Abstract
Gammaretroviruses, such as murine leukemia virus (MLV), are functionally distinguished from lentiviruses, such as human immunodeficiency virus, by their inability to infect nondividing cells. Attempts to engineer this property into MLV have been hindered by an incomplete understanding of early events in the viral life cycle. We utilized a transposon-based method to generate saturated peptide insertion libraries of MLV gag-pol variants with nuclear localization signals randomly incorporated throughout these overlapping genes. High-throughput selection of the libraries via iterative retroviral infection of nondividing cells led to the identification of a novel variant that successfully transduced growth-arrested cells. Vector packaging by cotransfection of the gag-pol.NLS variant with wild-type gag-pol produced high-titer virions capable of infecting neurons in vitro and in vivo. The capacity of mutant virions to transduce nondividing cells could help to elucidate incompletely understood mechanisms of the viral life cycle and greatly broaden the gene therapy applications of retroviral vectors. Furthermore, the ability to engineer key intracellular viral infection steps has potential implications for the understanding, design, and control of other postentry events. Finally, this method of library generation and selection for a desired phenotype directly in a mammalian system can be readily expanded to address other challenges in protein engineering.
The capability to modify or engineer novel viral phenotypes could enhance the use of retroviral vectors as tools for biology and medicine and further elucidate fundamental issues in retroviral biology. Such modifications to the physical and functional properties of retroviruses can be achieved through engineering one of the three genes, gag, pol, or env, from which all retroviral proteins are derived. However, retroviral protein engineering requires not only a consideration of how an individual folded protein will behave but also the intersubunit interactions necessary for assembly into a multimeric capsid and interprotein interactions necessary to construct the preintegration complex (PIC). Genetic modifications to a compact viral genome can be further complicated if the target sequence contributes to multiple gene products, such as in the overlapping gag-pol genes. Therefore, the development of efficient and high-throughput viral protein engineering approaches can aid in addressing questions and challenges in viral and vector biology, including mechanisms of intracellular viral transport.
Though the biochemical processes of reverse transcription and chromosomal integration are well described, the period of the retroviral life cycle between these two events remains incompletely understood (reviewed in references 1 and 21). The retroviral PIC is too large to passively diffuse through the nuclear membrane and must wait until membrane dissolution during mitosis or be actively transported through a nuclear pore complex (NPC) to gain access to chromosomal DNA for integration (45, 50). Several mechanisms for the active nuclear import of lentiviruses, such as human immunodeficiency virus (HIV), which are able to infect nondividing cells, have been proposed (reviewed in references 13 and 49). However, none of HIV's karyophilic signals has been shown to be definitively necessary or sufficient, and lentiviral vectors with deletions of all previously implicated karyophilic elements were recently shown to still infect nondividing cells (52). In contrast to lentiviruses, gammaretroviruses, such as murine leukemia virus (MLV), can infect only proliferating cells (34, 45). Engineering a simple retrovirus to be capable of infecting nondividing cells could shed light on possible mechanisms sufficient for active nuclear import. In addition, human gene therapy vectors based on MLV have had significant clinical successes, but these have been balanced by potential adverse effects that illustrate the need to develop new techniques for engineering improved vectors (7, 14, 25).
The overlapping MLV gag-pol genes encode Gag and Gag-Pol polyproteins, which are proteolytically cleaved to yield all of the structural and functional proteins present after cellular entry. gag encodes the structural proteins matrix (MA), p12, nucleocapsid (NC), and capsid (CA), and pol encodes the enzymatic proteins protease (PR), reverse transcriptase (RT), and integrase (IN). The exact structure and composition of the PIC are undetermined, so any of these proteins could potentially be engineered to mediate nuclear import. Previous attempts to confer nuclear import capabilities to MLV have relied on rationally inserting nuclear localization signals (NLSs) into the MLV genome or substituting wild-type (WT) MLV domains with analogous HIV regions (11, 35, 48, 51). Some of these modifications resulted in the nuclear localization of viral subcomponents but did not enable the successful infection of nondividing cells (35, 48). The insertion of a NLS into the MA protein of spleen necrosis virus, an avian retrovirus, was reported to mediate successful infection of arrested cells in vitro (43); however, recent attempts to duplicate this result suggest that the effect seen may be limited to the in vitro system utilized and that low titers could have possibly masked native WT ability (6, 28). These studies demonstrate how the complex details of viral intracellular trafficking and nuclear entry make it difficult to determine where to engineer novel functionality without disrupting the native ability of each viral component to properly assemble and infect a cell.
To overcome the need for mechanistic knowledge to modify an integral part of the viral life cycle, we developed a library-based approach to aid in identifying optimal sites for inserting new features or functions. We have applied this capability to generate two libraries that contain a sequence from HIV MA that has been shown to function as a NLS (4) or the simian virus 40 (SV40) NLS (29) inserted in random locations throughout the entire MLV gag-pol sequence. Selection of these libraries via retroviral infection of growth-arrested cells led to the identification of a novel variant capable of infecting nondividing cells in vitro and in vivo, which could be used to study the viral life cycle and to significantly expand the gene therapy applications of MLV. Furthermore, the gag-pol insertion library can be utilized to introduce other desirable viral modifications, and analysis of the optimal locations for new features could yield future insights into viral structure-function relationships. Finally, the library generation and selection method can be expanded to the investigation of other viral genomes and more generally to address other challenges in mammalian protein engineering.
