Abstract
The biologic aggressiveness of prostate tumors is an important indicator of prognosis. Chromosome 7q32—q33 was recently reported to show linkage to more aggressive prostate cancer, based on Gleason score, in a large sibling pair study. We report confirmation and narrowing of the linked region using finer-scale genotyping. We also report a high frequency of allelic imbalance (AI) defined within this locus in a series of 48 primary prostate tumors from men unselected for family history or disease status. The highest frequency of AI was observed with adjacent markers D7S2531 (52%) and D7S1804 (36%). These two markers delineated a common region of AI, with 24 tumors exhibiting interstitial AI involving one or both markers. The 1.1-Mb candidate region contains relatively few transcripts. Additionally, we observed positive associations between interstitial AI at D7S1804 and early age at diagnosis (P=.03) as well as a high combined Gleason score and tumor stage (P=.06). Interstitial AI at D7S2531 was associated with a positive family history of prostate cancer (P=.05). These data imply that we have localized a prostate cancer tumor aggressiveness loci to chromosome 7q32–q33 that is involved in familial and nonfamilial forms of prostate cancer.
Keywords: prostate, linkage, AI, aggressive, 7q
Introduction
Prostate cancer is the most commonly diagnosed neoplasm and the second leading cause of cancer mortality in men in the USA, with 30,200 deaths predicted in 2002 [1]. Despite this, little is known regarding the genetic etiology of this disease or why some prostate tumors are biologically more aggressive than others.
Studies to date indicate the presence of multiple prostate cancer susceptibility loci including: HPC1 located on 1q24–q25 [2], CAPB on 1p36 [3], PCAP on 1q42.2–q43 [4], 8p22–p23 [5], HPC2 on 17p [6], 16q23–q24 [7], 20q [8] and HPCX on Xq27–q28 [9]. Whereas there is some supporting evidence for many of these candidate loci [10–18], several studies have failed to verify any linkage to these regions [19–25]. These complex and apparently contradictory linkage data strongly suggest a heterogeneous nature of hereditary prostate cancer [26]. Four members of our group (D.V.C., W.J.C., B.K.S. and J.S.W.) have recently reported the identification of chromosomal regions at 5q31–q33, 7q32–q33, and 19q12 that exhibit linkage to more aggressive forms of prostate cancer [27], as determined by Gleason score [28].
We chose to further examine chromosome 7q32–q33 as reports indicate frequent deletions of the long arm of chromosome 7 in prostate cancer [29,30] and many other tumor types including breast [31], pancreas [32], stomach [33], thyroid [34], and ovary [35]. Allelic imbalance (AI) of 7q is also associated with poor outcome in patients with prostate cancer [36]. Specifically, a significant correlation has been reported between 7q AI and higher Gleason score, increased mortality and systemic progression of disease at follow-up [37], all considered markers of more aggressive disease. Loss of heterozygosity (LOH) at 7q31 is the most commonly reported alteration in many tumor types and is often observed in early-stage cancers [29,34,35]. Recent evidence suggests that ST7 is the tumor suppressor gene associated with LOH at 7q31 [38].
The present study was designed to further characterize the candidate prostate tumor aggressiveness locus on 7q32–q33 by utilizing linkage analysis and AI techniques. In this report, we confirm and narrow the previously reported linkage region on chromosome 7q32–q33 and report a high frequency of AI within this region. The current study provides strong evidence for the presence of a prostate cancer aggressiveness gene mapping to 7q32–q33 that may play a role in both familial and nonfamilial forms of prostate cancer.
Materials and Methods
Radiation Hybrid Mapping
The order of the 11 microsatellite markers across 7q used in the AI study was determined using the Genebridge 4 (GB4) low-resolution radiation hybrid panel (Research Genetics, Huntsville, AL). The GB4 panel [39] contains DNAs from 93 human fibroblast-derived human:hamster hybrids. Individual PCR reactions were performed for each hybrid with each of the 11 markers shown in Table 1. The PCR reactions were performed using a PCR thermal cycler (MWG Biotech, Highpoint, NC). Each 15-µl reaction contained 2 µl of DNA, 1.25 mM of each deoxynucleotide triphosphate, 0.5 µM reverse primer, 0.5 µM forward primer, 0.75 U of Taq DNA polymerase (Life Technologies, Rockville, MD), 67 mM Tris-HCl (pH 8.8), 67 mM magnesium chloride, 16.6 mM ammonium sulfate, 10 mM β-mercaptoethanol, and 10% DMSO. The DNA amplification cycle comprised a 5-minute denaturation step at 94°C, 35 cycles of 94°C for 45 seconds, annealing at the appropriate temperature for 1 minute, and extension for 1 minute at 72°C, followed by a final 5-minute extension at 72°C. Ten microliters of each PCR product was resolved on a 6% nondenaturing polyacrylamide gel and visualized following ethidium bromide staining. Each hybrid was scored for the presence or absence of the PCR product with all 11 markers. The results were analyzed using the Map Manager QT program [40] to determine the relative order and distance between markers (Table 1).
