | |||||||||||||||||||||||||||||||||||||||
| HSPV197 | 194331-195971 (547) | 204 | 134 | B19R | 557 | 94 | 215 | 557 | Kelch-like protein | |||||||||||||||||||||||||||||||
| HSPV198 | 196272-197342 (357) | C12L | 353 | 97 | 205 | 353 | C19L | 353 | 004L | 353 | 005 | 357 | B20R | 375 | 95 | 217 | 372 | Serpin, SPI-1 | ||||||||||||||||||||||
| HSPV199 | 197515-198090 (192) | C13L | 65 | 90 | 206 | 190 | 003L | 190 | 004 | 192 | B21R | 190 | 94 | 218 | 198 | Chemokine binding domain protein | ||||||||||||||||||||||||
| C14L | 82 | 98 | ||||||||||||||||||||||||||||||||||||||
| HSPV200 | 198347-204106 (1,920) | 188R | 70 | B22R | 1,033 | 97 | 219 | 1,019 | VARV B22R-like protein | |||||||||||||||||||||||||||||||
| HSPV201a | 204447-204806 (120) | B21R | 91 | 96 | 002L | 89 | 003 | 91 | K1R | 581 | 96 | 220 | 579 | Ankyrin repeat protein | ||||||||||||||||||||||||||
| HSPV201b | 204961-205161 (67) | 189R | 188 | K1R | 581 | 90 | 220 | 579 | ||||||||||||||||||||||||||||||||
| HSPV202 | 205215-205673 (153) | B22R | 181 | 97 | 189R | 188 | L01L | 147 | 002 | 184 | I1R | 153 | 97 | 222 | 153 | |||||||||||||||||||||||||
| HSPV203a | 205837-207027 (397) | B23R | 386 | 92 | 190R | 233 | L02L | 416 | 385 | I2R | 672 | 94 | 223 | 672 | Ankyrin repeat protein | |||||||||||||||||||||||||
| 191R | 102 | |||||||||||||||||||||||||||||||||||||||
| HSPV203b | 207104-207553 (150) | B24R | 150 | 98 | L03L | 98 | 163 | I2R | 672 | 92 | 223 | 672 | ||||||||||||||||||||||||||||
| HSPV203c | 207583-207855 (91) | L03L | 93 | I2R | 672 | 97 | 223 | 672 | ||||||||||||||||||||||||||||||||
| HSPV204 | 208064-209824 (587) | B25R | 259 | 90 | 211 | 112 | L04L | 100 | 140 | I3R | 586 | 95 | 225 | 619 | Ankyrin repeat protein | |||||||||||||||||||||||||
| B26R | 103 | 69 | 212 | 109 | L06L | 128 | 77 | |||||||||||||||||||||||||||||||||
| B27R | 113 | 100 | 213 | 64 | L07L | 48 | 109 | |||||||||||||||||||||||||||||||||
| 214 | 48 | 113 | ||||||||||||||||||||||||||||||||||||||
| HSPV205 | 209912-210958 (349) | B28R | 122 | 91 | 215 | 122 | 192R | 176 | L08L | 122 | 122 | I4R | 351 | 95 | 226 | 355 | TNFR II-like protein, CrmB | |||||||||||||||||||||||
| 217 | 61 | L09L | 34 | 63 | ||||||||||||||||||||||||||||||||||||
| HSPV206 | 211087-211830 (248) | B29R | 244 | 86 | 218 | 244 | B23R | 244 | 193R | 136 | L10L | 258 | 001 | 258 | I5R | 255 | 87 | 227 | 246 | Chemokine binding protein | ||||||||||||||||||||
| HSPV207 | 211946-212161 (72) | 229 | 64 | |||||||||||||||||||||||||||||||||||||
Boldface indicates ORFs >10% different in length from intact orthologues from CPXV GRI-90 or BRI. Names of ORF homologues have been abbreviated here for simplicity and lack the following prefixes for the following viruses: VACV WR, WR; T, Tian Tan; MVA, MVA; m0LTR, ORFs in the m0 long terminal repeat indicated here with prefix L; m0, unique m0 ORFs; RPXV, RPXV; CPXV, BRI.
All lengths are in amino acids.
VACV strains (accession numbers): CPN (M35027); WR (AY243312); Tian, Tian Tan (AF095689); MVA (U94848); m0, LC16m0 (AY678277). Larger ORFs matching multiple HSPV ORFs are VACV CPN ORFs C5L, C4L, M1L, A26L, A51R, A57R, and B6R; VACV WR ORFs 014, 023, 024, 030, 177, 182, and 188; VACV Tian Tan ORFs C5L, C4L, M1L, A27L, A62R, A67R, and B6R; VACV MVA ORFs 164R, 174R, and 189R; and VACV m0 ORFs L03L, 022L, 023L, 030L, 185L, and 244R.
RPXV strain Utrecht. RPXV ORF lengths lacking an ORF designation indicate ORFs lacking translation products annotated in sequence AY484669. Larger ORFs matching multiple HSPV ORFs are the 409-amino-acid-long ORF, ORF 015, ORF 016, ORF 022, the 233-amino-acid-long ORF, ORF 164, and ORF 168.
CPXV strains (accession numbers): GRI, GRI-90 (X94355); BRI (AF482758). Larger ORFs matching multiple HSPV ORFs are GRI-90 ORFs D4L, D14L, C3L, C4L, C9L, C15L, C16L, P1L, A26L, A54R, A59R, B2R, B5R, K1R, and 12R and BRI ORFs 008, 016, 019, 020, 025, 033, 039, 158, 189, 195, 197, 200, 220, and 223.
% Id, percent amino acid identity in local BLAST match.
Asterisks indicate ORFs resequenced/reannotated by Upton et al. (92) as present, intact, or fused to a subsequent ORF in the Tian Tan genome.
Abbreviations: IL-18, interleukin-18; TLR, Toll-like receptor; PKR, double-stranded RNA-dependent protein kinase; IEV, intracellular enveloped virion; IFN-γ, gamma interferon; MYXV, myxoma virus; dsRNA, double-stranded RNA.
TABLE 2.