MATERIALS AND METHODS
Cell lines and constructs.
HEK 293T cells were cultured in Iscore’s modified Dulbecco’s medium with 10% fetal bovine serum at 37°C and 5% CO2. The retroviral construct pCLGIT gag-pol was constructed to express enhanced green fluorescent protein (eGFP) from the viral long terminal repeat and to allow for regulation of gag-pol expression by doxycycline (22). The vector construct pCLPUGW expresses eGFP (BD Clontech, Mountain View, CA) from the human ubiquitin C promoter (gift from Carlos Lois) and contains the woodchuck hepadnavirus posttranscriptional regulatory element (37). The helper constructs pCMV gag-pol and pcDNA3 IVS VSV-G express MLV gag-pol and the vesicular stomatitis virus glycoprotein (VSV-G), respectively, from the cytomegalovirus (CMV) immediate-early promoter (53).
Construction of pCLGIT gag-pol.NLS libraries.
The gag-pol.NLS insertion libraries were constructed using a transposon-based approach we have previously described (53). Briefly, a modified transposon containing the kanamycin resistance gene (kanR) was randomly inserted into gag-pol using the MGS kit (Finnzymes, Espoo, Finland) to bookmark insertion sites. The gene library of gag-pol-kanR variants was then cloned into the pCLGIT construct before replacing kanR with the NLS sequences identified in HIV MA (NLSHIV) or SV40 (NLSSV40). The resulting pCLGIT gag-pol.NLS plasmid libraries thus contain MLV constructs that express gag-pol variants with the NLSHIV or NLSSV40 sequence incorporated in a random position. The NLS inserts were constructed by annealing oligonucleotides 5′-GGCCGGAAAGAAGAAGTATAAGTTAAAGCATGG-3′ and 5′-GGCCCCATGCTTTAACTTATACTTCTTCTTTCC-3′ for the NLSHIV insert and 5′-GGCCCCAAAGAAGAAGAGAAA-3′ and 5′-GGCCTTTCTCTTCTTCTTTGG-3′ for the NLSSV40 insert. The library sizes were estimated by colony counting of a dilution of each transformation.
Library production and selection.
The initial vector preparations of the gag-pol.NLS libraries were packaged by calcium phosphate transfection of the plasmid library, pcDNA3 IVS VSV-G, and pCMV gag-pol into 293T cells in the presence of 100 ng/ml doxycycline to suppress the expression of gag-pol.NLS and thereby allow packaging with WT gag-pol. Vector supernatant was harvested after 2 or 3 days and concentrated by ultracentrifugation. This viral library, containing pCLGIT gag-pol.NLS library genomes encapsulated by WT Gag-Pol proteins, was used to infect 293T cells at a multiplicity of infection (MOI) of <0.1. eGFP-positive (eGFP+) cells were fractionated by fluorescence-activated cell sorting on a MoFlo high-speed sorter (Dako, Denmark). Virus was rescued via the transfection of pcDNA3 IVS VSV-G, and harvested virus (round 1 of selection) was then used to infect naïve 293T cells at a MOI of <0.1. eGFP+ cells were again sorted, and the viral rescue process was repeated for each successive round of selection. eGFP expression was measured by flow cytometry, and MOIs were estimated assuming a Poisson distribution for infection. Titers were determined by a linear regression of at least three independent cell populations infected at MOIs between 0.05 and 1.
For initial growth arrest selection, target 293T cells were preincubated for 4 h with 2 μg/ml aphidicolin prior to a 4-h incubation with virus and aphidicolin. Cells were washed with phosphate-buffered saline and incubated in aphidicolin for an additional 16 h before release. Analysis of DNA content by propidium iodide staining at each stage of infection indicated that the majority of cells were arrested after the initial 4-h aphidicolin exposure (data not shown). Infected cells were propagated for at least 10 days before fluorescence-activated cell sorting. For higher-stringency arrests, the aphidicolin concentration was increased and/or the cells were serum starved (0.5% fetal bovine serum) for 48 h before infection. The titers of viral libraries in every round were determined on cycling cells as described above, and the titers were equivalent to the titers with WT gag-pol throughout the growth arrest selection (data not shown).
Clonal isolation and analysis.
After the final selection round, viral genomic RNA was isolated from vectors using the Qiamp viral RNA kit (QIAGEN, Palo Alto, CA). The 5.2-kbp gag-pol sequences were amplified by RT-PCR of five overlapping fragments from 800 bp to 1.2 kbp in size and inserted into a plasmid for DNA sequencing (primer sequences available upon request). Clonal pCMV gag-pol.NLS helper plasmids were constructed by swapping isolated NLS-containing fragments lacking any point mutations into pCMV gag-pol. Vectors were packaged by transient transfection of single or multiple gag-pol helper plasmids with pCLPUGW and pcDNA3 IVS VSV-G. Titers were determined by flow cytometry as described above.
293T cells were growth arrested with 2 μg/ml aphidicolin, 1 μg/ml mitomycin C, or 1 mM hydroxyurea for 24 h. Cells were then exposed to virus and the antimitotic agent for 4 h, washed, and incubated with each drug for an additional 48 h before flow cytometry analysis. Titers on arrested cells were determined by flow cytometry as for cycling cells. Growth arrest was confirmed by propidium iodide staining (data not shown).
Infection of neurons in vitro and in vivo.
