Abstract
The high biodiversity in tropical seas provides a long-standing challenge to allopatric speciation models. Physical barriers are few in the ocean and larval dispersal is often extensive, a combination that should reduce opportunities for speciation. Yet coral reefs are among the most species-rich habitats in the world, indicating evolutionary processes beyond conventional allopatry. In a survey of mtDNA sequences of five congeneric west Atlantic reef fishes (wrasses, genus Halichoeres) with similar dispersal potential, we observed phylogeographical patterns that contradict expectations of geographical isolation, and instead indicate a role for ecological speciation. In Halichoeres bivittatus and the species pair Halichoeres radiatus/brasiliensis, we observed strong partitions (3.4% and 2.3% divergence, respectively) between adjacent and ecologically distinct habitats, but high genetic connectivity between similar habitats separated by thousands of kilometres. This habitat partitioning is maintained even at a local scale where H. bivittatus lineages are segregated between cold- and warm-water habitats in both Bermuda and Florida. The concordance of evolutionary partitions with habitat types, rather than conventional biogeographical barriers, indicates parapatric ecological speciation, in which adaptation to alternative environmental conditions in adjacent locations overwhelms the homogenizing effect of dispersal. This mechanism can explain the long-standing enigma of high biodiversity in coral reef faunas.
Keywords: biodiversity, coral reef, marine biogeography, mtDNA, speciation
1. Introduction
Allopatric speciation models, initially proposed by Dobzhansky (1937) and Mayr (1942), have been a dominant paradigm in evolutionary biology for over half a century. However, Mayr (1954) recognized that evolution in the sea may operate under a different set of rules, given the difficulties with applying allopatric models to marine speciation. First, there are few opportunities for geographical isolation in a circumglobal aquatic medium. Second, most marine organisms have a pelagic larval stage that has tremendous potential for dispersal (Palumbi 1994; Mora & Sale 2002), a potential that is often realized (Lessios et al. 1998). However, while both of these factors would be expected to reduce opportunities for speciation, coral reefs host some of the most diverse species assemblages. Reinforcing these concerns about the importance of allopatry to speciation in the sea are an abundance of congeneric species whose distributions overlap and for which no apparent geographical barrier exists to explain the corresponding evolutionary partitions.
While allopatry is undoubtedly an important factor in marine speciation, it cannot account for all coral reef biodiversity because it requires complete isolation between incipient species, a condition difficult to meet in a fluid medium where most organisms have a pelagic dispersing phase. A neglected alternative is parapatric ecological speciation, in which natural selection in alternative environments at adjacent locations overrides gene flow, and drive populations along separate evolutionary pathways (Schluter 2001; Coyne & Orr 2004). It is now widely recognized that divergent selection in contrasting environments can lead directly or indirectly to reproductive isolation, and examples of ecological speciation have been identified in terrestrial (Smith et al. 1997; Ogden & Thorpe 2002) and freshwater habitats (Lu & Bernatchez 1999; Morell 1999; Schluter 2001). However, ecological speciation has not been previously proposed for tropical reef fishes, mainly because of the assumption that a highly dispersive larval stage precludes habitat-specific adaptation (Warner 1997). This assumption has eroded in recent years, with observations of local retention of reef-fish larvae (Jones et al. 1999; Swearer et al. 2002), active habitat choice by larvae (Bierne et al. 2003) and concordant evidence of reduced gene flow over short geographical distances (Taylor & Hellberg 2003). The findings of larval retention and reduced gene flow resurrect the possibility that ecological partitions can drive speciation, especially when contrasting environments are in geographically separated but potentially connected locations (parapatry), and thereby may help explain the high biodiversity in tropical seas.
General expectations of parapatric ecological speciation are that: (i) evolutionary partitions align with habitat distributions rather than geographical and oceanographic barriers; and (ii) contact zones between different lineages are often located in areas with mixed environmental conditions. To resolve evolutionary patterns in reef fishes, we analysed mitochondrial DNA sequences (707 bp of mtDNA cytochrome b; n=508) in five congeneric wrasses that inhabit the tropical western Atlantic. To assess the degree of corresponding habitat differences among species, habitat-association data for all available species were obtained from three locations in the Caribbean, and three in Brazil.