Table 1.
Radiation Hybrid Map Order of 11 Microsatellite Markers on 7q.
| Marker | Intermarker Distance* (cR-3000) |
| D7S3061 | |
| 34.44 | |
| D7S1875 | |
| 27.44 | |
| D7S530 | |
| 8.06 | |
| D7S2519 | |
| 14.45 | |
| D7S2531 | |
| 30.62 | |
| D7S1804 | |
| 12.68 | |
| D7S2452 | |
| 8.92 | |
| D7S640 | |
| 63.15 | |
| D7S684 | |
| 11.02 | |
| D7S495 | |
| 28.44 | |
| D7S1824 | |
Map distances are based on D7S3061 as 0.
Genotyping Methods
A higher density linkage analysis across the 7q32–q34 region was performed on 513 men, from the equivalent of 326 concordant sibships, from the original sibling pair study [27]. Five microsatellite markers were chosen; one of these markers (D7S530) lies distal and the remaining four (D7S2452, D7S640, D7S684, and D7S495) lie proximal to the marker exhibiting the highest degree of linkage (D7S1804) in the previous study [27]. PCR was carried out for each marker using blood lymphocyte DNA from each individual in the study. Each forward primer was designed to include a fluorophore at the 5′ end to enable detection and analysis on an ABI 373 XL DNA sequencer. Before loading onto a 6% denaturing polyacrylamide gel, PCR products were diluted in water and multiplexed according to marker size and fluorophore. One microliter of multiplexed product was then added to 3 µl of formamide loading dye, containing a 350 base pair 6-carboxytetramethylrhodamine size standard (Applied Biosystems, Foster City, CA) and denatured at 95°C for 5 minutes. ABI Collection and ABI Genescan version 3.1 software packages (Applied Biosystems) were used to process each electrophoresis run. Marker allele sizes and intensity for each sample was assessed using Genotyper software (Applied Biosystems).
Patient Selection and Evaluation of Tissue
A consecutive series of 51 prostate patients, for whom we had both tumor tissue and comprehensive clinical data available, were identified through the tumor registry at the Cleveland Clinic Foundation. This study was approved by the Institutional Review Board of the Cleveland Clinic Foundation. Each tumor was staged at the time of surgery and graded, using the Gleason system, following assessment of microscopic sections of the surgical specimen. Normal and tumor tissue from each case was micro-dissected from 5-µm unstained paraffin-embedded tissue sections, as previously described [16], following the review and annotation of a corresponding hematoxylin and eosin stained section by a pathologist (H.L.). DNA was extracted from the microdissected tissue using the Qiagen Tissue Kit (Qiagen, Valencia, CA) and eluted in 100 µl of Tris buffer (pH 9). Final tumor DNA content was estimated to be at least 70%. Three patients were subsequently removed from the study due to insufficient DNA quality for PCR amplification. A total of 48 patients were included in our genetic analysis. Clinical information for all patients is noted in Table 2.
Table 2.
Selected Clinical Parameters for the 48 Prostate Cancer Patients in the Study.
| Patient ID | Age at Diagnosis | Prostate Cancer Family History | PSA at Diagnosis | Pathology Stage | Surgical Grade |
| 3-104 | 63 | Y | 5.4 | T2 | 6 |
| 3-130 | 66 | N | 6.0 | T2 | 5 |
| 3-249 | 51 | Y | 5.0 | T2 | 6 |
| 3-342 | 64 | N | 5.0 | T3 | 6 |
| 4-188 | 54 | Y | 4.6 | T3 | 7 |
| 5-121 | 55 | N | 5.7 | T3 | 7 |
| 5-187 | 63 | Y | 5.7 | T3 | 7 |
| 5-189 | 62 | N | 3.5 | T2 | 6 |
| 5-350 | 60 | Y | 6.3 | T2 | 6 |
| 5-369 | 66 | N | 5.6 | T2 | 7 |
| 5-436 | 63 | Y | 6.0 | T2 | 6 |
| 5-665 | 65 | Y | 6.0 | T3 | 5 |
| 5-905 | 63 | N | 5.4 | T2 | 6 |
| 6-201 | 64 | Y | 5.4 | T2 | 5 |
| 6-322 | 56 | Y | 5.6 | T2 | 6 |
| 6-350 | 58 | N | 5.6 | T2 | 7 |
| 6-425 | 61 | N | 6.2 | T2 | 6 |
| 6-452 | 64 | Y | 6.3 | T3 | 7 |
| 7-155 | 65 | Y | 5.5 | T3 | 7 |
| 7-187 | 63 | Y | 5.3 | T3 | 7 |
| 7-206 | 71 | Y | 5.5 | T2 | 7 |
| 7-220 | 56 | Y | 6.0 | T2 | 6 |