Rat hippocampal neuronal cultures (gift from Lu Chen) were isolated as previously described (8). Cytosine arabinoside (5 μM) was added 7 days after plating to inhibit glial division. Two days later, cells were incubated with equal volumes of titer-matched vector stocks for 24 h and cultured for an additional 4 days before fixation and immunofluorescence staining with rabbit anti-GFP (1:2,500; Molecular Probes, Eugene, OR), mouse anti-microtubule-associated protein 2 (a and b forms) [anti-MAP-2(a+b)] (1:250; Sigma, St. Louis, MO), and guinea pig anti-glial fibrillary acidic protein (anti-GFAP) (1:1,000; Advanced Immunochemical, Long Beach, CA) primary antibodies (Abs) and corresponding Alexafluor 488 and 546 (1:250, Molecular Probes)- or Cy3 (1:250; Jackson Immunoresearch, West Grove, PA)-conjugated secondary Abs. Nuclei were counterstained with 4,6-diamidine-2-phenylindole dihydrochloride (DAPI). eGFP+ cells in wells infected with 2 or 5 μl concentrated, high-titer virus were counted.
Animal protocols were approved by the University of California, Berkeley Animal Care and Use Committee in accordance with NIH guidelines. High-titer CLPUGW vectors (∼1010 IU/ml) packaged with pCMV gag-pol or a combination of pCMV gag-pol and pCMV gag-pol.p12.NLS were stereotaxically injected into anesthetized adult female Fischer-344 rats. Three microliters was administered bilaterally into the striatum (n = 6 striata per vector) or hippocampus (n = 4) as we have previously described (32, 53). After 1 week, the brains were harvested, postfixed, sectioned (40 μm), and analyzed by fluorescence confocal microscopy after staining with rabbit anti-GFP (1:2,500), guinea pig anti-GFAP (1:1,000), and mouse anti-NeuN (1:200; Chemicon, Temecula, CA) Abs and the corresponding Alexafluor 488 and 633 (1:250)- and Cy3 (1:250)-conjugated secondary Abs.
RESULTS
Construction and selection of gag-pol.NLS libraries.
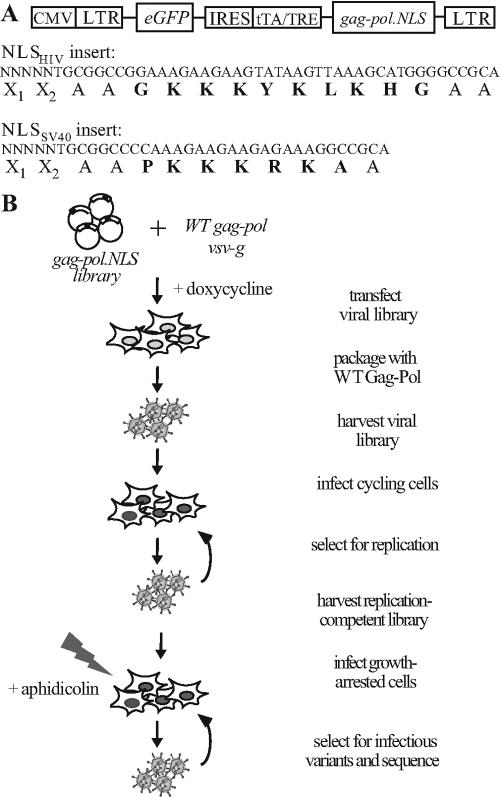
Modified transposons were used to introduce NLSHIV and NLSSV40 into random positions throughout Moloney MLV gag-pol via a library generation method we have recently described (53). While the sequence from SV40 represents a classical, bipartite NLS, it should be noted that there are conflicting reports on the ability of the sequence from HIV MA to direct nuclear localization (4, 19, 29). The resulting gag-pol.NLSHIV or gag-pol.NLSSV40 libraries were inserted into the retroviral construct pCLGIT gag-pol, which expresses eGFP from the viral long terminal repeat and gag-pol under tetracycline control (Fig. 1A). The plasmid libraries were approximately 1 × 105 and 1.4 × 105 in size for the NLSHIV and NLSSV40 libraries, respectively, and thus contained insertions in most or all of the 5,214 internucleotide positions in gag-pol.
FIG. 1.
Library construction and selection. (A) Vector construct and NLS insert sequences. The postinsertion DNA sequences are shown with the amino acid sequence resulting from the desired frame of insertion and the NLS is shown in bold type. X1 and X2 will depend on the 5 host nucleotides (N) duplicated during insertion. LTR, long terminal repeat; IRES, internal ribosome entry site; tTA, tetrecycline-controlled transactivator; TRE, tetracycline response element. (B) Schematic of library selection.
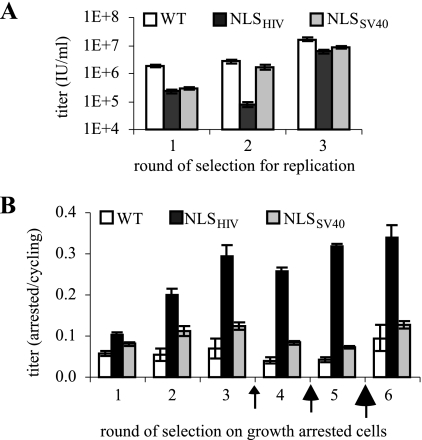
Library and WT vectors were packaged with VSV-G and serially replicated on 293T cells to isolate variants that could yield infectious virions (Fig. 1B). As expected, the initial viral titers were low compared to the WT virus titers, indicating that the majority of the insertions into gag-pol were deleterious. However, by the third round of selection, the library titers reached the same level as the WT virus did (Fig. 2A).