The primary biogeographical barrier for these and other reef-associated organisms in the western Atlantic is the outflow of the Amazon and Oronoco Rivers (Briggs 1974; Rocha 2003). This ‘Amazon barrier’ includes 2300 km of muddy coastline, which lacks coral reefs and separates the reef fauna of the Greater Caribbean (Trinidad to Bermuda) from that of its Brazilian counterpart, which includes the mainland south of the Equator and adjacent oceanic islands (figure 1; Rocha 2003). It is important to note, however, that a wedge of salt water with sponge habitat and a corresponding community of fishes, which includes many reef species, exists under the freshwater plume of the Amazon outflow (Collette & Rützler 1977).
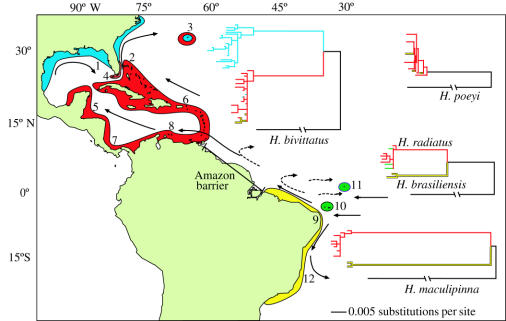
Figure 1.
Western Atlantic Ocean with sampled locations, main oceanic currents, ecological regions discussed in the text (blue for subtropical; red and green for tropical, mostly insular and clear water; yellow for tropical continental and turbid water) and simplified phylogenetic trees for the habitat generalists Halichoeres bivittatus, H. poeyi and the reef specialists H. radiatus, H. brasiliensis and H. maculipinna (complete trees in Electronic Appendix; figures 3–6). Branch lengths are according to indicated scale; the branches leading to out-groups (in black) were reduced by 75%. Colours of branches in trees correspond to colours of locations on map. Solid arrows indicate direction of mean oceanic currents; broken arrows indicate direction of seasonal currents. Sampling locations: 1, Gulf of Mexico; 2, Bahamas; 3, Bermuda; 4, Florida Keys; 5, Belize; 6, St Croix USVI; 7, Panama; 8, Venezuela; 9, northeastern Brazil; 10, Fernando de Noronha; 11, St Paul's Rocks; 12, southeastern Brazil.
Reef habitats vary significantly within and between Brazil and the Caribbean. The Brazilian continental coast is characterized by turbid waters, heavy freshwater runoff and fine terrigenous substrata (Leao & Dominguez 2000). By contrast, nearby oceanic islands have clear waters and lower primary productivity, and are rich in calcareous sediments. Likewise, the subtropical coast of North America has continental conditions (Avise 1992), while adjacent offshore locations in the Bahamas and Caribbean have typical tropical reef characteristics (Robins 1971). Thus, the Amazon barrier defines northern and southern biogeographical provinces, within each of which there is a strong ecological distinction between continental and insular reef habitats, and a steep temperature decline towards higher latitudes. Samples were collected at continental and insular locations (also tropical and subtropical), on both sides of the Amazon barrier.
2. Material and methods
(a) Specimen collection and DNA extraction
A total of 508 tissue samples was collected from five widely distributed wrasses (family Labridae) across their entire geographical range: Halichoeres bivittatus (n=225); Halichoeres radiatus (n=116) and its Brazilian sister Halichoeres brasiliensis (n=22); Halichoeres maculipinna (n=86) and Halichoeres poeyi (n=59; see figure 1). Fishes were collected with pole-spears or hand nets while scuba diving or snorkelling. Larvae were collected with light traps. Tissue samples were stored in a saturated salt–DMSO buffer. DNA was extracted by using standard protocols (Hillis et al. 1996).
(b) PCR and sequencing
Initial cytochrome b sequences were obtained using the primers L14725 (5′ GTG ACT TGA AAA ACC ACC GTT G 3′) and H15573 (5′ AAT AGG AAG TAT CAT TCG GGT TTG ATG 3′), which were also successfully used in other fishes (Meyer 1993). These primers did not work consistently among species, so the internal primers L14768 (5′ ACC CAC CCA CTC CTT AAA ATC 3′) and H15496 (5′ GTG AAA TTA TCT GGG TCT CCA A 3′) were designed and worked well in Halichoeres spp. Thermal cycling in polymerase chain reactions and sequencing reactions followed steps described in detail elsewhere (Rocha et al. 2002). All samples were sequenced in the forward direction. To ensure accuracy of nucleotide designations, rare and questionable haplotypes were sequenced in both directions. The sequences reported in this paper have been deposited in the GenBank database with accession numbers AY591354–AY591365 and AY823558–AY823581.