| 7-286 | 57 | N | 7.2 | T3 | 7 |
| 7-293 | 62 | N | 5.9 | T2 | 7 |
| 7-297 | 57 | N | 10.9 | T3 | 6 |
| 7-309 | 68 | N | 4.4 | T2 | 7 |
| 7-310 | 63 | N | 7.1 | T2 | 7 |
| 7-311 | 62 | N | 6.1 | T2 | 6 |
| 7-324 | 58 | N | 8.0 | T2 | 5 |
| 7-341 | 62 | N | 12.9 | T3 | 7 |
| 7-348 | 60 | Y | 17.0 | T3 | 7 |
| 7-353 | 56 | N | 5.9 | T2 | 7 |
| 7-375 | 57 | N | 4.8 | T2 | 6 |
| 7-392 | 60 | N | 13.0 | T2 | 9 |
| 7-393 | 58 | N | 3.4 | T2 | 7 |
| 7-401 | 59 | N | 25.0 | T3 | 7 |
| 7-404 | 60 | Y | 8.0 | T2 | 7 |
| 7-410 | 73 | N | 5.0 | T2 | 6 |
| 7-433 | 66 | N | 9.8 | T3 | 7 |
| 7-441 | 69 | N | 10.0 | T3 | 8 |
| 7-451 | 68 | N | 5.3 | T2 | 7 |
| 7-475 | 55 | Y | 6.6 | T2 | 6 |
| 7-484 | 62 | N | 8.2 | T2 | 7 |
| 7-485 | 63 | N | 7.6 | T2 | 7 |
| 7-491 | 63 | Y | 4.3 | T2 | 7 |
| 7-684 | 48 | Y | 5.9 | T2 | 7 |
| 7-923 | 71 | Y | 5.0 | T3 | 7 |
| 8-501 | 47 | N | 18.2 | T3 | 9 |
AI Study
Eleven microsatellite markers were used in the AI study (Table 3). Of these, three (D7S3061, D7S1804, and D7S1824) had shown significant linkage to aggressive disease in our prostate cancer sibling linkage study [27]. Information regarding primer sequence was obtained from the Genome Database (http://www.gdb.org). Separate PCR reactions were performed using DNA from microdissected normal and tumor tissue. PCR conditions were as described above, but using a γ-32P end-labeled forward primer. End-labeling with [γ-32P]dATP was carried out using T4 polynucleotide kinase (USB, Cleveland, OH). Two microliters of PCR product was combined with 4 µl of 95% formamide loading dye containing 20 mM EDTA, 0.5 mg/ml bromophenol blue, and 0.5 mg/ml xylene cyanol. The sample mixtures were denatured at 95°C for 5 minutes and immediately cooled on ice before loading onto a 6% denaturing polyacrylamide gel. Gels were run for approximately 3 hours at 65 W and analyzed following autoradiographic band detection using a STORM phosphorimager (Molecular Dynamics, Sunnyvale, CA). AI was determined by visual examination by two investigators as previously reported [41] and samples that defined AI breakpoints were verified using PCR products from a separate PCR amplification reaction.
Table 3.
AI on Chromosome 7q31–q35 Loci in 48 Primary Prostate Tumors.
| Marker | Name | AI/Informative (% of AI) | Cytogenetic Location* |
| D7S3061 | CHCL.GGAA6D03 | 12/43 (28%) | 7q31.32 |
| D7S1875 | AFMa345wc9 | 5/30 (17%) | 7q31.33 |
| D7S530 | AFM249×f9 | 4/42 (10%) | 7q32.1 |
| D7S2519 | AFMc024we9 | 7/35 (20%) | 7q32.2 |
| D7S2531 | AFM338wd5 | 16/31 (52%) | 7q32.2 |
| D7S1804 | CHCL.GATA43C11 | 12/33 (36%) | 7q33 |
| D7S2452 | AFMa282wf9 | 7/39 (18%) | 7q33 |
| D7S640 | sWSS1204 | 6/37 (16%) | 7q33 |
| D7S684 | AFM312wb5 | 9/42 (21%) | 7q34 |
| D7S495 | AFM168×c3 | 6/25 (24%) | 7q34 |
| D7S1824 | CHLC.GATA32C12 | 8/41 (20%) | 7q35 |
According to the NCBI Entrez Genome web site
Statistical Analyses
Linkage was statistically evaluated using a multipoint generalized Haseman-Elston (HE) linkage test [42] as described previously [27]. To determine whether AI at any marker was associated with clinical characteristics of the men with prostate cancer (e.g., high Gleason score) Fischer's exact χ2 test was used. Analyses were carried out using the statistical software SAS (SAS Institute, Cary, NC).
Results
Radiation Hybrid Mapping of 7q Microsatellite Markers
To confirm the order of the 11 microsatellite markers used in this study, the low-resolution GB4 radiation hybrid panel [39] was used. We found the mapping order of the markers to be in agreement with that given for the Marshfield linkage map with the exception of marker D7S640, which our data places distal to D7S2452, and marker D7S495, which lies distal to D7S684 according to our study. The chromosomal order of these markers, as determined by analysis with Map Manager QT is shown in Table 1.