FIG. 2.
Library selection of replication-competent virions that infect growth-arrested cells. (A) Titers of concentrated stocks for each round of selection for replication. (B) Titers for each round of selection on growth-arrested cells, normalized by titers on cycling cells. Arrows indicate an increase in the stringency of cell arrest. Error bars reflect the standard errors of the linear regression used to determine titers.
The resulting replication-competent vector library was then selected on 293T cells incubated with aphidicolin, which arrests cells in G1/S (27). A treatment protocol of 4 h of preincubation, 4 h of viral exposure, and 16 h of postincubation with 2 μg/ml aphidicolin was sufficient to reduce the titer of virus containing WT gag-pol to less than 5% of that on cycling cells. Furthermore, limiting the aphidicolin incubation time to 24 h maintained the viability needed for cell propagation and viral recovery (via the introduction of VSV-G) (3). Both libraries initially showed a modest improvement over WT in their ability to infect arrested cells but steadily improved over subsequent rounds of selection, especially the NLSHIV library (Fig. 2B). The aphidicolin concentration was elevated to 4 μg/ml in round 4 and 6 μg/ml in round 5 to escalate the selective pressure on the libraries. To explore the possibility for multiple NLSs to improve nuclear import (possibly the case for HIV), cells in round 5 were infected at a higher MOI to potentially allow some variants to acquire multiple gag-pol.NLS sequences. That is, the resulting viral progeny could result from recombination between distinct RNA genomes and thereby contain multiple NLS sequences. The final high-stringency selection involved 6 μg/ml aphidicolin after 48 h of serum starvation and infection at an MOI of <0.1.
Isolation of gag-pol.NLS clones.
Viral genomic RNA was extracted from virus rescued from the final selection round, and overlapping fragments of gag-pol were amplified by RT-PCR. NLSHIV insertions were found in two primary locations. The majority of the amplicons with the first 800 bp of gag-pol had a NLSHIV insertion before amino acid (aa) 50 in the viral protein p12 (aa 181 of the polyprotein) in the desired frame. The transposition reaction results in the duplication of 5 host nucleotides at the site of insertion (Fig. 1A), so this could also represent an insertion at aa 52. Additionally, amplification of the first half of pol revealed an in-frame NLSHIV inserted before aa 196 of RT in some clones. Sequencing of remaining regions encoding NC, CA, the latter portion of RT, and IN did not reveal any other insertions.
The NLSSV40 library yielded variable results. The majority of the isolated sequences were identical to the WT sequence, but a few unique insertions were detected in p12 and RT. These were in three different frames and encoded an Arg-rich sequence resulting from a double insert before aa 53 in p12, a Ser-rich sequence before aa 71 in p12, and a Glu-rich sequence before aa 36 in RT. While the Arg-rich insertion is basic, none of these inserts encodes a known NLS; however, they confirm that these sites in gag-pol are permissive to insertion. Amino acid sequences at each insertion are detailed in Table 1.
TABLE 1.
Dominant gp.NLS sequences
| Protein | Amino acid sequencea |
|---|---|
| WT p12 | 41PSDRDGNGE51ATPAGEAPDP61SPMASRLRGR71REPPVADSTT |
| p12.NLSHIV | 41PSDRDGNGEAAAGKKKYKLKHGAAEATPAGEAP |
| p12.NLSSV40-1 | 41PSDRDGNGEATPVRPQRRRERPQRRRERPHPAGEAP |
| p12.NLSSV40-2 | 61SPMASRLRGRRVRPFSSSLGPQREPPVADSTT |
| WT RT | 31QAWAETGGMG…191GFKNSPTLFD |
| RT.NLSHIV | 191GFKNSPTAAGKKKYKLKHGAAPTLFD |
| RT.NLSSV40 | 31QAWAETGCGPKEEEKGRTGGMG |
Newly introduced amino acids are shown in bold type and underlined, and residue positions in mature WT proteins are indicated.
Though the libraries were independently constructed and propagated, there was an insertion in the NLSSV40 library in a position nearly identical to that of the dominant p12 insert in the NLSHIV library. Both identified sites in p12 agree with previous analyses of gag subregions (2, 55), which indicates that the region around aa 50 is particularly tolerant to modification. The identified sites in RT occur in the N-terminal portion and the finger domain of the polymerase region near four highly conserved residues that participate in DNA binding (42). These areas have been previously modified by deletions and site-specific mutations, and this study now identifies them as sites that allow peptide insertions of 12 to 16 aa. While there are likely other permissive sites, such as in the C terminus of IN (46), the emergence of these sites from our high-throughput vector selection indicates they may be particularly able to participate in the production of high-titer virus.
Since the libraries were selected by iterative retroviral replication, point mutations could conceivably have arisen to compensate for or contribute to the performance of the mutants. However, while many clones had several point mutations, none was consistently present in every sequence. Furthermore, individual variants containing only the NLS insert were constructed and analyzed to determine whether the NLS insertion alone was sufficient to confer the ability to infect nondividing cells.
Packaging with gag-pol.NLSHIV clones.