(c) Data analysis
Population structure was assessed with an analysis of molecular variance (AMOVA; Excoffier et al. 1992) using the program Arlequin v. 2.0 (Schneider et al. 2000) using sequence divergence estimates (d-values) calculated with the method of Tamura & Nei (1993). For each species, a Mantel test of isolation by distance was conducted with the program IBD (Bohonak 2002) and selected chi-squared-tests (χ2) were performed with Microsoft Excel. The nucleotide substitution models (TrN+I, with I=0.8157 for H. radiatus/brasiliensis, and TrN+gama, with gama=0.01 for H. bivittatus, H. poeyi and H. maculipinna) for all datasets were optimized using an Akaike information criterion approach and hierarchical likelihood ratio tests (Posada & Crandall 1998). Trees in figure 2 were initiated with the neighbour-joining method (Saitou & Nei 1987) and were further searched by 107 tree bisection and reconnection iterations under the minimum evolution criterion (Swofford 2002). Additionally, maximum-parsimony analyses were performed and topological confidence was evaluated with 1000 bootstrap replicates (Felsenstein 1985). Equal weighting of all three codon positions was used.
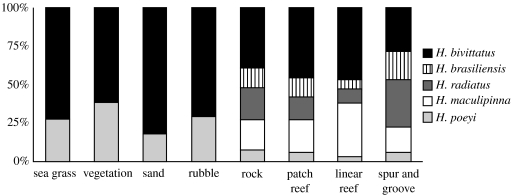
Figure 2.
Proportional abundances of five Halichoeres species in various habitat types in and around coral and rocky reefs at six locations. Although the density of H. bivittatus and H. poeyi was lower in sand/seagrass versus reef, they were the only species consistently observed in non-reef habitats. The species that occur only in reefs (the pair H. brasiliensis/radiatus, and H. maculipinna) show phylogenetic breaks between continental and insular reefs (see figure 1 and supporting figures 3–6).
(d) Habitat preferences
The presence and abundance of wrasses at different habitat types were determined using the stationary visual census technique (Bohnsack & Bannerot 1986). A tape-measure 15 m long was placed at the point to be counted and all fishes observed within or passing through a 7.5 m radius from the centre of the tape, extending upwards in a cylinder above the bottom, were counted during a 15 min period. The censuses were done during the day at depths between 1 m and 50 m. Locations were randomly chosen, but only one census was performed at each point. Data from a total of 638 censuses carried out between 1998 and 2003 at the Florida Keys, the US Virgin Islands (St Croix and St John), and Brazilian coastal (Parcel Manuel Luiz and Paraiba) and oceanic sites (Atol das Rocas) were combined in figure 2. All censuses were carried out by teams with the participation of or led by L.A.R., except for censuses at St Croix, which were carried out by a NOAA team led by Chris Caldow.
3. Results
Four factors are routinely used to explain genetic structure in marine biota: biogeographical barriers, ocean currents, oceanic expanses (distance) and pelagic larval duration (Shulman & Bermingham 1995; Lessios et al. 1998; Barber et al. 2000; Planes 2002). Based on these four factors, we would expect congruent genetic patterns in western Atlantic wrasses because they are distributed across the same biogeographical provinces, are subject to the same ocean currents that disperse their larvae, and have similar pelagic larval durations of 25–30 days (Sponaugle & Cowen 1997; Wellington & Robertson 2001). Contrary to these expectations, we observed patterns ranging from extensive sharing of haplotypes among locations in H. poeyi and H. bivittatus, to a phylogenetic break in H. radiatus/brasiliensis (d=0.024), and an ancient Caribbean/Brazil partition in H. maculipinna (d=0.065). This diversity of outcomes is matched by a diversity of habitat preferences. H. poeyi and H. bivittatus were observed in all sampled habitats (which ranged from sea-grass to coral reef), whereas the pair H. radiatus/brasiliensis and H. maculipinna are reef specialists (figure 2).