HSPV ORFs in central genomic regions compared to orthologues annotated in VACV CPNaa
| HSPV ORF | Position (lengthc) | VACV CPN
|
Putative function/similarity | |
|---|---|---|---|---|
| ORF | Length | |||
| HSPV063 | 61662-63362 (567) | E6R | 567 | |
| HSPV064 | 63447-63944 (166) | E7R | 166 | |
| HSPV065 | 64072-64890 (273) | E8R | 273 | Virion core protein |
| HSPV066 | 67919-64902 (1,006) | E9L | 1,006 | DNA polymerase |
| HSPV067 | 67951-68235 (95) | E10R | 96 | IMV redox protein |
| HSPV068 | 68622-68236 (129) | E11L | 129 | Virion core protein |
| HSPV069 | 70609-68612 (666) | O1L | 666 | |
| HSPV070 | 70983-70660 (108) | O2L | 108 | Glutaredoxin |
| HSPV071 | 72067-71132 (312) | I1L | 312 | DNA binding virion core protein |
| HSPV072 | 72304-72077 (76) | I2L | 73 | |
| HSPV073 | 73114-72308 (269) | I3L | 269 | DNA binding phosphoprotein |
| HSPV074a | 73439-73200 (80) | I4Lb | 771 | Ribonucleotide reductase large subunit |
| HSPV074b | 74885-73566 (440) | I4L | 771 | |
| HSPV074c | 75213-74842 (124) | I4L | 771 | |
| HSPV074d | 75503-75216 (96) | I4L | 771 | |
| HSPV075 | 75770-75534 (79) | I5L | 79 | IMV membrane protein |
| HSPV076 | 76937-75792 (382) | I6L | 382 | Telomere binding protein |
| HSPV077 | 78201-76933 (423) | I7L | 423 | Virion core proteinase |
| HSPV078 | 78207-80234 (676) | I8R | 676 | RNA helicase NPH-II |
| HSPV079 | 82016-80244 (591) | G1L | 591 | Metalloprotease |
| HSPV080 | 82342-83001 (220) | G3L | 220 | |
| HSPV081 | 82348-82016 (111) | G2R | 111 | Transcriptional elongation factor |
| HSPV082 | 83348-82977 (124) | G4L | 124 | Glutaredoxin 2 |
| HSPV083 | 83351-84652 (434) | G5R | 434 | Virion core protein |
| HSPV084 | 84663-84851 (63) | G5.5R | 63 | RNA polymerase subunit RPO7 |
| HSPV085 | 84856-85350 (165) | G6R | 166 | |
| HSPV086 | 86433-85321 (371) | G7L | 371 | Virion core protein |
| HSPV087 | 86464-87243 (260) | G8R | 260 | Late transcription factor VLTF-1 |
| HSPV088 | 87266-88285 (340) | G9R | 340 | Myristylated protein |
| HSPV089 | 88289-89038 (250) | L1R | 250 | Myristylated IMV envelope protein |
| HSPV090 | 89073-89333 (87) | L2R | 87 | |
| HSPV091 | 90378-89329 (350) | L3L | 350 | |
| HSPV092 | 90403-91155 (251) | L4R | 251 | DNA binding virion core protein |
| HSPV093 | 91168-91551 (128) | L5R | 128 | IMV membrane protein |
| HSPV094 | 91511-91969 (153) | J1R | 153 | IMV membrane protein |
| HSPV095 | 91988-92518 (177) | J2R | 177 | Thymidine kinase |
| HSPV096 | 92587-93585 (333) | J3R | 333 | Poly(A) polymerase small subunit |
| HSPV097 | 93503-94057 (185) | J4R | 185 | RNA polymerase subunit RPO22 |
| HSPV098 | 94585-94187 (133) | J5L | 133 | |
| HSPV099 | 94692-98549 (1,286) | J6R | 1,286 | RNA polymerase subunit RPO147 |
| HSPV100 | 99064-98552 (171) | H1L | 171 | Tyr/Ser protein phosphatase |
| HSPV101 | 99078-99644 (189) | H2R | 189 | IMV membrane protein |
| HSPV102 | 100624-99653 (324) | H3L | 324 | IMV envelope protein |
| HSPV103 | 103012-100628 (795) | H4L | 795 | RNA polymerase-associated protein |
| HSPV104 | 103198-103830 (211) | H5R | 203 | Late transcription factor VLTF-4 |
| HSPV105 | 103834-104775 (314) | H6R | 314 | DNA topoisomerase IB |
| HSPV106 | 104815-105252 (146) | H7R | 146 | |
| HSPV107 | 105299-107830 (844) | D1R | 844 | mRNA capping enzyme large subunit |
| HSPV108 | 108225-108935 (237) | D3R | 237 | Virion core protein |
| HSPV109 | 108232-107795 (146) | D2L | 146 | Virion core protein |
| HSPV110 | 108938-109591 (218) | D4R | 218 | Uracil DNA glycosylase |
| HSPV111 | 109626-111980 (785) | D5R | 785 | NTPase, DNA replication |
| HSPV112 | 112024-113934 (637) | D6R | 637 | Early transcription factor small subunit |
| HSPV113 | 113964-114446 (161) | D7R | 161 | RNA polymerase subunit RPO18 |
| HSPV114 | 115326-114415 (304) | D8L | 304 | IMV membrane protein, cell binding |
| HSPV115 | 115368-116006 (213) | D9R | 213 | MutT motif |
| HSPV116 | 116006-116749 (248) | D10R | 248 | MutT motif |
| HSPV117 | 118648-116756 (631) | D11L | 631 | NPH-I, transcription termination factor |
| HSPV118 | 119546-118686 (287) | D12L | 287 | mRNA capping enzyme small subunit |
| HSPV119 | 121232-119580 (551) | D13L | 551 | Rifampin resistance protein |
| HSPV120 | 121708-121259 (150) | A1L | 150 | Late transcription factor VLTF-2 |
| HSPV121 | 122403-121732 (224) | A2L | 224 | Late transcription factor VLTF-3 |
| HSPV122 | 122630-122403 (76) | A2.5L | 76 | Virion redox protein |
| HSPV123 | 124579-122648 (644) | A3L | 644 | Virion core protein P4b |
| HSPV124 | 125477-124635 (281) | A4L | 281 | Virion core protein |
| HSPV125 | 125515-126006 (164) | A5R | 164 | RNA polymerase subunit RPO19 |
| HSPV126 | 127124-126009 (372) | A6L | 372 | |
| HSPV127 | 129280-127151 (710) | A7L | 710 | Early transcription factor large subunit |
| HSPV128 | 129334-130197 (288) | A8R | 288 | Intermediate transcription factor VITF-3 |
| HSPV129 | 130501-130196 (102) | A9L | 99 | IMV membrane protein |
| HSPV130 | 133177-130505 (891) | A10L | 891 | Virion core protein P4a |
| HSPV131 | 133192-134145 (318) | A11R | 318 | Nonstructural protein |
| HSPV132 | 134725-134153 (191) | A12L | 192 | Virion core protein |
| HSPV133 | 134961-134752 (70) | A13L | 70 | IMV membrane protein |
| HSPV134 | 135341-135072 (90) | A14L | 90 | IMV membrane protein |
| HSPV135 | 135519-135361 (53) | A14.5L | 53 | IMV membrane protein |
| HSPV136 | 135793-135512 (94) | A15L | 94 | Virion core protein |
| HSPV137 | 136913-135780 (378) | A16L | 378 | Myristylated IMV membrane protein |
| HSPV138 | 137527-136919 (203) | A17L | 203 | Phosphorylated IMV membrane protein |
| HSPV139 | 137542-139020 (493) | A18R | 493 | DNA helicase, transcriptional elongation |
| HSPV140 | 139237-139007 (77) | A19L | 77 | |
| HSPV141 | 139590-140867 (426) | A21L | 426 | DNA polymerase processivity factor |
| HSPV142 | 139591-139241 (117) | A20R | 117 | IMV membrane protein |
| HSPV143 | 140833-141360 (176) | A22R | 176 | Holliday junction resolvase |
| HSPV144 | 141383-142528 (382) | A23R | 382 | Intermediate transcription factor VITF-3 |
| HSPV145 | 142528-146019 (1,164) | A24R | 1,164 | RNA polymerase subunit RPO132 |
Boldface indicates ORFs >10% different in length from intact orthologues from CPXV GRI-90 or Brighton Red.
I4L is a larger ORF matching multiple HSPV ORFs.
Lengths are in amino acids.
HSPV contained 236 ORFs potentially encoding proteins of 53 to 1,920 amino acids and sharing similarity with those in previously described OPV genomes (Tables 1 and 2). Of these 236 annotated ORFs, 54 were significantly smaller or fragmented forms of 25 larger ORFs present in other OPVs, leaving 182 potentially full-length OPV gene homologues. The HSPV central genomic region contained genes colinear and highly conserved among other OPV genomes, with ORFs HSPV041 to HSPV145 sharing an average 98% amino acid identity with VACV CPN ORFs F1L to A24R and with CPXV GRI-90 ORFs G1L to A25R (Table 2 and data not shown). Genes in this conserved region included those involved in basic replicative functions such as viral transcription and transcript modification, DNA replication, and assembly of intracellular mature and extracellular enveloped virions (IMVs and EEVs, respectively), indicating that HSPV is similar to other OPVs in these functions (59) (Table 2).