Helper constructs pCMV gp.p12.NLS and pCMV gp.RT.NLS that drive expression of clonal gag-pol.NLSHIV sequences from the CMV promoter were constructed by site-directed insertion of NLSHIV into the novel locations in p12 and RT, respectively. A third clone, pCMV gp.dbl mut, with both insertions was also constructed. As the performance of the NLSSV40 library in the growth arrest assays, as well as the isolated mutant sequences, indicated no significant ability to infect nondividing cells, these variants were not pursued.
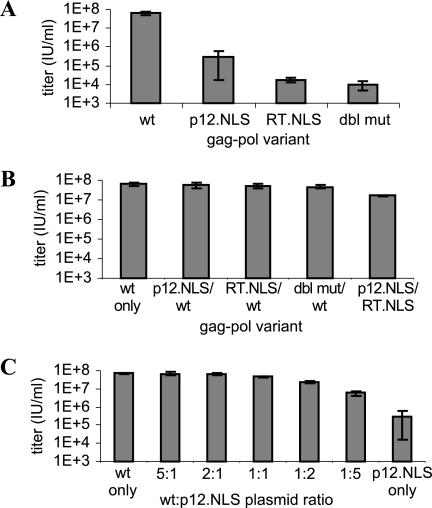
The NLSHIV clones were used to package a retroviral construct expressing eGFP (but no viral proteins), and vectors packaged with mutant gag-pol.NLSHIV in place of WT gag-pol exhibited significantly reduced titers. When vectors were packaged by cotransfection of both the mutant and WT gag-pol plasmids to create mixed Gag-Pol virions, however, the resulting titers were equivalent to vectors packaged with WT plasmid only. Furthermore, packaging by cotransfection of pCMV gag-pol and pCMV gp.p12.NLS at various ratios showed that the titer increased proportionately with the amount of WT plasmid present (Fig. 3).
FIG. 3.
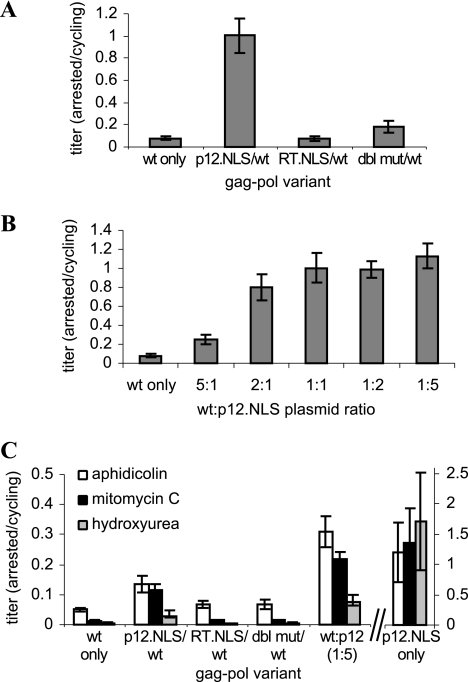
Supernatant titers of vectors packaged with gag-pol.NLSHIV clones. (A) Titers of vectors packaged with individual gag-pol variants. (B) Titers of vectors packaged by transfection of equal amounts of WT gag-pol and mutant gag-pol.NLSHIV plasmids. (C) Titers of vectors packaged by transfection of different ratios of WT gag-pol and gp.p12.NLS plasmids. The total amount of gag-pol plasmid was kept constant for all vector packaging. Error bars represent the standard deviations of two to five biological replicates.
The low titers of vectors packaged with individual mutants seemingly contradict the WT-equivalent titers seen in the library propagation. Low-MOI infections during selection were intended to increase the likelihood that infected cells would contain only one provirus, such that subsequently rescued virus would contain a gag-pol gene packaged inside the Gag-Pol proteins it encodes. However, depending on promoter usage, eGFP expression can underestimate MOI, and infections may have been at higher MOIs than predicted (23, 47). Therefore, copropagation with WT virus (generated through recombination of NLS mutants) may have contributed to high library titers analogous to the copackaging of clonal variants (Fig. 2A and 3B).
Infection of growth-arrested cells with mixed WT/Gag-Pol.NLSHIV virions.
Mixed WT/Gag-Pol.NLSHIV virions were used to infect growth-arrested cells, which were subsequently expanded for 8 days after aphidicolin release to confirm that expression was the result of stable integration events. The mixed WT/RT.NLS vector did not exhibit significant improvement on arrested cells; however, the mixed WT/p12.NLS vector strikingly exhibited the same infectivity on arrested cells as on dividing cells, representing a greater than 12-fold improvement over the WT vector (Fig. 4A). Mixed Gag-Pol vectors packaged at different ratios of WT and mutant plasmids revealed that the enhanced ability to infect growth-arrested cells directly correlated to the levels of gp.p12.NLS plasmid present in the transfection (Fig. 4B).
FIG. 4.
Infectivity of mutant vectors on growth-arrested cells. (A) Infectivity of copackaged variants on cells arrested with aphidicolin for 4 h before, 4 h during, and 16 h after infection. (B) Infectivity of mixed WT/p12.NLS vectors on cells arrested with aphidicolin for 4 h before, 4 h during, and 16 h after infection. (C) Infectivity of mutant vectors on cells arrested with aphidicolin, mitomycin C, or hydroxyurea for 24 h before, 4 h during, and 48 h after infection. To maintain viability, cells were analyzed 48 h after infection, so expression levels may not have reached steady state. Vector titers on arrested cells have been normalized by their titer on cycling cells (Fig. 3). Error bars reflect the standard errors of the linear regression used to determine titers.