In both H. bivittatus and H. radiatus/brasiliensis, the strongest genetic partitions were within, rather than between, biogeographical provinces. Although population genetic separations of Φst=0.14–0.62 were observed across the Amazon barrier in H. radiatus (table 1), the populations at Fernando de Noronha (oceanic island off Brazil) and St Croix (oceanic island in the central Caribbean) were not significantly different. Despite being separated by 4200 km, most specimens collected at Fernando de Noronha and St Croix had an identical mtDNA sequence. By contrast, this offshore Brazil population was distinguished by d=2.3% from its look-alike sister species H. brasiliensis on the adjacent Brazilian coastline, only 360 km away (figure 1). Similarly, H. bivittatus maintains genetic homogeneity between the offshore reefs of Belize and St Croix in the Lesser Antilles, separated by 2430 km, but displays a divergence of 3.4% between the tropical Bahamas and subtropical Florida, separated by only 300 km (table 1). Hence, biogeographical provinces did not forecast genetic separations in either of these cases. To assess whether distance is a predictor of divergence we used Mantel tests of population separations (Φst values) versus geographical distance. Only one out of four species, H. maculipinna (a reef specialist with d=0.065 across the Amazon) showed a significant relationship between genetic and geographical distance (r=0.805, p=0.029). Thus, in three out of four cases, both distance and biogeographical barriers failed to predict patterns of genetic divergence.
Table 1.
Population genetic subdivision among selected geographical locations in five species of Halichoeres. (Asterisk (*) represents comparisons of localities in different environments (tropical versus subtropical in H. bivittatus; inshore versus offshore in H. radiatus/brasiliensis, H. maculipinna and H. poeyi). Symbols used: PLD, mean pelagic larval duration in days; distance, straight line distance in km; ΦST, genetic separation between populations (negative values indicate very high gene flow); TN, mean sequence divergence calculated using the Tamura–Nei substitution model (a dash indicates that locations share one or more haplotypes); **significant population structure at α=0.05.)
| pairwise comparisons | PLD (days) | distance (km) | ΦST | TN distance |
|---|---|---|---|---|
| Halichoeres bivittatus | 24.2 | |||
| Bermuda inshore–Bermuda offshore* | 2 | 0.57** | 0.02–3.6% | |
| Florida inshore–Florida offshore* | 10 | 0.65** | 0.02–3.6% | |
| Gulf of Mexico–Bahamas* | 350 | 0.91** | 3.5% | |
| Gulf of Mexico–Belize* | 1200 | 0.94** | 3.6% | |
| Bahamas–St Croix (USVI) | 1580 | 0.001 | — | |
| Panama–Bahamas | 1860 | 0.004 | — | |
| Gulf of Mexico–Panama* | 2000 | 0.93** | 3.5% | |
| Gulf of Mexico–Venezuela* | 2100 | 0.94** | 3.6% | |
| St Croix–Belize | 2430 | 0.04 | — | |
| St Croix–NE Brazil | 3100 | 0.59** | 0.04% | |
| Florida inshore–NE Brazil* | 4800 | 0.80** | 3.6% | |
| Florida offshore–NE Brazil | 4800 | 0.25** | 0.02–3.6% | |
| Belize–NE Brazil | 5100 | 0.50** | — | |
| Halichoeres radiatus and brasiliensis | 26 | |||
| F. de Noronha–NE Brazil* | 340 | 0.96** | 2.4% | |
| St Paul's Rocks–NE Brazil* | 1000 | 0.98** | 2.4% | |
| St Croix–Bermuda | 1550 | 0.01 | — | |
| St Paul's Rocks–St Croix | 3900 | 0.62** | — | |
| F. de Noronha–St Croix | 4200 | 0.14 | — | |
| Halichoeres maculipinna | 29 | |||
| Florida Keys–Bahamas | 100 | −0.10 | — | |
| Florida Keys–Belize | 980 | −0.07 | — | |
| Florida Keys–Venezuela | 1780 | −0.10 | — | |
| NE Brazil–SE Brazil | 3000 | 0.01 | — | |
| St Croix–NE Brazil* | 3100 | 0.97** | 6.6% | |
| Venezuela–NE Brazil* | 3200 | 0.97** | 6.5% | |
| Halichoeres poeyi | 25 | |||
| St Croix–Belize | 2430 | 0.05 | — | |
| NE Brazil–SE Brazil | 3000 | −0.04 | — | |
| St Croix–NE Brazil* | 3100 | 0.32** | — | |
| Belize–NE Brazil* | 5100 | 0.34** | — |
4. Discussion
Could oceanic currents isolate continents and offshore island populations of wrasses? Briggs (1974) suggested that the Gulf Stream, which flows between Florida and the Bahamas, may prevent larvae from colonizing across the Florida Strait. However, our data show that there is no such isolating mechanism in two cases. First, the two lineages of H. bivittatus co-occur in south Florida and Bermuda (with mixed tropical and subtropical characteristics). Second, the sister species pair H. radiatus/brasiliensis co-occur on outer-shelf reefs (with mixed insular and continental characteristics) on the northeastern Brazilian coast. Moreover, it is exceedingly unlikely that these species can maintain genetic connectivity across thousands of kilometres of open-ocean (table 1) and yet be incapable of crossing much shorter distances between offshore islands and continental coastline.