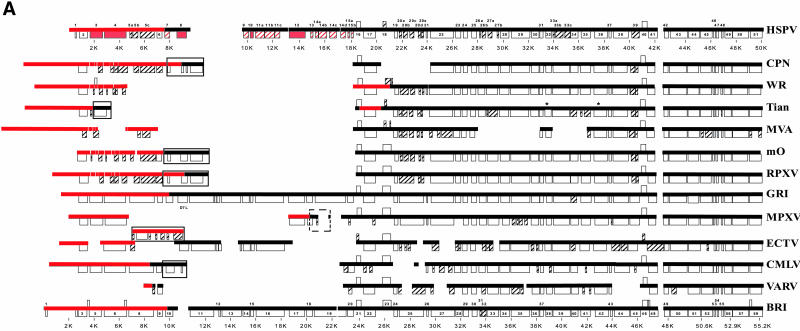
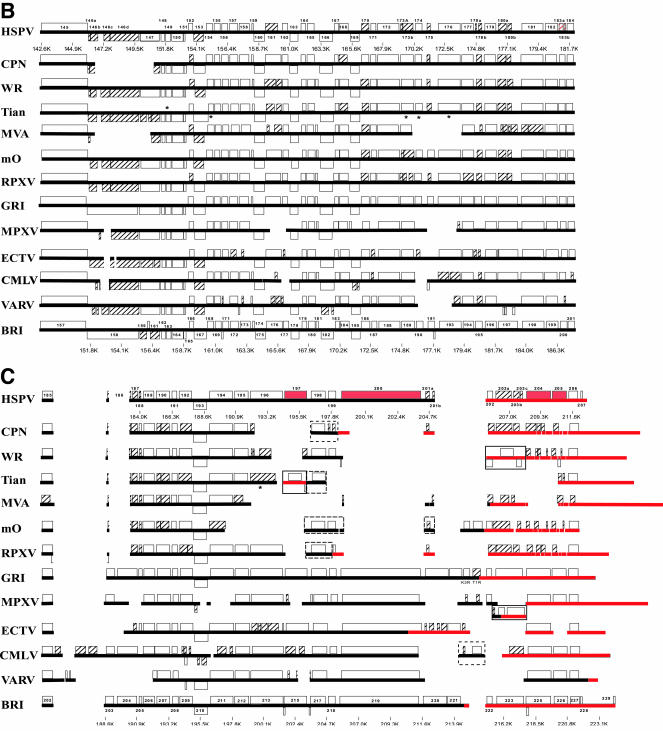
HSPV terminal genomic regions were similar to other OPVs in that they contained a homologous subset of the sequence and intact ORFs present in various strains of CPXV, viruses found to contain a relatively complete OPV genotype and thus thought to be viruses from which other OPV lineages are derived following gene fragmentation and loss (Table 1; Fig. 1) (75, 79). Many of these ORFs have been characterized in other OPVs as affecting viral virulence, host range, and modification of host responses, including apoptosis and innate and adaptive immune mechanisms (59, 60, 82). However, the specific subset of genes present in HSPV was unique relative to other OPVs, containing terminal genomic sequences not characteristic of currently known OPVs and including approximately 1.4 kb of sequence found only in CPXV (located between positions 15453 and 16985) (Fig. 1).
FIG. 1.
Schematic comparison of HSPV left (A) and right (B and C) terminal genomic regions to those of other orthopoxviruses. Virus names were abbreviated as follows and correspond to sequences from the following GenBank accession numbers in parentheses: VACV CPN (M35027); VACV WR (AY243312); Tian, VACV Tian Tan (AF095689); VACV MVA (U94848); m0, VACV Lister isolate LC16m0 (AY678277); RPXV, RPXV Utrecht (AY484669); GRI, CPXV GRI-90 (X94355); MPXV, MPXV Zaire-96-I-16 (AF380138); ECTV, ECTV Moscow (AF012825); CMLV, CMLV M-96 (AF438165); VARV, VARV Bangladesh-1975 (L22579); CPXV BRI (AF482758). Heavy lines indicate nucleotide sequences; boxes indicate ORFs matching those annotated in HSPV and those in genomic regions absent in HSPV. ORF names and genomic positions in kilobase pairs (K) are indicated for HSPV and CPXV Brighton Red, as are names of ORFs absent in these two species. Hatching indicates ORFs different in length (>10%) from intact orthologues from CPXV GRI-90 or Brighton Red. Red ORFs indicate HSPV ORFs intact or carried on sequences that are absent relative to VACV-like viruses. Asterisks indicate ORFs resequenced/reannotated by Upton et al. (92) as present or intact in the Tian Tan genome. Large solid-lined boxes indicate sequences matching the global alignment only at the opposite genomic terminus; dashed boxes indicate where sequences located in the opposite genomic terminus match the global alignment. Red lines indicate ITR sequence in each virus and are unaligned on the terminal side of HSPV002/HPSV206. Panel A is presented at a different scale relative to panels B and C.
Phylogenetic analysis.
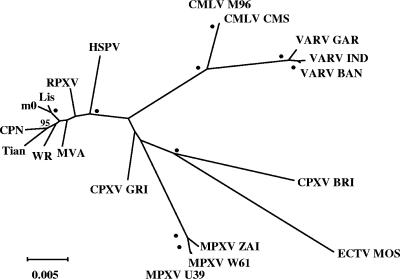
Phylogenetic analysis of OPV genomic regions, including the highly conserved central region and parts of the more variable terminal regions, indicated that HSPV is closely related to sequenced strains of VACV and RPXV, falling very close to or within this VACV subgroup (referred to here as VACV-like viruses) relative to other OPVs (Fig. 2). These results are consistent with those obtained previously for OPVs, with VACV-like viruses closely related to each other compared to other OPV species, and they indicated that HSPV is a VACV-like virus (21, 38, 51). As a VACV-like virus, HSPV also shares a closer relationship with CPXV strain GRI-90 than with CPXV strain Brighton Red (BRI), consistent with previous OPV phylogenetic analyses and indicating the distinct nature of CPXV species despite the relative conservation in gene content (Fig. 1 and 2) (21, 38, 51). Similarly, a close relationship was observed between HSPV and VACV using concatenated right terminal OPV gene sequences used previously for OPV phylogenetic analysis (HSPV177, HSPV179, HSPV182, and HSPV191; data not shown) (38). These results indicate that HSPV and VACV are very similar phylogenetically and share a relatively recent common ancestor. Notably, HSPV had a slightly greater estimated distance to VACV-like isolates than they demonstrated to each other, with HSPV tending to fall outside the rest of the VACV-like cluster (Fig. 2). These data suggested that, while very closely related, HSPV is phylogenetically distinct from other characterized VACV-like viruses.
FIG. 2.
Phylogenetic analysis of HSPV central genomic regions. Conserved HSPV central genomic nucleotide sequences (positions 26800 to 170171) corresponding to regions used previously for OPV phylogenetic analysis (51) were aligned with homologous OPV sequences using DIALIGN, and gapped regions were realigned with CLUSTAL W and trimmed with Gblocks. The unrooted tree for 124,677 aligned characters was generated using maximum likelihood with general time reversible correction for multiple substitutions, four-category discrete gamma model, estimation for proportion of invariant residues, and 100 bootstrap replicates as implemented in PHYML. Bootstrap values greater than 70 are indicated at appropriate nodes; dots indicate values of 100. Homologous nucleotide sequences from the following viruses and accession numbers were compared: VACV strain CPN, M35027; VACV WR, AY243312; VACV Lister (Elstree) vaccine consensus (Lis), AY678276; VACV Lister-derived LC16m0 (m0), AY678277; VACV Tian Tan (Tian), AF095689; VACV MVA, U94848; RPXV Utrecht (RPXV), AY484669; CPXV strain GRI-90 (X94355); CPXV BRI, AF482758; MPXV strain Zaire-96-I-16 (MPXV ZAI), AF380138; MPXV WRAIR7-61 (MPXV W61), AY603973; MPXV USA_2003_039 (MPXV U39), DQ011157; CMLV strain M-96 (CMLV M96), AF438165; CMLV CMS, AY009089; VARV strain Bangladesh-1975 (VARV BAN), L22579; VARV India-1967 (VARV IND), X69198; VARV Garcia-1966 (VARV GAR); Y16780; ECTV strain Moscow (ECTV MOS), AF012825. The scale indicates estimated distance. Identical topologies at supported nodes were obtained using additional maximum likelihood analyses as implemented in TREE-PUZZLE, using neighbor-joining and maximum parsimony as implemented in PHYLO_WIN and PHYLIP, respectively, and using an unedited alignment (146,439 characters) (data not shown). Similar topologies were also obtained using similar analyses on whole-genomic alignments (data not shown).