Linkage Analysis
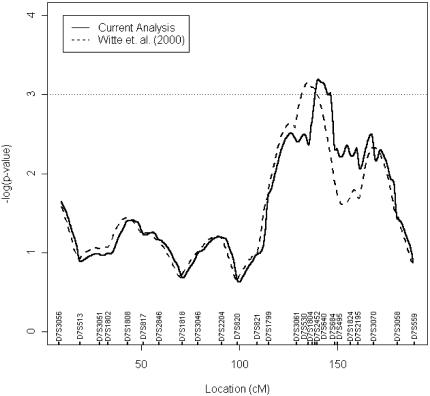
Incorporating the data from the five additional markers into our original linkage analysis strengthened evidence of linkage (Figure 1). We observed a broad region of linkage for which P<.005 between markers D7S3061 and D7S495. Within this region, a 6-cM peak (P<.001) encompassing markers D7S2452 and D7S640 was identified, supporting the presence of a gene involved in the development of aggressive forms of prostate cancer on 7q32–q33.
Figure 1.
Results from linkage analysis of prostate cancer aggressiveness on chromosome 7. Broken lines indicate results from the original analysis; solid line denotes results from new analysis. Values on the x-axis show marker positions according to Marshfield Medical Research Foundation, Human Genetics web site and our radiation hybrid mapping data.
AI at 7q32–q33
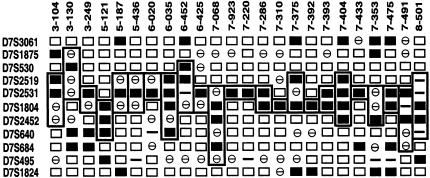
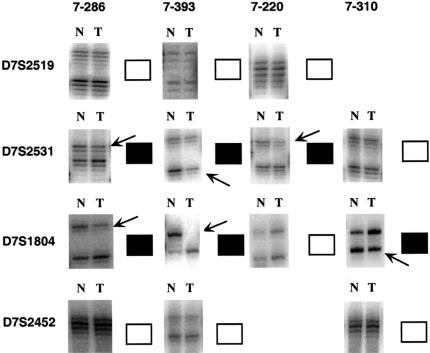
Overall, 38 of the 48 (79%) samples studied showed AI with at least one marker. The AI frequencies for each marker are shown in Table 3. The highest frequencies of AI were exhibited by adjacent markers D7S2531 (52%) and D7S1804 (36%). Twenty-seven of the 48 samples studied displayed AI involving D7S2531 and/or D7S1804, with 24 tumors showing interstitial deletions across this region (Figure 2). Representative autoradiographs for four of the samples showing interstitial AI across D7S2531/D7S1804 are shown in Figure 3.
Figure 2.
Summary of 24 primary prostate tumor samples showing interstitial AI on 7q involving D7S2531 and D7S1804. Markers are listed on the left; sample numbers are shown across the top. (□) Informative samples with no AI; (■) informative samples demonstrating AI; (⊖) noninformative (homozygous) samples; (—) did not work. Boxes define maximum extent of AI for each sample.
Figure 3.
Examples of autoradiographs from AI studies on matched normal (N) and prostate tumor (T) pairs. Samples 7-286 and 7-393 both exhibit AI at markers D7S2531 and D7S1804 but retain D7S2519 and D7S2452. Tumor 7-220 shows AI at D7S2531 but no AI at any neighboring marker. Sample 7-310 demonstrates AI at D7S1804 but not at any flanking marker. (■) AI; (□) no AI; arrows indicate AI.
Clinical Associations
We examined any potential clinical associations between tumors showing interstitial AI involving D78S2531 and/or D7S1804 compared with 21 samples that showed no AI at either marker. This analysis was designed to remove any bias from surrounding unrelated AI. Parameters such as family history (self-reported and defined as having at least one first-degree affected relative), PSA at diagnosis, Gleason score, and tumor stage were studied (Table 2). A statistically significant association (P=.03) was seen between early age at diagnosis (<60 years of age) and interstitial AI at marker D7S1804. A weaker, but still noteworthy, association was observed between interstitial AI involving D7S1804 and a high combined Gleason score (>7) and tumor stage (≥T2c) (P=.06). An association between AI with marker D7S2531 and positive family history for prostate cancer (P=.05) was also observed.