In the absence of successful predictions based on geography, oceanography and pelagic larval duration, two observations provide powerful evidence that ecological factors guide evolutionary processes in this genus. First, the genetic lineages were distributed according to environmental differences: phylogenetic breaks were observed between adjacent but ecologically distinct habitats, whereas high genetic connectivity was observed between similar habitats separated by thousands of kilometres. The partitions observed in H. bivittatus (between tropical and subtropical habitats) and in H. radiatus/brasiliensis (between continental and oceanic island habitats) persist even at a local scale. In Bermuda, offshore reefs rarely cool below 18 °C, a temperature tolerated by most tropical reef fishes. However, temperatures in inshore channels and bays there can drop below 10 °C (Smith-Vaniz et al. 1999). At that site, all H. bivittatus collected inshore were of the subtropical type, whereas half of the specimens collected on offshore reefs were of the tropical type (table 2). Similarly, in the Florida Keys, the subtropical type was abundant in inshore channels, whereas the tropical dominated on warmer offshore reefs (table 2). This distribution of lineages is significantly different between inshore and offshore habitats, which are separated by less than 5 km in Bermuda and less than 15 km in Florida. Thus, the hypothesis of a random distribution of lineages between those habitats was rejected; χ2=23.14, p<0.01 for Florida and χ2=9.11, p<0.01 for Bermuda. At offshore reefs in the northern Gulf of Mexico (Cedar Key), which endure subtropical temperatures in the winter (14 °C), all specimens were the subtropical type. The habitat segregation by cryptic lineages observed in H. bivittatus is the first such example described for any reef fishes not accompanied by obvious morphological differences.
Table 2.
Frequency distribution of lineage types of H. bivittatus in inshore channels with cold winter water versus offshore reefs with constant warm water.
| habitat type | n tropical lineage | n subtropical lineage |
|---|---|---|
| Florida inshore | 1 | 15 |
| Florida offshore | 16 | 4 |
| Bermuda inshore | 0 | 14 |
| Bermuda offshore | 6 | 6 |
A parallel situation occurs with the H. radiatus/brasiliensis lineage. On the continental shelf of northeastern Brazil, the insular species H. radiatus, which can be distinguished from H. brasiliensis by colour pattern (Rocha & Rosa 2001), was never recorded on near-shore reefs, although two individuals were collected at deeper reefs (more than 30 km from the coast) on the continental shelf. By contrast, the continental species (H. brasiliensis) is abundant on near-shore reefs (from tidal pools to 20 km from the coast), but rare at the insular-like offshore reefs.
A second indication of ecological influences on the evolution of western Atlantic Halichoeres is the positive relationship between genetic divergence and the degree of specialization, a pattern also observed in other taxa (Smith & Fujio 1982; Lu & Bernatchez 1999). Although H. bivittatus shows a sharp break between tropical and subtropical habitat, we observed no phylogenetic breaks across tropical habitats for this generalist species. Similarly, there are no phylogenetic breaks (only modest population structure) in the other generalist (H. poeyi; figure 2), which occurs on reefs as well as in sea-grass, sand, rubble and ‘continental-like’ near-shore reefs at insular locations. By contrast, deep breaks occur in the H. radiatus/brasiliensis pair and H. maculipinna, which were observed only on reefs (figure 2). The deepest genetic partition (d=6.5%) was observed in H. maculipinna (figure 1), a reef-dweller with the most specialized diet and feeding apparatus among the surveyed species (Wainwright 1988).