Comparison of HSPV with VACV-like viruses.
Given the close phylogenetic relationship between HSPV and VACV-like viruses, HSPV ORFs were compared to VACV-like homologues in the more variable terminal genomic regions which tend to contain genes dispensable for basic replicative processes but important for specific virus-host interaction and aspects of virulence and host range (Fig. 1; Table 1). While HSPV maintained a high level of amino acid identity where homologous terminal region ORFs were present (average of 95% amino acid identity to CPN), we focused here on comparison of HSPV and VACV in genes likely fragmented relative to CPXV and other OPVs. Overall, these differences often involved genes that are members of multigene families and/or homologues of genes shown or thought to affect OPV virulence or host range, among them those that code for ankyrin repeat proteins, kelch-like proteins, and tumor necrosis factor receptors (TNFRs) (4, 45, 77). While terminal-region genotypes vary both among OPVs and between known VACV-like viruses, HSPV contained features similar to known VACV-like viruses relative to other OPVs and features that were quite novel (Table 1; Fig. 1).
HSPV genetic features similar to VACV.
Genotypic similarity between HSPV and other VACV-like viruses included a number of genes that were fragmented relative to CPXV and occasionally relative to other OPVs. These genes included several which were fragmented or arranged in a similar fashion between HSPV and VACV-like viruses, commensurate with their close phylogenetic relationship (Table 1; Fig. 2). HSPV genes sharing similar ORF fragments with those in certain VACVs include HSPV005/HSPV203 and HSPV020, genes encoding ankyrin proteins and fragmented or missing in most OPVs (Fig. 1A). HSPV005b/HSPV203b in the ITR represents the same fragment of GRI-90 D4L/I2R as CPN C18L/B24R. HSPV020a to -e and similar ORFs in VACV are homologous fragments of CPXV CHOhr, a gene which enables replication of VACV in the normally nonpermissive CHO cell line and affects eukaryotic initiation factor 2α (eIF2α) phosphorylation in HeLa cells (41, 85). Other HSPV ORFs with similar VACV fragments included HSPV146d, HSPV180, and HSPV186. HSPV146d encodes the same 725-amino-acid amino-terminal fragment of the A-type inclusion (ATI) protein present in several VACV-like viruses and expressed in some as a soluble 94-kDa protein (26). HSPV186 is a VACV-like ORF fragment homologous to the amino-terminal region of the OPV homologue of myxoma virus M-T4, a protein important for virulence and infection of lymphocytes by myxoma virus (12). The HSPV186 homologue is expressed in VACV strain Western Reserve (WR); however, deletion mutants were not affected for viral growth in vitro or virulence in mice (68). While amino-terminal M-T4-like fragments are also present in certain strains of MPXV (22, 52), the large nucleotide deletion affecting HSPV186 was characteristic of VACV (Fig. 1C). Also characteristic of VACV are homologues of HSPV180a and HSPV180b (CPN B2R and B3R, respectively), apparent fragments of a larger ORF intact in all OPV species other than VACV and VARV and previously annotated as similar to cellular Schlafen, a family of variably sized proteins with the prototypical 337-amino-acid murine Schlafen 1 recently shown to target cyclin D1 pathways during induction of cellular mid-G1 cell cycle arrest (15, 39). Notably, HSPV180a and HSPV180b revealed the bipartite nature of the larger OPV homologue, with Schlafen similarity present in the HSPV180b-like (carboxyl-terminal) region and the HSPV180a-like (amino-terminal) region sharing similarity with the putative B2R homologue of Melanoplus sanquinipes entomopoxvirus (MSV237) and limited similarity with ORFs of unknown function (p26) from nucleopolyhedrosis viruses (data not shown). While maintenance of these two domains as separate ORFs in HSPV and VACV conceivably suggests function, HSPV180b and VACV orthologues lack carboxyl-terminal sequences both present in the intact OPV ORF and similar to the carboxyl terminus of cellular Schlafen. Overall, similar fragmentation patterns between HSPV and VACV potentially represent shared, derived characters.
Several genes fragmented in HSPV were also fragmented in certain VACV-like isolates but intact in others (Table 1). HSPV ORF fragments with intact homologues in certain VACVs included HSPV018, HSPV161, HSPV173a and -b, and HSPV175. HSPV018 is an amino-terminal fragment homologue of the ECTV p28 ubiquitin ligase, a protein critical for ECTV virulence and macrophage host range and having intact homologues in all other OPV species (74, 87) (Fig. 1). While this gene is also fragmented in several VACV strains, intact homologues have been identified in VACV strains IHD-W and Lister and in RPXV (51, 58, 91). Similarly, HSPV173a and -b resembled homologous ORFs in VACV Lister and RPXV and fragments of the CPN A51R gene intact in other VACV strains and all other OPVs. HSPV161 was a homologue of CPN A39R, a secreted semaphorin affecting viral virulence and host inflammatory responses during infection, but, similarly to homologues in WR and other VACV strains, contained a carboxyl-terminal truncation that may predict a nonfunctional product (35). HSPV175, similar to several VACV-like viruses, encoded a truncated copy of the intact CrmC TNFR-like protein encoded by VACV strains Lister, Evans, and USSR (5). HSPV039 and HSPV187 were fragmented genes with homologues fragmented in all VACV-like viruses but with VACV-like homologues fragmented in a pattern distinct from those in HSPV. HSPV039 was similar to both CPN K5L and K6L fragments of the OPV monoglyceride lipase-like gene but was much closer in size to the intact CPXV homologue, and HSPV187 was a smaller fragment of the CPXV GRI-90 B9R kelch-like protein. While HSPV175, HSPV039, and HSPV187 homologues were fragmented in both HSPV and most VACV-like viruses, these genes were also disrupted in most other OPVs (Fig. 1).