Discussion
The first aim of this project was to provide further evidence for a prostate cancer tumor aggressiveness locus on chromosome 7q32–q33. In our original genome-wide multipoint linkage study [27] we found three microsatellite markers that defined linkage to Gleason score (P<.01) across 7q31-q35 covering a 28 cM region. The linkage region with P<.0001 spanned approximately 8 cM and included the marker D7S1804. In the present study, we undertook finer-scale genotyping in this region using five additional markers (D7S530, D7S2452, D7S640, D7S684, and D7S495) with the same sibling population used in the original study. We found that linkage was maintained on chromosome 7q32–q33, when data were analyzed with respect to Gleason score (Figure 1) and strengthened (P<.005) across the 28 cM region. Furthermore, the highest linkage peak was reduced in size to approximately 6 cM (P<.001) between markers D7S2452 and D7S684. These data imply that this region harbors a gene involved in increased risk for developing more aggressive forms of prostate cancer.
Hereditary chordoma was recently mapped to this region on chromosome 7q32–q33 [43]. Chordomas are rare, locally invasive primary tumors of the bone believed to develop from notochordal remnants. Although these tumors are slow growing, they frequently recur following surgery or radiotherapy [44], implying some aggressive biologic behavior. Three families affected by chordoma showed linkage between markers D7S512 and D7S684, which span the region observed in the present study [43]. Furthermore, another form of familial chordoma has been mapped to 1p36 [45], also reported as a prostate cancer susceptibility locus CAPB [3]. Although there is a moderate male predominance (1.7:1) for this disease [43], there are no reports of an association with prostate cancer. Reported analysis of four chordoma tumor samples from affected family members did not reveal any LOH suggesting that this gene and the prostate cancer aggressiveness gene may not be the same [43] or do not undergo the same forms of gene inactivation. However, additional studies would be needed to determine this.
A second goal of the study was to provide evidence of a tumor suppressor gene in this region by applying AI analyses to 48 prostate tumors from patients unselected for family history or clinical status of disease. These studies demonstrated a high frequency of AI within this region. Overall, 79% (38 of 48) of the tumors showed AI of at least one marker. The highest frequency of AI was found at markers D7S2531 (52%) and D7S1804 (36%). Furthermore, 24 tumors (50%) showed interstitial AI involving D7S2531 and/or D7S1804 (Figure 2) and defined a common region of deletion between these two markers of approximately ∼1.1 Mb in size, based on build 28 of the public genome sequencing database at NCBI. This pattern of interstitial AI also suggests that the region we have defined in this study is distinct from that containing the ST7 candidate tumor suppressor gene on 7q31.3 (Figure 4).
Figure 4.
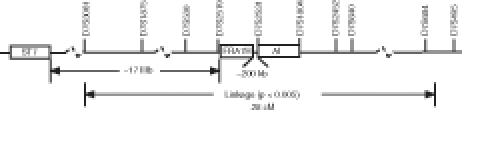
Schematic representation of 7q32-q33 region showing positions of linked region, AI, fragile site and ST7 tumor suppressor gene. Not shown to scale. ST7=supression of tumorigenicity 7 gene; FRA7H=fragile site 7H; AI=common region of allelic imbalance.
The common fragile site FRA7H maps between markers D7S2519 and D7S2531 [46,47]. Fragile sites are regions of the genome particularly prone to breakage and often are associated with regions of AI. Several candidate tumor suppressor genes have been mapped to fragile sites including FHIT on 3p14.2 (FRA3B) [48], the multiple candidate tumor suppressor ST7 on 7q31 (FRA7G) [38,49] and the putative prostate cancer susceptibility locus at 16q23 (FRA16D) [16,50,51].
We also investigated any clinical associations with occurrence of AI at markers D7S2531 and D7S1804 by comparing tumors with interstitial AI at D7S2531 and/or D7S1804 and those without AI at either marker. A statistically significant association (P=.05) was observed between AI at D7S2531 and positive family history. AI at D7S1804 was associated with early age at diagnosis (P=.03) and a high combined Gleason score and tumor stage (P=.06). A similar trend was seen when Gleason score (P=.14) or tumor stage (P=.16) were analyzed separately. These data strongly suggest that this region contains a gene associated with prostate tumor aggressiveness that is implicated in the etiology of both hereditary and nonfamilial forms of prostate cancer.
Sequence coverage of the chromosome 7q32–q33 region is almost complete with three small gaps in the NCBI database. The region contains relatively few transcripts, but contains some intriguing candidates for a prostate cancer tumor aggressiveness gene, including two known genes, muskelin-1 (MKLN1) [52] and podocalyxin-like protein (PODXL), and four hypothetical genes, KIAA1550, LOC91583, LOC91584 (similar to mouse plexin 3), and LOC96016 (similar to Eukaryotic Translation Elongation Factor 1 Beta 2).
Muskelin-1 is a novel intracellular protein that acts as a positive mediator of cell-spreading, adhesion, and cytoskeletal responses to the extracellular matrix component thrombospondin-1 (TSP-1) [53]. TSP-1 is a potent antiangiogenic molecule [54] and has been shown to inhibit tumor growth and metastasis [55]. TSP-1 is upregulated by p53 and depleted in primary prostate tumors that express mutant p53 [56]. Evidence suggests that muskelin (MKLN1) is required for cellular responses to TSP-1 [53], implying a role for this protein in the regulation of the biologic aggressiveness of tumors.