The ecogeographical partitions described here are not unique to wrasses. Notably, a similar relationship between genetic divergence and habitat specialization has been observed among west Atlantic surgeon-fishes that occur on both sides of the Amazon barrier (Rocha et al. 2002). Moreover, many tropical reef fishes have distributions that do not extend across the narrow straits between continental and offshore reefs (Robins 1971; Williams 1991; Rocha 2003), and at least one other west Atlantic reef fish surveyed by our team has a phylogeographical pattern nearly identical to that of H. bivittatus (Carlin et al. 2003). Ecological differentiation may also explain patterns recently reported in phylogeographical studies of marine species. These cases include sympatric speciation in sea horses (Jones et al. 2003) and gobies (Dawson et al. 2002), phylogenetic divergence between parrotfish in seagrass and coral reef habitat (Streelman et al. 2002), and genetic partitions among habitat-segregated colour morphs of hamlets (McCartney et al. 2003) and cleaner gobies (Taylor & Hellberg 2003).
In the case of cleaner gobies, Taylor & Hellberg (2003) document limited larval dispersal and abrupt genetic discontinuities among Caribbean populations separated by only 20 km. Genetic breaks are coupled with coloration differences that are probably under intense selection. As noted by Palumbi & Warner (2003), colour morphs that are not recognized as cleaners would be regarded as prey, such that larvae of a blue colour morph may disperse to some limited extent, but would fare poorly in an area where white colour morphs predominate. Parapatric ecological speciation, facilitated by limited larval dispersal, may well play a role in diversification of these cleaner gobies.
Mayr (1954) ultimately decided that while mechanisms of speciation in the sea are the same as those on land, they operate on different geographical and temporal scales in those two environments. With the proposal of ecological speciation in reef fishes, the entire suite of evolutionary mechanisms recognized for the terrestrial fauna (including isolation, hybridization, drift, mutation and selection) would apply to the marine realm, potentially paving the way for a synthesis of speciation in both spheres.
Acknowledgments
We thank A. L. Bass, H. Choat, B. B. Collette, M. Coura, B. M. Feitoza, C. E. Ferreira, S. R. Floeter, J. L. Gasparini, Z. Hillis-Star, S. Karl, D. Murie, D. Parkyn, B. Philips, J. Pitt, C. Rocha, I. L. Rosa, R. S. Rosa, S. Sponaugle, B. Victor and D. Weaver for help with sampling and laboratory work. Discussions with D. Bellwood, R. Chapman, M. Courtney, S. A. Karl, N. Knowlton, C. Gilbert, W. S. Grant, H. Lessios, G. Paulay, C. St Mary, W. F. Smith-Vaniz and P. Taylor greatly improved the manuscript. Visual census data from the US Virgin Islands were provided by C. Caldow. This research was supported by CAPES Brazilian Ministry of Education, the Brazilian Navy, PADI Project Aware, the Smithsonian Tropical Research Institute and the US National Science Foundation (DEB 9727048).
Footnotes
As this paper exceeds the maximum length normally permitted, the authors have agreed to contribute to production costs.
The supplementary Electronic Appendix is available at http://dx.doi.org/rspb.2004.3003 or via http://www.journals.royalsoc.ac.uk.