The cell growth arrest mechanism and stringency that had been used for library selection were limited to reversible treatments, which left a small percentage of cells permissive to infection by WT virus (Fig. 2B and 4A). We therefore investigated vector performance in high-stringency growth arrest via incubation in aphidicolin 24 h before infection and until the time of analysis. We also infected cells treated with mitomycin C, which arrests cells in G2, and hydroxyurea, which arrests cells in G1/S through a different mechanism than aphidicolin, but which can also reduce infection by inhibiting reverse transcription (27). With all treatments, virions containing some or all gp.p12.NLS-derived proteins exhibited significantly enhanced infectivity compared to WT virions, indicating the ability to infect cells arrested at multiple points in the cell cycle (Fig. 4C). Notably, the low-titer virus packaged exclusively with the gag-pol.p12.NLS helper plasmid exhibited the same level of infectivity on growth-arrested cells as on cycling cells.
Infection of neurons in vitro and in vivo.
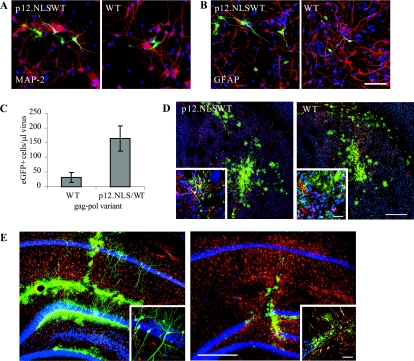
Because antimitotic agents could conceivably influence other aspects of retroviral infection, we further tested the ability of the mixed WT/p12.NLS vector to infect terminally differentiated cells. Primary hippocampal cultures were isolated and infected with high-titer viral stocks. Though cells were cultured in cytosine arabinoside to prevent glial division, eGFP+ cells were visible 2 days later in every well. Immunofluorescence staining 4 days postinfection revealed a small number of eGFP+ cells in wells infected with WT virions that colocalized with the astrocytic marker GFAP. However, a significantly greater number of eGFP+ cells were present in wells infected with mixed WT/p12.NLS virions. The majority of these colocalized with GFAP, but several infected cells also stained positively for MAP-2(a+b), a neuronal marker (Fig. 5A to C).
FIG. 5.
Infection of primary neurons in vitro and in vivo. (A, B) Colocalization of p12.NLS/WT- or WT virus-infected hippocampal cells (eGFP+ [green]) with (A) MAP-2+ neurons (red) and/or (B) GFAP+ astrocytes (red). Cells are counterstained with DAPI (blue). Bar, 40 μm (C) Mean titers and standard deviations (error bars) on hippocampal cultures (values were significantly different [P < 0.005]) (n = 4). Titers on 293T cells were equivalent to each other (P > 0.05 [data not shown]). (D, E) eGFP expression (green) in (D) striatal and (E) hippocampal sections of animals injected with WT/p12.NLS or WT vector. Cells were stained for NeuN (blue) and GFAP (red) to detect neurons and astrocytes, respectively. Bars, 400 μm. Bars in insets, 40 μm.
To determine whether the enhanced infection of terminally differentiated cells was restricted to in vitro culture conditions, we injected high-titer WT or mixed WT/p12.NLS vectors into the striatum and hippocampus of adult rats. A large number of dividing cells, such as GFAP+ glia, were infected by both vector preparations. However, NeuN+ eGFP+ cells were also present in the striata and hippocampal CA1 regions of animals injected with the mixed WT/p12.NLS vector, indicating successful infection of mature neurons, whereas none were observed in animals injected with the WT vector (Fig. 5D and E).
DISCUSSION
To our knowledge, this is the first demonstration of successful infection of nondividing cells both in vitro and in vivo by a gammaretrovirus. Two models have been proposed to explain the discrepancy between the ability of HIV and MLV to infect nondividing cells. The predominant hypothesis is that, unlike HIV, MLV lacks a NLS required for active nuclear import (34). The presence of a NLS has also been implicated in the in vitro infection of nondividing cells by alpharetroviruses, but this has not been confirmed in vivo (24, 26, 30, 31, 44). However, previous MLV variants with NLSs engineered at fixed locations in IN or proviral DNA were unable to infect growth-arrested cells (35, 48). We therefore developed a transposon-based approach to comprehensively probe NLS insertions at possibly every site in the entire gag-pol sequence for the optimal location. Selection of the libraries via low-MOI retroviral infection of growth-arrested cells led to the generation of vectors exhibiting an enhanced phenotype while still retaining WT virus titers, a result previously unachievable using rational design. Specifically, these vectors demonstrated that a single NLS insertion into MLV p12 was sufficient to confer the novel ability to infect nondividing cells.
Although several NLSs have been identified in HIV, there is little consensus as to which, if any, are necessary or sufficient for active nuclear import (13, 49). Whereas studies with HIV have had to rely on loss-of-function deletions, our gain-of-function study with MLV may enable a future complementary analysis of viral nuclear entry mechanisms. For instance, a detailed investigation of the physical composition of mixed Gag-Pol virions packaged by transfection of various ratios of gag-pol plasmids could help identify the amount of p12.NLS protein necessary to impart the novel functionality and strong trends to MLV (Fig. 4B). Notably, though the sequence in HIV MA has been shown to mediate the nuclear localization of conjugated proteins, there is considerable evidence that it does not confer these capabilities to HIV MA itself, suggesting the importance of the specific context of a given sequence (4, 12, 19, 41, 48). Furthermore, as the identified HIV NLSs are believed to interact with different host factors (18, 36, 41), additional or alternate NLSs can be incorporated into MLV via this random insertion approach to explore whether accessing multiple, unique import pathways can further enhance functionality.