Podocalyxin-like protein is a transmembrane sialomucin involved in adhesion in renal glomerular podocytes and vascular endothelium [57]. PODXL was recently shown to be identical to the testicular germ cell tumor marker, gp200, which is associated with highly malignant tumors [58].
Several regions of homology to both mouse and human plexins are identified within the hypothetical genes including PSI (domain found in plexins, semaphorins, and integrins) and IPT (immunoglobulin-like fold shared by plexins and transcription factors) domains common to the plexin family of proteins [59]. Plexins have been reported to act as cell surface receptors for semaphorins [60], and are implicated in axon repulsion, angiogenesis regulation, and tumor growth and metastasis [61]. The PSI domain found within the extracellular regions of both plexins and semaphorins shows homology to part of the oncoprotein MET. MET and other scatter factor receptors have been shown to play a role in cell motility and invasion [62].
To provide evidence of a role in prostate tumor aggressiveness, we are currently performing germline mutation analyses of candidate genes in men with prostate cancer that show linkage to this region. In addition, we hope to identify more linked families for which there is tumor tissue available and perform AI studies to further confirm and narrow this region.
In summary, we have further refined a region of linkage on chromosome 7q32–q33 associated with prostate cancer tumor aggressiveness. We have also demonstrated a high frequency of AI within this region and have mapped the smallest region of AI to approximately 1.1 Mb between markers D7S2531 and D7S1804. Furthermore, we found that AI at marker D7S1804 was associated with early age of onset of prostate cancer and high combined Gleason score/tumor stage and that marker D7S2531 was associated with family history of prostate cancer in an unselected series of prostate cancer patients. These data support the mapping of a gene to 7q32–q33 associated with aggressive forms of both familial and nonfamilial prostate cancer.
References
- 1.American Cancer Society, Atlanta, GA, author. Cancer Facts and Figures 2002. 2002:1–41. [Google Scholar]
- 2.Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, Schuster EF, Buckley VA, Miller EL, Brandzel S, Li S, Hood L, Ostrander EA. Evidence for a rare prostate cancer-susceptibility locus at chromosome 1p36. Am J Hum Genet. 1999;64:776–787. doi: 10.1086/302287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, Wohr G, Latil A, Millasseau P, Mellah I, Cohen N, Blanche H, Bellane-Chantelot C, Demenais F, Teillac P, Le Duc A, de Petriconi R, Hautmann R, Chumakov I, Bachner L, Maitland NJ, Lidereau R, Vogel W, Fournier G, Mangin P, Cohen D, Cussenot O. Predisposing gene for early-onset prostate cancer, localized on chromosome 1q42.2–43. Am J Hum Genet. 1998;62:1416–1424. doi: 10.1086/301879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Zheng SL, Hawkins GA, Faith DA, Kelly B, Isaacs SD, Wiley KE, Chang B, Ewing CM, Bujnovszky P, Carpten JD, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB. Linkage and association studies of prostate cancer susceptibility: evidence for linkage at 8p22–23. Am J Hum Genet. 2001;69:341–350. doi: 10.1086/321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, Desrochers M, Dumont M, Farnham JM, Frank D, Frye C, Ghaffari S, Gupte JS, Hu R, Iliev D, Janecki T, Kort EN, Laity KE, Leavitt A, Leblanc G, McArthur-Morrison J, Pederson A, Penn B, Peterson KT, Reid JE, Richards S, Schroeder M, Smith R, Snyder SC, Swedlund B, Swensen J, Thomas A, Tranchant M, Woodland AM, Labrie F, Skolnick MH, Neuhausen S, Rommens J, Cannon-Albright LA. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 7.Suarez BK, Lin J, Burmester JK, Broman KW, Weber JL, Banerjee TK, Goddard KA, Witte JS, Elston RC, Catalona WJ. A genome screen of multiplex sibships with prostate cancer. Am J Hum Genet. 2000;66:933–944. doi: 10.1086/302818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry R, Schroeder JJ, French AJ, McDonnell SK, Peterson BJ, Cunningham JM, Thibodeau SN, Schaid DJ. Evidence for a prostate cancer-susceptibility locus on chromosome 20. Am J Hum Genet. 2000;67:82–91. doi: 10.1086/302994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleutker J, Matikainen M, Smith J, Koivisto P, Baffoe-Bonnie A, Kainu T, Gillanders E, Sankila R, Pukkala E, Carpten J, Stephan D, Tammela T, Brownstein M, Bailey-Wilson J, Trent J, Kallioniemi OP. A genetic epidemiological study of hereditary prostate cancer (HPC) in Finland: frequent HPCX linkage in families with late-onset disease. Clin Cancer Res. 2000;6:4810–4815. [PubMed] [Google Scholar]