HSPV also contained intact genes whose homologues were intact in certain VACV-like viruses but disrupted in others, similar to genes recently described in the RPXV genome (Table 1) (51). HSPV002/HSPV206 in the ITR encoded the OPV 35-kDa secreted chemokine binding protein and, similarly to the functional, full-length protein expressed by VACV Lister and other OPVs, lacked the amino-terminal mutation preventing expression of functional protein in CPN, WR, and VACV strain Tian Tan (6). HSPV147 was an intact copy of the gene encoding P4c, a protein involved with direction of IMV to insoluble ATIs but with homologues fragmented or absent in CPN, Tian Tan, and modified vaccinia Ankara (MVA). HSPV190 was only the third intact VACV-like orthologue of the serine proteinase inhibitor (serpin) 2 (SPI-2) to be identified, and HSPV198 was an intact orthologue of the SPI-1 gene intact in most VACV-like viruses but transposed to the opposite terminus in RPXV and VACV CPN, Tian Tan, and Lister and absent in MVA (Fig. 1C). Intact SPI-1 and SPI-2 exhibit antiapoptotic and/or anti-inflammatory activity through inhibition of caspases and have been shown to affect viral virulence and/or host range (48, 59, 82, 87). HSPV196 encodes an intact ankyrin repeat protein truncated by deletion in all VACV-like viruses except RPXV, where the homologue was recently identified as unique among VACV-like viruses in that the entire nucleotide region encompassing the gene was present (51). Similarly, HSPV199 encodes an intact homologue of the BRI CPXV218 chemokine binding protein, with intact homologues also encoded in the right terminus of WR and in the left terminus of VACV Lister and RPXV (Fig. 1) (7). Overall, different fragmentation patterns or gene loss between HSPV genes and VACV homologues may indicate sequence divergence after functional gene loss or, alternatively, could conceivably reflect independent loss of gene function in different VACV-like lineages during convergent adaptation toward similar virulence or host range phenotypes. Gene loss near ITR boundaries may reflect loss during terminal transposition events (47, 61). These phenomena would help explain gene fragmentation that is variable both within the VACV-like lineage and between OPV species.
HSPV genetic features distinct from VACV.
Despite sharing specific genomic and genotypic features with some or all known VACV-like viruses within the range of VACV-like genotypic heterogeneity, HSPV contained many features that were unique. These included genes uniquely intact in HSPV but for which homologous nucleotide sequence was present in other VACVs, and they included HSPV genes, both intact and fragmented, that were associated with nucleotide sequences completely novel among VACV-like viruses, resulting in terminal genomic regions encoding additional proteins and protein fragments resembling those in CPXV (94% average amino acid identity to CPXV GRI-90 orthologues) (Fig. 1; Table 1). Finally, HSPV demonstrated unique fragmentation of several genes, including those that were intact in all or most other known VACV-like viruses.
HSPV contained in the ITRs intact genes that are fragmented or absent in all other VACVs (Table 1). HSPV003/HSPV205 is an intact homologue of the secreted CPXV Brighton Red CrmB TNFR II-like protein (CPXV GRI-90 D2L/I2R), a protein which interacts with and inhibits TNF and lymphotoxin alpha and whose orthologue in VARV has been recently shown to contain a novel carboxyl-terminal chemokine binding domain also present and active in several other OPV proteins (4, 7, 42). HSPV004/HSPV204 encodes an intact homologue of the ankyrin repeat protein encoded by CPXV GRI D3L/I3R and intact homologues in MPXV, ECTV, and CMLV (Fig. 1).
HSPV contains approximately 17 kbp of sequence in three distinct genomic regions (positions 7527 to 18195 in the left terminal region and 194379 to 195517 and 198775 to 204285 in the right terminal region) absent in known VACV-like viruses but homologous to sequences in sequenced strains of CPXV and other OPVs (Fig. 1). HSPV also contains approximately 1.4 kbp of sequence absent not only in VACV but also in all known OPVs except CPXV. For this region, located between positions 15453 and 16985, only MPXV contains a fragment (approximately 75 bp) of homologous sequence. Notably, sequences near this region reflect ITR and/or terminal translocations in several OPVs (Fig. 1), and repetitive sequence near this locus in ECTV has been suggested to be a dynamic genomic region (21). Conceivably, the presence of this 1.4-kbp sequence in HSPV is consistent with retention of adjacent and relatively significant amounts of CPXV-like sequence in this left terminal region relative to other OPVs (Fig. 1).
HSPV sequence in the left terminal region absent in other VACV-like viruses corresponds to the D7L loci of CPXV GRI-90 and the CPXV014 to CPXV020 region of CPXV BRI (Fig. 1A). These sequences relative to other VACV-like viruses essentially extend from the ITR boundary region to the region upstream of the HSPV016 viral growth factor homologue (CPN C11R), replacing the OPV-like sequence that is transposed from the right terminal region to the left terminal region in other VACV-like viruses. HSPV sequences in this region include 15 ORFs representing three intact OPV genes (HSPV008, HSPV010, and HSPV012) and six potentially truncated or fragmented genes (HSPV007, HSPV009, HSPV011a to -c, HSPV013, HSPV014a to -d, and HSPV015a and -b) (Table 1; Fig. 1). HSPV008 encodes an intact protein orthologous only to CPXV GRI-90 D7L and ECTV strain Moscow EVM004 (21, 79). These proteins contain amino-terminal BTB/POZ domains, evolutionarily conserved domains important for oligomerization and ordering of protein complexes and often present in amino-terminal regions of both cellular and poxviral kelch-like proteins, but in these smaller HSPV008 orthologues the BTB/POZ domain is not associated with kelch repeat domains (3, 75). HSPV009 encodes a truncated orthologue of CPXV GRI-90 D12L product, a protein similar to the CrmB carboxyl terminus and whose orthologue in ECTV was recently characterized as a secreted chemokine binding protein (7). HSPV010 encodes an intact orthologue of CD30 TNFR-like proteins present in CPXV and ECTV, proteins able to bind CD30 ligands and/or have immunomodulatory effects (63, 72). HSPV left-end sequences also contain genes for three ankyrin repeat proteins absent in VACV. While HSPV012 encodes an intact ankyrin repeat protein also intact in CPXV and MPXV, HSPV011a to -c and HSPV014a to -d encode fragments of intact ankyrin repeat proteins encoded only in ECTV and/or CPXV, with HSPV014b and -c encoded within the region containing 1.4 kbp of sequence found only in CPXV. Finally, HSPV015a and -b appeared to encode fragments of a paralogue of CPN C7L, a VACV host range protein which enables viral replication in human cells (67). While all OPVs appear to encode intact C7L orthologues (HSPV024), intact HSPV015 orthologues are encoded only in CPXV and CMLV, with fragmented ORFs annotated in MPXV and VARV (Table 1).
HSPV sequence in the right terminal region absent in other VACV-like viruses essentially bound the region homologous to the VACV WR SPI-1 (HSPV198) locus, a region transposed to the opposite terminus in several other VACVs (Fig. 1). Unique sequence upstream of HSPV198 includes HSPV197, an intact kelch-like protein also intact in CPXV and ECTV but fragmented or absent in MPXV, CMLV, and VARV. Unique sequences downstream of HSPV198 contain an intact orthologue of the VARV strain Bangladesh B22R gene (HSPV200). B22R homologues represent the largest poxviral genes, encoding proteins of approximately 2,000 amino acids and with no known function but predicted to contain carboxyl-terminal transmembrane domains and cysteine residues which conceivably mediate disulfide bond formation (54, 56, 76). B22R homologues are intact in all OPV species except VACV-like viruses, making the presence of HSPV200 notable (Fig. 1).
Despite containing additional sequence not present in other VACV-like viruses, HSPV did lack sequences homologous to several larger regions in other OPVs. These include from GRI-90 the D8L to D11L locus, a region encoding ankyrin repeat, kelch-like, and lectin-like proteins with homologous sequence only in ECTV (79) (Table 1), and most of the K1R to S1R/T1R locus, a region encoding ankyrin repeat, CrmD TNFR, and CrmE TNFR proteins and with homologous sequence present in MPXV, ECTV, and CMLV and, notably, in VACV Lister (Fig. 1C). HSPV also lacks any remnant of the second VARV B22R-like gene identified in certain strains of CPXV and of which remnants remain in VARV and CMLV lineages (HSPV185-HSPV186 locus [Fig. 1C]) (56).