A second explanation that has been proposed for the inability of MLV to infect nondividing cells is that, whereas CA rapidly dissociates from HIV after cellular entry, the continued association of MLV CA either makes the PIC too large to efficiently pass through a NPC or shields a possible MLV NLS (16, 17, 45, 51). The successful infection of neurons in vivo by the novel variant (Fig. 5D and E) indicates that the MLV PIC is physically capable of passing through a NPC. While no NLS has yet been identified in WT MLV, we and others have seen low but detectable levels of WT MLV infection on terminally differentiated cells under certain in vitro culture conditions (24, 28, 40). If the rate of CA dissociation were the limiting factor in nuclear entry, a small number of PICs with an uncoated and yet to be identified WT NLS could explain the active nuclear import implied by these results. The significant increase in neuronal infection due to the addition of a novel NLS into p12 does not contradict this possibility, as it likely provides an alternate, accessible avenue for active nuclear entry. Our identification of a permissive insertion site that can influence nuclear entry could also aid the development of novel tools to further delineate whether the limiting factor in MLV nuclear entry is the absence of a native active import mechanism or possibly an indirect result of another postentry event, such as uncoating (52).
The identification of p12 as a successful mediator of intracellular trafficking could serve to address additional questions in MLV biology. Little is known about the structure and intravirion location of p12, but recent genetic studies have suggested a role in nuclear entry and/or sequestration (2, 33, 54-56). Our results strongly imply that p12 is present in the PIC in an exposed location that can influence nuclear transport and potentially support the model that p12 mediates transport of the WT MLV PIC to the proper nuclear location for integration (54). The introduction of a marker (38) into our identified location in p12 could help illuminate its role in WT MLV infection as well as analyze interactions with additional cellular factors that participate in nuclear import and chromosomal attachment of the MLV PIC (reviewed in reference 20). Such information could further aid the study of integration, as differences in host factor associations with the individual NLSs in HIV and chromosomal state of the target cell have both been implicated in integration site determination (5, 9, 10, 15, 18, 36). Interestingly, we found that the steady-state expression levels (i.e., mean fluorescence intensity) in cells infected with virions packaged exclusively with gp.p12.NLS were slightly lower than with virions packaged with WT plasmid, suggesting a possible difference in chromosomal location, since the integrated proviruses are otherwise identical (data not shown). Further analysis with our and other NLS insertions into gag-pol may help determine the influence of nuclear factors and chromosomal state on the integration site preferences of different retroviruses (39).
We have used a high-throughput library generation and selection method to isolate a MLV variant that is capable of infecting growth-arrested cells. Virions packaged by transfection of a mixture of gag-pol.NLS and WT gag-pol plasmids could be generated at a high titer and were able to transduce mature neurons in vitro and in vivo. This novel functionality could help to elucidate details of viral intracellular trafficking and also broadens the possible applications for MLV gene delivery vectors. Future work with these mutants and identified insertion sites may be useful for engineering other desirable vector features, such as site-specific integration. The method utilized to generate the libraries and select for desirable phenotypes directly in mammalian cells could be generally applied to investigate other postentry mechanisms of retroviral restriction, and, more broadly, to the engineering of other mammalian proteins.
Acknowledgments
We thank Lu Chen (UC Berkeley) for providing the primary neuronal cultures and Carlos Lois (Caltech) for pFUGW.
This work was supported by the Whitaker Foundation Graduate Fellowship to J.H.Y. and an NSF career award and NIH NS048248 to D.V.S.
REFERENCES
- 1.Anderson, J. L., and T. J. Hope. 2005. Intracellular trafficking of retroviral vectors: obstacles and advances. Gene Ther. 12:1667-1678. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach, M. R., C. Shu, A. Kaplan, and I. R. Singh. 2003. Functional characterization of a portion of the Moloney murine leukemia virus gag gene by genetic footprinting. Proc. Natl. Acad. Sci. USA 100:11678-11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borel, F., F. B. Lacroix, and R. L. Margolis. 2002. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J. Cell Sci. 115:2829-2838. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. [DOI] [PubMed] [Google Scholar]
- 6.Caron, M. C., and M. Caruso. 2005. A nuclear localization signal in the matrix of spleen necrosis virus (SNV) does not allow efficient gene transfer into quiescent cells with SNV-derived vectors. Virology 338:292-296. [DOI] [PubMed] [Google Scholar]
- 7.Cavazzana-Calvo, M., S. Hacein-Bey, G. de Saint Basile, F. Gross, E. Yvon, P. Nusbaum, F. Selz, C. Hue, S. Certain, J. L. Casanova, P. Bousso, F. L. Deist, and A. Fischer. 2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288:669-672. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., D. M. Chetkovich, R. S. Petralia, N. T. Sweeney, Y. Kawasaki, R. J. Wenthold, D. S. Bredt, and R. A. Nicoll. 2000. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408:936-943. [DOI] [PubMed] [Google Scholar]
- 9.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffi, A., R. S. Mitchell, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. D. Bushman. 2006. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol. Ther. 13:366-373. [DOI] [PubMed] [Google Scholar]
- 11.Deminie, C. A., and M. Emerman. 1994. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J. Virol. 68:4442-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 13.Dvorin, J. D., and M. H. Malim. 2003. Intracellular trafficking of HIV-1 cores: journey to the center of the cell. Curr. Top. Microbiol. Immunol. 281:179-208. [DOI] [PubMed] [Google Scholar]
- 14.Edelstein, M. L., M. R. Abedi, J. Wixon, and R. M. Edelstein. 2004. Gene therapy clinical trials worldwide 1989-2004—an overview. J. Gene Med. 6:597-602. [DOI] [PubMed] [Google Scholar]
- 15.Engelman, A. 2003. The roles of cellular factors in retroviral integration. Curr. Top. Microbiol. Immunol. 281:209-238. [DOI] [PubMed] [Google Scholar]
- 16.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 38:61-85. [DOI] [PubMed] [Google Scholar]
- 21.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 22.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg, K. P., S. F. Gellar, D. V. Schaffer, and J. G. Flannery. Submitted for publication.