- 10.Cancel-Tassin G, Latil A, Valeri A, Mangin P, Fournier G, Berthon P, Cussenot O. PCAP is the major known prostate cancer predisposing locus in families from south and west Europe. Eur J Hum Genet. 2001;9:135–142. doi: 10.1038/sj.ejhg.5200592. [DOI] [PubMed] [Google Scholar]
- 11.Rebbeck TR, Walker AH, Zeigler-Johnson C, Weisburg S, Martin AM, Nathanson KL, Wein AJ, Malkowicz SB. Association of HPC2/ELAC2 genotypes and prostate cancer. Am J Hum Genet. 2000;67:1014–1019. doi: 10.1086/303096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooney KA, McCarthy JD, Lange E, Huang L, Miesfeldt S, Montie JE, Oesterling JE, Sandler HM, Lange K. Prostate cancer susceptibility locus on chromosome 1q: a confirmatory study. J Natl Cancer Inst. 1997;89:955–959. doi: 10.1093/jnci/89.13.955. [DOI] [PubMed] [Google Scholar]
- 13.Gronberg H, Xu J, Smith JR, Carpten JD, Isaacs SD, Freije D, Bova GS, Danber JE, Bergh A, Walsh PC, Collins FS, Trent JM, Meyers DA, Isaacs WB. Early age at diagnosis in families providing evidence of linkage to the hereditary prostate cancer locus (HPC1) on chromosome 1. Cancer Res. 1997;57:4707–4709. [PubMed] [Google Scholar]
- 14.Gronberg H, Smith J, Emanuelsson M, Jonsson BA, Bergh A, Carpten J, Isaacs W, Xu J, Meyers D, Trent J, Damber JE. In Swedish families with hereditary prostate cancer, linkage to the HPC1 locus on chromosome 1q24–25 is restricted to families with early-onset prostate cancer. Am J Hum Genet. 1999;65:134–140. doi: 10.1086/302447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuhausen SL, Farnham JM, Kort E, Tavtigian SV, Skolnick MH, Cannon-Albright LA. Prostate cancer susceptibility locus HPC1 in Utah high-risk pedigrees. Hum Mol Genet. 1999;8:2437–2442. doi: 10.1093/hmg/8.13.2437. [DOI] [PubMed] [Google Scholar]
- 16.Paris PL, Witte JS, Kupelian PA, Levin H, Klein EA, Catalona WJ, Casey G. Identification and fine mapping of a region showing a high frequency of allelic imbalance on chromosome 16q23.2 that corresponds to a prostate cancer susceptibility locus. Cancer Res. 2000;60:3645–3649. [PubMed] [Google Scholar]
- 17.Xu J. Combined analysis of hereditary prostate cancer linkage to 1q24–25: results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet. 2000;66:945–957. doi: 10.1086/302807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng SL, Xu J, Isaacs SD, Wiley K, Chang B, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB. Evidence for a prostate cancer linkage to chromosome 20 in 159 hereditary prostate cancer families. Hum Genet. 2001;108:430–435. doi: 10.1007/s004390100513. [DOI] [PubMed] [Google Scholar]
- 19.Bergthorsson JT, Johannesdottir G, Arason A, Benediktsdottir KR, Agnarsson BA, Bailey-Wilson JE, Gillanders E, Smith J, Trent J, Barkardottir RB. Analysis of HPC1, HPCX, and PCaP in Icelandic hereditary prostate cancer. Hum Genet. 2000;107:372–375. doi: 10.1007/s004390000384. [DOI] [PubMed] [Google Scholar]
- 20.Bock CH, Cunningham JM, McDonnell SK, Schaid DJ, Peterson BJ, Pavlic RJ, Schroeder JJ, Klein J, French AJ, Marks A, Thibodeau SN, Lange EM, Cooney KA. Analysis of the prostate cancer-susceptibility locus HPC20 in 172 families affected by prostate cancer. Am J Hum Genet. 2001;68:795–801. doi: 10.1086/318797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goode EL, Stanford JL, Chakrabarti L, Gibbs M, Kolb S, McIndoe RA, Buckley VA, Schuster EF, Neal CL, Miller EL, Brandzel S, Hood L, Ostrander EA, Jarvik GP. Linkage analysis of 150 high-risk prostate cancer families at 1q24–25. Genet Epidemiol. 2000;18:251–275. doi: 10.1002/(SICI)1098-2272(200003)18:3<251::AID-GEPI5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.McIndoe RA, Stanford JL, Gibbs M, Jarvik GP, Brandzel S, Neal CL, Li S, Gammack JT, Gay AA, Goode EL, Hood L, Ostrander EA. Linkage analysis of 49 high-risk families does not support a common familial prostate cancer-susceptibility gene at 1q24–25. Am J Hum Genet. 