Finally, HSPV contains fragmented genes intact in all or nearly all other VACV-like viruses. Within the central conserved region, HSPV074a to -d represented fragments of the CPN I4L ribonucleotide reductase large subunit gene, while HSPV044 encoded an intact small subunit (Table 2). Ribonucleotide reductase is a heterodimeric protein involved in redox reactions that are key to synthesis of deoxyribonucleotides, an activity for which various poxviruses encode different enzyme complements, potentially adapted to replication in specific host cell types lacking adequate nucleotide pools (59). Experimental disruption of the VACV ribonucleotide reductase large subunit has been shown previously to have no effect on virus replication in vitro and a mild effect on virulence in mice (23). Although I4L homologues are not encoded in all other poxviral genera, to our knowledge this is the first example of its natural disruption in an OPV genome. Similarly, HSPV183 is unique among VACV-like homologues (CPN B6R) as the only form of the gene to be fragmented, although a fragmented form is also found in VARV (Fig. 1B) and an isolate of MPXV (accession no. AAY97373). Notably, HSPV contained fragmented genes intact in all VACVs except MVA, a virus that has accumulated numerous mutations and extensive nucleotide deletions through extensive passage in vitro and concomitant attenuation and restriction of host range (9). These include HSPV026, orthologue of CPN C5L BTB domain protein, and the HSPV033 ankyrin repeat protein. In addition, HSPV178, similar to MVA, demonstrates a smaller fragmented form of the guanylate kinase gene than do other VACV-like viruses.
Perspective on relationship of HSPV to VACV.
Genomic sequence analysis of HSPV MNR-76 indicates that it is a novel VACV-like OPV that contains unique features not present in known VACVs. Although MNR-76 is unique in the complement of OPV genes remaining intact in HSPV, the pattern of terminal gene loss/fragmentation is commensurate with genotypes observed in other VACV-like viruses. Notably, the majority of left terminal HSPV sequence absent in VACV appears to contain gene fragments, with HSPV conceivably in the process of losing this sequence similarly to other VACV-like viruses.
The close phylogenetic and genotypic relationship between HSPV and other VACV-like viruses and the presence of additional CPXV-like sequences in HSPV are notable given previous speculations involving horsepox and the origins of VACV (14). While the origins of current VACV-like strains have been heavily debated and remain obscure, current knowledge affirms that VACV-like viruses constitute an OPV lineage independent of known CPXV and VARV species from which VACV has been speculated to be derived (14, 32, 33, 38) (Fig. 2). It is likely that a once naturally circulating but now rare VACV-like virus(s) from which current strains are derived was introduced as a vaccine virus, and the agent of horsepox has been surmised as a likely candidate (14). Indeed, apparently Edward Jenner believed that his vaccine originated from the “grease” infection found in the heels of horses, and the use of horse-derived material for use as vaccines is documented (14, 33). In addition, phenotypic similarity of certain vaccines transmitted between cows, humans, and horses has been noted, and experimental infection of horses with VACV can produce clinical signs of horsepox (14, 44, 86). The data presented here indicate that the HSPV MNR-76 genome contains features consistent with such a hypothesis, a phylogenetically VACV-like virus isolated from a horse and containing additional OPV-like terminal sequences, sequences likely ancestral and absent in other VACV-like viruses yet in certain regions appearing to be undergoing gene fragmentation and loss commensurate with transition toward a VACV-like genotype.
Despite speculation as to what role horsepox played in the development of smallpox vaccines, it is clear that HSPV MNR-76 does not represent a direct ancestral genotype to all known VACVs, given the disruption of many HSPV genes intact in certain VACV isolates (Table 1). It is unclear what constitutes the genotypic diversity of all the viruses historically used for smallpox vaccine, especially considering the potential for disparate source material and passage histories of VACV-like vaccine viruses (14, 33). Indeed, phenotypic and genotypic diversity is observed between and within strains of VACV (14, 33, 58) (Fig. 1). This diversity does include sequence unique to a given strain, such as the presence of CPXV GRI K3R and S1R/T1R-like genes in the historically important Lister vaccine strain (Fig. 1C), making the presence of HSPV MNR-76-like sequences in uncharacterized vaccine strains a possibility. Isolated in 1976, HSPV was causing disease in horses while smallpox vaccines were still being distributed during the World Health Organization global smallpox eradication program (32). Conceivably, local or a currently uncharacterized vaccine could have been introduced into the horse population, as contact with vaccinated persons is known to have been a source of OPV disease in animals (33). Vaccine escape has been hypothesized to account for other VACV-like viruses occasionally isolated from domestic and sentinel animals, including RPXV, buffalopox in India, and viruses associated with zoonosis in South America; however, unique biological properties and/or inability to associate the isolate with vaccine virus has also led to suggestions that they are natural VACV isolates or VACV subspecies (19, 24, 25, 27, 33, 46, 90). Similarly, HSPV MNR-76 may represent a novel, naturally circulating virus and perhaps one for which the horse was an incidental host, just as other domestic and captive animals are not thought to be the reservoir for CPXV infection despite being susceptible to infection (13, 33). Unfortunately, little is known of the prevalence of disease associated with HSPV MNR-76 in Mongolia, either in horse or in human populations. Conceivably, MNR-76 may represent a naturally circulating member of the VACV lineage, as were viruses circulating among domestic animals in the era in which current VACV-like viruses were collected as vaccine. Whatever the historical relationship between HSPV MNR-76 and characterized VACV-like viruses may be, genomic sequence analysis of other VACV-like virus isolates may add perspective to the novel nature of HSPV relative to other viruses within the VACV lineage.
ADDENDUM IN PROOF
Since completion of the analyses presented here, the genome sequences of several VACV clones derived from the Dryvax vaccine have become available. Preliminary analysis indicates that while most of the HSPV sequence reported here as absent in VACV was also absent in these clones, one (GenBank accession no. AY313848) contained nucleotide sequence and ORF fragments at the HSPV 197 locus, stressing the need for additional genomic sequence and analyses in examining the nature of VACV-like virus variability.
Acknowledgments
We thank A. Lakowitz and A. Waite Lund for excellent technical assistance.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genome of camelpox virus. Virology 295:1-9. [DOI] [PubMed] [Google Scholar]
- 3.Albagli, O., P. Dhordain, C. Deweindt, G. Lecocq, and D. Leprince. 1995. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 6:1193-1198. [PubMed] [Google Scholar]
- 4.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. [DOI] [PubMed] [Google Scholar]
- 5.Alcami, A., A. Khanna, N. L. Paul, and G. L. Smith. 1999. Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J. Gen. Virol. 80:949-959. [DOI] [PubMed] [Google Scholar]
- 6.Alcami, A., J. A. Symons, P. D. Collins, T. J. Williams, and G. L. Smith. 1998. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J. Immunol. 160:624-633. [PubMed] [Google Scholar]
- 7.Alejo, A., M. B. Ruiz-Arguello, Y. Ho, V. P. Smith, M. Saraiva, and A. Alcami. 2006. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. USA 103:5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 10.Baroudy, B. M., and B. Moss. 1982. Sequence homologies of diverse length tandem repetitions near ends of vaccinia virus genome suggest unequal crossing over. Nucleic Acids Res. 10:5673-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baroudy, B. M., S. Venkatensan, and B. Moss. 1982. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 28:315-324. [DOI] [PubMed] [Google Scholar]
- 12.Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360-377. [DOI] [PubMed] [Google Scholar]
- 13.Baxby, D. 1977. Is cowpox misnamed? A review of 10 human cases. Br. Med. J. 1:1379-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baxby, D. 1981. Jenner's smallpox vaccine: the riddle of vaccinia virus and its origin. Heinemann Educational Books Ltd., London, United Kingdom.