References
- Avise J.C. Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos. 1992;63:62–76. [Google Scholar]
- Barber P.H, Palumbi S.R, Erdmann M.V, Moosa M.K. A marine Wallace's line? Nature. 2000;406:406–407. doi: 10.1038/35021135. [DOI] [PubMed] [Google Scholar]
- Bierne N, Bonhomme F, David P. Habitat preference and the marine-speciation paradox. Proc. R. Soc. B. 2003;270:1399–1406. doi: 10.1098/rspb.2003.2404. http://dx.doi.org/doi:10.1098/rspb.2003.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack J.A, Bannerot S.P. A stationary visual census technique for quantitavively assessing community structure of coral reef fishes. NOAA Tech. Rep. 1986;41:1–15. [Google Scholar]
- Bohonak A.J. IBD (isolation by distance): a program for analyses of isolation by distance. J. Hered. 2002;93:153–154. doi: 10.1093/jhered/93.2.153. [DOI] [PubMed] [Google Scholar]
- Briggs J.C. McGraw-Hill; New York: 1974. Marine zoogeography. [Google Scholar]
- Carlin J.L, Robertson D.R, Bowen B.W. Ancient divergences and recent connections in two tropical Atlantic reef fishes: Epinephelus adscensionis and Rypticus saponaceous (Percoidei: Serranidae) Mar. Biol. 2003;143:1057–1069. [Google Scholar]
- Collette B.B, Rützler K. Reef fishes over sponge bottoms off the mouth of the Amazon River. Proc. 3rd Int. Coral Reef Symp. 1977;1:305–310. [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer Associates; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Dawson M.N, Louie K.D, Barlow M, Jacobs D.K, Swift C.C. Comparative phylogeography of sympatric sister species, Clevelandia ios and Eucyclogobius newberryi (Teleostei, Gobiidae), across the California transition zone. Mol. Ecol. 2002;11:1065–1075. doi: 10.1046/j.1365-294x.2002.01503.x. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Columbia University Press; New York: 1937. Genetics and the origin of species. [Google Scholar]
- Excoffier L, Smouse P.E, Quatro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Hillis D.M, Mable B.K, Larson A, Davis S.K, Zimmer E.A. Nucleic acids. IV. Sequencing and cloning. In: Hillis D.M, Moritz C, Mable B.K, editors. Molecular systematics. Sinauer Associates; Sunderland, MA: 1996. pp. 321–381. [Google Scholar]
- Jones G.P, Milicich M.J, Emslie M.J, Lunow C. Self-recruitment in a coral reef fish population. Nature. 1999;402:802–804. [Google Scholar]
- Jones A.G, Moore G.I, Kvarnemo C, Walker D, Avise J.C. Sympatric speciation as a consequence of male pregnancy in seahorses. Proc. Natl Acad. Sci. USA. 2003;100:6598–6603. doi: 10.1073/pnas.1131969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao Z.M.A.N, Dominguez J.M.L. Tropical coast of Brazil. Mar. Pollut. Bull. 2000;41:112–122. [Google Scholar]
- Lessios H.A, Kessing B.D, Robertson D.R. Massive gene flow across the world's most potent marine biogeographic barrier. Proc. R. Soc. B. 1998;265:583–588. http://dx.doi.org/doi:10.1098/rspb.1998.0334 [Google Scholar]
- Lu G, Bernatchez L. Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): support for the ecological speciation hypothesis. Evolution. 1999;53:1491–1505. doi: 10.1111/j.1558-5646.1999.tb05413.x. [DOI] [PubMed] [Google Scholar]
- McCartney M.A, Acevedo J, Heredia C, Rico C, Quenoville B, Bermingham E, McMillan W.O. Genetic mosaic in a marine species flock. Mol. Ecol. 2003;12:2963–2973. doi: 10.1046/j.1365-294x.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- Mayr E. Columbia University Press; New York: 1942. Systematics and the origin of species. [Google Scholar]
- Mayr E. Geographic speciation in tropical echinoids. Evolution. 1954;8:1–18. [Google Scholar]
- Meyer A. Evolution of mitochondrial DNA in fishes. In: Hochanchka P.W, Mommsen T.P, editors. Biochemistry and molecular biology of fishes. vol. 2. Elsevier; New York: 1993. pp. 1–38. [Google Scholar]
- Mora C, Sale P.F. Are populations of coral reef fish open or closed? Trends Ecol. Evol. 2002;17:422–428. [Google Scholar]
- Morell V. Ecology returns to speciation studies. Science. 1999;284:2106–2108. doi: 10.1126/science.284.5423.2106. [DOI] [PubMed] [Google Scholar]
- Ogden R, Thorpe R.S. Molecular evidence for ecological speciation in tropical habitats. Proc. Natl Acad. Sci. USA. 2002;99:13 612–13 615. doi: 10.1073/pnas.212248499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi S.R. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 1994;25:547–572. [Google Scholar]