- 24.Greger, J. G., R. A. Katz, K. Taganov, G. F. Rall, and A. M. Skalka. 2004. Transduction of terminally differentiated neurons by avian sarcoma virus. J. Virol. 78:4902-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo, and A. Fischer. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348:255-256. [DOI] [PubMed] [Google Scholar]
- 26.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung, D. T., T. F. Jamison, and S. L. Schreiber. 1996. Understanding and controlling the cell cycle with natural products. Chem. Biol. 3:623-639. [DOI] [PubMed] [Google Scholar]
- 28.Jarrosson-Wuilleme, L., C. Goujon, J. Bernaud, D. Rigal, J. L. Darlix, and A. Cimarelli. 2006. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 80:1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 30.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kukolj, G., R. A. Katz, and A. M. Skalka. 1998. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene 223:157-163. [DOI] [PubMed] [Google Scholar]
- 32.Lai, K., B. K. Kaspar, F. H. Gage, and D. V. Schaffer. 2003. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 6:21-27. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. K., K. Nagashima, and W. S. Hu. 2005. Cooperative effect of Gag proteins p12 and capsid during early events of murine leukemia virus replication. J. Virol. 79:4159-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieber, A., M. A. Kay, and Z. Y. Li. 2000. Nuclear import of Moloney murine leukemia virus DNA mediated by adenovirus preterminal protein is not sufficient for efficient retroviral transduction in nondividing cells. J. Virol. 74:721-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llano, M., M. Vanegas, O. Fregoso, D. Saenz, S. Chung, M. Peretz, and E. M. Poeschla. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78:9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868-872. [DOI] [PubMed] [Google Scholar]
- 38.Martin, B. R., B. N. Giepmans, S. R. Adams, and R. Y. Tsien. 2005. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 23:1308-1314. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitrophanous, K., S. Yoon, J. Rohll, D. Patil, F. Wilkes, V. Kim, S. Kingsman, A. Kingsman, and N. Mazarakis. 1999. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 6:1808-1818. [DOI] [PubMed] [Google Scholar]
- 41.Nadler, S. G., D. Tritschler, O. K. Haffar, J. Blake, A. G. Bruce, and J. S. Cleaveland. 1997. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 272:4310-4315. [DOI] [PubMed] [Google Scholar]
- 42.Najmudin, S., M. L. Cote, D. Sun, S. Yohannan, S. P. Montano, J. Gu, and M. M. Georgiadis. 2000. Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain. J. Mol. Biol. 296:613-632. [DOI] [PubMed] [Google Scholar]
- 43.Parveen, Z., A. Krupetsky, M. Engelstadter, K. Cichutek, R. J. Pomerantz, and R. Dornburg. 2000. Spleen necrosis virus-derived C-type retroviral vectors for gene transfer to quiescent cells. Nat. Biotechnol. 18:623-629. [DOI] [PubMed] [Google Scholar]
- 44.Parveen, Z., M. Mukhtar, M. Rafi, D. A. Wenger, K. M. Siddiqui, C. A. Siler, B. Dietzschold, R. J. Pomerantz, M. J. Schnell, and R. Dornburg. 2003. Cell-type-specific gene delivery into neuronal cells in vitro and in vivo. Virology 314:74-83. [DOI] [PubMed] [Google Scholar]
- 45.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth, M. J. 1991. Mutational analysis of the carboxyl terminus of the Moloney murine leukemia virus integration protein. J. Virol. 65:2141-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sastry, L., T. Johnson, M. J. Hobson, B. Smucker, and K. Cornetta. 2002. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 9:1155-1162. [DOI] [PubMed] [Google Scholar]
- 48.Seamon, J. A., K. S. Jones, C. Miller, and M. J. Roth. 2002. Inserting a nuclear targeting signal into a replication-competent Moloney murine leukemia virus affects viral export and is not sufficient for cell cycle-independent infection. J. Virol. 76:8475-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 50.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 51.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita, M., and M. Emerman. 2005. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, J. H., and D. V. Schaffer. 2006. Engineering retroviral and lentiviral vectors by selection of a novel peptide insertion library for enhanced purification. J. Virol. 80:3285-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, B., A. Fassati, A. Yueh, and S. P. Goff. 2002. Characterization of Moloney murine leukemia virus p12 mutants blocked during early events of infection. J. Virol. 76:10801-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yueh, A., and S. P. Goff. 2003. Phosphorylated serine residues and an arginine-rich domain of the Moloney murine leukemia virus p12 protein are required for early events of viral infection. J. Virol. 77:1820-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]