- 15.Brady, G., L. Boggan, A. Bowie, and L. A. O'Neill. 2005. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J. Biol. Chem. 280:30723-30734. [DOI] [PubMed] [Google Scholar]
- 16.Breman, J. G., and D. A. Henderson. 2002. Diagnosis and management of smallpox. N. Engl. J. Med. 346:1300-1308. [DOI] [PubMed] [Google Scholar]
- 17.Brudno, M., M. Chapman, B. Gottgens, S. Batzoglou, and B. Morgenstern. 2003. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics 4:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brudno, M., C. B. Do, G. M. Cooper, M. F. Kim, E. Davydov, E. D. Green, A. Sidow, and S. Batzoglou. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buller, R. M., and G. J. Palumbo. 1991. Poxvirus pathogenesis. Microbiol. Rev. 55:80-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 21.Chen, N., M. I. Danila, Z. Feng, R. M. Buller, C. Wang, X. Han, E. J. Lefkowitz, and C. Upton. 2003. The genomic sequence of ectromelia virus, the causative agent of mousepox. Virology 317:165-186. [DOI] [PubMed] [Google Scholar]
- 22.Chen, N., G. Li, M. K. Liszewski, J. P. Atkinson, P. B. Jahrling, Z. Feng, J. Schriewer, C. Buck, C. Wang, E. J. Lefkowitz, J. J. Esposito, T. Harms, I. K. Damon, R. L. Roper, C. Upton, and R. M. Buller. 2005. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 340:46-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Child, S. J., G. J. Palumbo, R. M. Buller, and D. E. Hruby. 1990. Insertional inactivation of the large subunit of ribonucleotide reductase encoded by vaccinia virus is associated with reduced virulence in vivo. Virology 174:625-629. [DOI] [PubMed] [Google Scholar]
- 24.da Fonseca, F. G., G. S. Trindade, R. L. Silva, C. A. Bonjardim, P. C. Ferreira, and E. G. Kroon. 2002. Characterization of a vaccinia-like virus isolated in a Brazilian forest. J. Gen. Virol. 83:223-228. [DOI] [PubMed] [Google Scholar]
- 25.Damaso, C. R., J. J. Esposito, R. C. Condit, and N. Moussatche. 2000. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology 277:439-449. [DOI] [PubMed] [Google Scholar]
- 26.de Carlos, A., and E. Paez. 1991. Isolation and characterization of mutants of vaccinia virus with a modified 94-kDa inclusion protein. Virology 185:768-778. [DOI] [PubMed] [Google Scholar]
- 27.de Souza Trindade, G., F. G. da Fonseca, J. T. Marques, M. L. Nogueira, L. C. Mendes, A. S. Borges, J. R. Peiro, E. M. Pituco, C. A. Bonjardim, P. C. Ferreira, and E. G. Kroon. 2003. Aracatuba virus: a vaccinialike virus associated with infection in humans and cattle. Emerg. Infect. Dis. 9:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essajee, S., and H. L. Kaufman. 2004. Poxvirus vaccines for cancer and HIV therapy. Expert Opin. Biol. Ther. 4:575-588. [DOI] [PubMed] [Google Scholar]
- 29.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 30.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 32.Fenner, F., D. A. Henderson, I. Arita, Z. Jezek, and I. D. Ladnyi. 1988. Smallpox and its eradication. World Health Organization, Geneva, Switzerland.
- 33.Fenner, F., R. Wittek, and K. Dumbell. 1989. The orthopoxviruses. Academic Press, Inc., San Diego, Calif.
- 34.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biol. Sci. 12:543-554. [DOI] [PubMed] [Google Scholar]
- 35.Gardner, J. D., D. C. Tscharke, P. C. Reading, and G. L. Smith. 2001. Vaccinia virus semaphorin A39R is a 50-55 kDa secreted glycoprotein that affects the outcome of infection in a murine intradermal model. J. Gen. Virol. 82:2083-2093. [DOI] [PubMed] [Google Scholar]
- 36.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 37.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 38.Gubser, C., S. Hue, P. Kellam, and G. L. Smith. 2004. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 85:105-117. [DOI] [PubMed] [Google Scholar]
- 39.Gubser, C., and G. L. Smith. 2002. The sequence of camelpox virus shows it is most closely related to variola virus, the cause of smallpox. J. Gen. Virol. 83:855-872. [DOI] [PubMed] [Google Scholar]
- 40.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 41.Hsiao, J. C., C. S. Chung, R. Drillien, and W. Chang. 2004. The cowpox virus host range gene, CP77, affects phosphorylation of eIF2 alpha and vaccinia viral translation in apoptotic HeLa cells. Virology 329:199-212. [DOI] [PubMed] [Google Scholar]
- 42.Hu, F. Q., C. A. Smith, and D. J. Pickup. 1994. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology 204:343-356. [DOI] [PubMed] [Google Scholar]
- 43.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutyra, F., J. Marek, and R. Manninger. 1949. Pox (variola), p. 348-393. In J. R. Greig (ed.), Special pathology and therapeutics of the diseases of domestic animals, vol. 1. Bailliere, Tindall and Cox, London, United Kingdom. [Google Scholar]
- 45.Kochneva, G., I. Kolosova, T. Maksyutova, E. Ryabchikova, and S. Shchelkunov. 2005. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch. Virol. 150:1857-1870. [DOI] [PubMed] [Google Scholar]
- 46.Kolhapure, R. M., R. P. Deolankar, C. D. Tupe, C. G. Raut, A. Basu, B. M. Dama, S. D. Pawar, M. V. Joshi, V. S. Padbidri, M. K. Goverdhan, and K. Banerjee. 1997. Investigation of buffalopox outbreaks in Maharashtra State during 1992-1996. Indian J. Med. Res. 106:441-446. [PubMed] [Google Scholar]
- 47.Kotwal, G. J., and B. Moss. 1988. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology 167:524-537. [PubMed] [Google Scholar]
- 48.Legrand, F. A., P. H. Verardi, L. A. Jones, K. S. Chan, Y. Peng, and T. D. Yilma. 2004. Induction of potent humoral and cell-mediated immune responses by attenuated vaccinia virus vectors with deleted serpin genes. J. Virol. 78:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leite, J. A., B. P. Drumond, G. S. Trindade, Z. I. Lobato, F. G. da Fonseca, S. J. Dos, M. C. Madureira, M. I. Guedes, J. M. Ferreira, C. A. Bonjardim, P. C. Ferreira, and E. G. Kroon. 2005. Passatempo virus, a vaccinia virus strain, Brazil. Emerg. Infect. Dis. 11:1935-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis-Jones, S. 2004. Zoonotic poxvirus infections in humans. Curr. Opin. Infect. Dis. 17:81-89. [DOI] [PubMed] [Google Scholar]
- 51.Li, G., N. Chen, R. L. Roper, Z. Feng, A. Hunter, M. Danila, E. J. Lefkowitz, R. M. Buller, and C. Upton. 2005. Complete coding sequences of the rabbitpox virus genome. J. Gen. Virol. 86:2969-2977. [DOI] [PubMed] [Google Scholar]
- 52.Likos, A. M., S. A. Sammons, V. A. Olson, A. M. Frace, Y. Li, M. Olsen-Rasmussen, W. Davidson, R. Galloway, M. L. Khristova, M. G. Reynolds, H. Zhao, D. S. Carroll, A. Curns, P. Formenty, J. J. Esposito, R. L. Regnery, and I. K. Damon. 2005. A tale of two clades: monkeypox viruses. J. Gen. Virol. 86:2661-2672. [DOI] [PubMed] [Google Scholar]
- 53.Massung, R. F., J. C. Knight, and J. J. Esposito. 1995. Topography of variola smallpox virus inverted terminal repeats. Virology 211:350-355. [DOI] [PubMed] [Google Scholar]
- 54.Massung, R. F., L.-I. Liu, J. Qi, J. C. Knight, T. E. Yuran, A. R. Kerlavage, J. M. Parsons, J. C. Venter, and J. J. Esposito. 1994. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology 201:215-240. [DOI] [PubMed] [Google Scholar]