- Palumbi S.R, Warner R.R. Why gobies are like hobbits. Science. 2003;299:51–52. doi: 10.1126/science.1080775. [DOI] [PubMed] [Google Scholar]
- Planes S. Biogeography and larval dispersal inferred from population genetic analysis. In: Sale P.F, editor. Coral reef fishes. Dynamics and diversity on a complex ecosystem. Academic Press; New York: 2002. pp. 201–220. [Google Scholar]
- Posada D, Crandall K.A. Model test: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Robins C.R. Distributional patterns of fishes from coastal and shelf waters of the tropical western Atlantic. FAO Fish. Rep. 1971;71,72:249–255. [Google Scholar]
- Rocha L.A. Patterns of distribution and processes of speciation in Brazilian reef fishes. J. Biogeogr. 2003;30:1161–1171. [Google Scholar]
- Rocha L.A, Rosa R.S. Halichoeres brasiliensis (Bloch, 1791), a valid wrasse species (Teleostei: Labridae) from Brazil, with notes on the Caribbean species Halichoeres radiatus (Linnaeus, 1758) Aqua J. Ichthyol. Aquat. Biol. 2001;4:161–166. [Google Scholar]
- Rocha L.A, Bass A.L, Robertson D.R, Bowen B.W. Adult habitat preferences, larval dispersal, and the comparative phylogeography of three Atlantic surgeonfishes (Teleostei: Acanthuridae) Mol. Ecol. 2002;11:243–252. doi: 10.1046/j.0962-1083.2001.01431.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends Ecol. Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Laboratory; University of Geneva, Switzerland: 2000. Arlequin, Version 2.0: a software for population genetics data analysis. [Google Scholar]
- Shulman M.J, Bermingham E. Early life histories, ocean currents and the population genetics of Caribbean reef fishes. Evolution. 1995;49:1041–1061. doi: 10.1111/j.1558-5646.1995.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Smith P.J, Fujio Y. Genetic variation in marine teleosts: high variability in habitat specialist and low variability in habitat generalists. Mar. Biol. 1982;69:7–20. [Google Scholar]
- Smith T.B, Wayne R.K, Girman D.J, Bruford M.W. A role for ecotones in generating rainforest biodiversity. Science. 1997;276:1855–1857. [Google Scholar]
- Smith-Vaniz W.F, Collette B.B, Luckhurst B.E. Special publication No. 4. Allen Press, Inc; Lawrence, KS: 1999. Fishes of Bermuda: history, zoogeography, annotated checklist, and identification keys. [Google Scholar]
- Sponaugle S, Cowen R.K. Early life history traits and recruitment patterns of Caribbean wrasses (Labridae) Ecol. Monogr. 1997;67:177–202. [Google Scholar]
- Streelman J.T, Alfaro M, Westneat M.W, Bellwood D.R, Karl S.A. Evolutionary history of the parrotfishes: biogeography, ecomorphology, and comparative diversity. Evolution. 2002;56:961–971. doi: 10.1111/j.0014-3820.2002.tb01408.x. [DOI] [PubMed] [Google Scholar]
- Swearer S.E, Shima J.S, Hellberg M.E, Thurrold S.R, Jones G.P, Robertson D.R, Morgan S.G, Selkoe K.A, Ruiz G.M, Warner R.R. Evidence of self-recruitment in demersal marine populations. Bull. Mar. Sci. 2002;70:251–271. [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 2002. Phylogenetic Analysis Using Parsimony (* and other methods). Version 4.0b10. [Google Scholar]
- Tamura K, Nei M. Estimating the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;15:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Taylor M.S, Hellberg M.E. Genetic evidence for local recruitment of pelagic larvae in a Caribbean reef fish. Science. 2003;299:107–109. doi: 10.1126/science.1079365. [DOI] [PubMed] [Google Scholar]
- Wainwright P.C. Morphology and ecology: functional basis of feeding constraints in Caribbean labrid fishes. Ecology. 1988;69:635–645. [Google Scholar]
- Warner R.R. Evolutionary ecology: how to reconcile pelagic dispersal with local adaptation. Coral Reefs. 1997;16:S115–S120. [Google Scholar]
- Wellington G.M, Robertson D.R. Variation in larval life-history traits among reef fishes across the Isthmus of Panama. Mar. Biol. 2001;138:11–22. [Google Scholar]
- Williams D.M. Patterns and processes in the distribution of coral reef fishes. In: Sale P.F, editor. The ecology of fishes on coral reefs. Academic Press; San Diego: 1991. pp. 437–474. [Google Scholar]