- 55.Merchlinsky, M., and B. Moss. 1989. Nucleotide sequence required for resolution of the concatemer junction of vaccinia virus DNA. J. Virol. 63:4354-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer, H., A. Totmenin, E. Gavrilova, and S. Shchelkunov. 2005. Variola and camelpox virus-specific sequences are part of a single large open reading frame identified in two German cowpox virus strains. Virus Res. 108:39-43. [DOI] [PubMed] [Google Scholar]
- 57.Morgenstern, B., K. Frech, A. Dress, and T. Werner. 1998. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics 14:290-294. [DOI] [PubMed] [Google Scholar]
- 58.Morikawa, S., T. Sakiyama, H. Hasegawa, M. Saijo, A. Maeda, I. Kurane, G. Maeno, J. Kimura, C. Hirama, T. Yoshida, Y. Asahi-Ozaki, T. Sata, T. Kurata, and A. Kojima. 2005. An attenuated LC16m8 smallpox vaccine: analysis of full-genome sequence and induction of immune protection. J. Virol. 79:11873-11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 60.Moss, B., and J. L. Shisler. 2001. Immunology 101 at poxvirus U: immune evasion genes. Semin. Immunol. 13:59-66. [DOI] [PubMed] [Google Scholar]
- 61.Moyer, R. W., and R. L. Graves. 1981. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell 27:391-401. [DOI] [PubMed] [Google Scholar]
- 62.Nylander, J. A. A. 2004. MrModeltest 2.2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 63.Panus, J. F., C. A. Smith, C. A. Ray, T. D. Smith, D. D. Patel, and D. J. Pickup. 2002. Cowpox virus encodes a fifth member of the tumor necrosis factor receptor family: a soluble, secreted CD30 homologue. Proc. Natl. Acad. Sci. USA 99:8348-8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 65.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 67.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 68.Price, N., D. C. Tscharke, and G. L. Smith. 2002. The vaccinia virus B9R protein is a 6 kDa intracellular protein that is non-essential for virus replication and virulence. J. Gen. Virol. 83:873-878. [DOI] [PubMed] [Google Scholar]
- 69.Reed, K. D., J. W. Melski, M. B. Graham, R. L. Regnery, M. J. Sotir, M. V. Wegner, J. J. Kazmierczak, E. J. Stratman, Y. Li, J. A. Fairley, G. R. Swain, V. A. Olson, E. K. Sargent, S. C. Kehl, M. A. Frace, R. Kline, S. L. Foldy, J. P. Davis, and I. K. Damon. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342-350. [DOI] [PubMed] [Google Scholar]
- 70.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 71.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saraiva, M., P. Smith, P. G. Fallon, and A. Alcami. 2002. Inhibition of type 1 cytokine-mediated inflammation by a soluble CD30 homologue encoded by ectromelia (mousepox) virus. J. Exp. Med. 196:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 74.Senkevich, T. G., E. J. Wolffe, and R. M. L. Buller. 1995. Ectromelia virus RING finger protein is localized in virus factories and is required for virus replication in macrophages. J. Virol. 69:4103-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shchelkunov, S., A. Totmenin, and I. Kolosova. 2002. Species-specific differences in organization of orthopoxvirus kelch-like proteins. Virus Genes 24:157-162. [DOI] [PubMed] [Google Scholar]
- 76.Shchelkunov, S. N., V. M. Blinov, S. M. Resenchuk, A. V. Totmenin, L. V. Olenina, G. B. Chirikova, and L. S. Sandakhchiev. 1994. Analysis of the nucleotide sequence of 53 kbp from the right terminus of the genome of variola major virus strain India-1967. Virus Res. 34:207-236. [DOI] [PubMed] [Google Scholar]
- 77.Shchelkunov, S. N., V. M. Blinov, and L. S. Sandakhchiev. 1993. Ankyrin-like proteins of variola and vaccinia viruses. FEBS Lett. 319:163-165. [DOI] [PubMed] [Google Scholar]
- 78.Shchelkunov, S. N., R. F. Massung, and J. J. Esposito. 1995. Comparison of the genome DNA sequences of Bangladesh-1975 and India-1967 variola viruses. Virus Res. 36:107-118. [DOI] [PubMed] [Google Scholar]
- 79.Shchelkunov, S. N., P. F. Safronov, A. V. Totmenin, N. A. Petrov, O. I. Ryazankina, V. V. Gutorov, and G. J. Kotwal. 1998. The genome sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology 243:432-460. [DOI] [PubMed] [Google Scholar]
- 80.Shchelkunov, S. N., A. V. Totmenin, V. N. Loparev, P. F. Safronov, V. V. Gutorov, V. E. Chizhikov, J. C. Knight, J. M. Parsons, R. F. Massung, and J. J. Esposito. 2000. Alastrim smallpox variola minor virus genome DNA sequences. Virology 266:361-386. [DOI] [PubMed] [Google Scholar]
- 81.Shchelkunov, S. N., A. V. Totmenin, P. F. Safronov, M. V. Mikheev, V. V. Gutorov, O. I. Ryazankina, N. A. Petrov, I. V. Babkin, E. A. Uvarova, L. S. Sandakhchiev, J. R. Sisler, J. J. Esposito, I. K. Damon, P. B. Jahrling, and B. Moss. 2002. Analysis of the monkeypox virus genome. Virology 297:172-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shisler, J. L., and B. Moss. 2001. Immunology 102 at poxvirus U: avoiding apoptosis. Semin. Immunol. 13:67-72. [DOI] [PubMed] [Google Scholar]
- 83.Smith, G. L., and G. McFadden. 2002. Smallpox: anything to declare? Nat. Rev. Immunol. 2:521-527. [DOI] [PubMed] [Google Scholar]
- 84.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1-GC10. [DOI] [PubMed] [Google Scholar]
- 85.Spehner, D., S. Gillard, R. Drillien, and A. Kirn. 1988. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J. Virol. 62:1297-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Studdert, M. J. 1989. Experimental vaccinia virus infection of horses. Aust. Vet. J. 66:157-159. [DOI] [PubMed] [Google Scholar]
- 87.Taylor, J. M., and M. Barry. 2006. Near death experiences: poxvirus regulation of apoptotic death. Virology 344:139-150. [DOI] [PubMed] [Google Scholar]
- 88.Thompson, C. H., J. A. Yager, and I. B. Van Rensburg. 1998. Close relationship between equine and human molluscum contagiosum virus demonstrated by in situ hybridisation. Res. Vet. Sci. 64:157-161. [DOI] [PubMed] [Google Scholar]
- 89.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trindade, G. S., F. G. da Fonseca, J. T. Marques, S. Diniz, J. A. Leite, S. De Bodt, Y. Van der Peer, C. A. Bonjardim, P. C. Ferreira, and E. G. Kroon. 2004. Belo Horizonte virus: a vaccinia-like virus lacking the A-type inclusion body gene isolated from infected mice. J. Gen. Virol. 85:2015-2021. [DOI] [PubMed] [Google Scholar]
- 91.Upton, C., L. Schiff, S. A. Rice, T. Dowdeswell, X. Yang, and G. McFadden. 1994. A poxvirus protein with a RING finger motif binds zinc and localizes in virus factories. J. Virol. 68:4186-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Upton, C., S. Slack, A. L. Hunter, A. Ehlers, and R. L. Roper. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 77:7590-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wesley, R. D., and A. E. Tuthill. 1984. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev. Vet. Med. 2:53-62. [Google Scholar]
- 94.Yager, J. A., D. W. Scott, and B. P. Wilcock. 1993. The skin and appendages, p. 531-738. In K. V. F. Jubb, P. C. Kennedy, and N. Palmer (ed.), Pathology of domestic animals, 4th ed., vol. 1. Academic Press, San Diego, Calif. [Google Scholar]