1997;61:347–353. doi: 10.1086/514853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suarez BK, Lin J, Witte JS, Conti DV, Resnick MI, Klein EA, Burmester JK, Vaske DA, Banerjee TK, Catalona WJ. Replication linkage study for prostate cancer susceptibility genes. Prostate. 2000;45:106–114. doi: 10.1002/1097-0045(20001001)45:2<106::aid-pros4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 24.Whittemore AS, Lin IG, Oakley-Girvan I, Gallagher RP, Halpern J, Kolonel LN, Wu AH, Hsieh CL. No evidence of linkage for chromosome 1q42.2–43 in prostate cancer. Am J Hum Genet. 1999;65:254–256. doi: 10.1086/302457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Zheng SL, Carpten JD, Nupponen NN, Robbins CM, Mestre J, Moses TY, Faith DA, Kelly BD, Isaacs SD, Wiley KE, Ewing CM, Bujnovszky P, Chang B, Bailey-Wilson J, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB. Evaluation of linkage and association of HPC2/ELAC2 in patients with familial or sporadic prostate cancer. Am J Hum Genet. 2001;68:901–911. doi: 10.1086/319513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrander EA, Stanford JL. Genetics of prostate cancer: too many loci, too few genes. Am J Hum Genet. 2000;67:1367–1375. doi: 10.1086/316916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witte JS, Goddard KA, Conti DV, Elston RC, Lin J, Suarez BK, Broman KW, Burmester JK, Weber JL, Catalona WJ. Genomewide scan for prostate cancer-aggressiveness loci. Am J Hum Genet. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gleason DF. Histologic grading of prostate cancer: a perspective. Hum Pathol. 1992;23:273–279. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- 29.Latil A, Cussenot O, Fournier G, Baron JC, Lidereau R. Loss of heterozygosity at 7q31 is a frequent and early event in prostate cancer. Clin Cancer Res. 1995;1:1385–1389. [PubMed] [Google Scholar]
- 30.Zenklusen JC, Thompson JC, Troncoso P, Kagan J, Conti CJ. Loss of heterozygosity in human primary prostate carcinomas: a possible tumor suppressor gene at 7q31.1. Cancer Res. 1994;54:6370–6373. [PubMed] [Google Scholar]
- 31.Zenklusen JC, Bieche I, Lidereau R, Conti CJ. (C-A)n microsatellite repeat D7S522 is the most commonly deleted region in human primary breast cancer. Proc Natl Acad Sci USA. 1994;91:12155–12158. doi: 10.1073/pnas.91.25.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- 33.Nishizuka S, Tamura G, Terashima M, Satodate R. Commonly deleted region on the long arm of chromosome 7 in differentiated adenocarcinoma of the stomach. Br J Cancer. 1997;76:1567–1571. doi: 10.1038/bjc.1997.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang JS, Nelson M, McIver B, Hay ID, Goellner JR, Grant CS, Eberhardt NL, Smith DI. Differential loss of heterozygosity at 7q31.2 in follicular and papillary thyroid tumors. Oncogene. 1998;17:789–793. doi: 10.1038/sj.onc.1201996. [DOI] [PubMed] [Google Scholar]
- 35.Watson RH, Neville PJ, Roy WJ Jr, Hitchcock A, Campbell IG. Loss of heterozygosity on chromosomes 7p, 7q, 9p and 11q is an early event in ovarian tumorigenesis. Oncogene. 1998;17:207–212. doi: 10.1038/sj.onc.1201945. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi S, Shan AL, Ritland SR, Delacey KA, Bostwick DG, Lieber MM, Thibodeau SN, Jenkins RB. Frequent loss of heterozygosity at 7q31.1 in primary prostate cancer is associated with tumor aggressiveness and progression. Cancer Res. 1995;55:4114–4119. [PubMed] [Google Scholar]
- 37.Jenkins R, Takahashi S, DeLacey K, Bergstralh E, Lieber M. Prognostic significance of allelic imbalance of chromosome arms 7q, 8p, 16q, and 18q in stage T3N0M0 prostate cancer. Genes Chromosomes Cancer. 1998;21:131–143. [PubMed] [Google Scholar]
- 38.Zenklusen JC, Conti CJ, Green ED. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet. 2001;27:392–398. doi: 10.1038/86891. [DOI] [PubMed] [Google Scholar]
- 39.Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud'Homme JF, Dib C, Auffray C, Morissette J, Weissenbach J, Goodfellow PN. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- 40.Manly KF, Olson JM. Overview of QTL mapping software and introduction to map manager QT. Mamm Genome. 1999;10:327–334. doi: 10.1007/s003359900997. [DOI] [PubMed] [Google Scholar]
- 41.Plummer SJ, Paris MJ, Myles J, Tubbs R, Crowe J, Casey G. Four regions of allelic imbalance on 17q12-qter associated with high-grade breast tumors. Genes Chromosomes Cancer. 1997;20:354–362. [PubMed] [Google Scholar]
- 42.Elston RC, Buxbaum S, Jacobs KB, Olson JM. Haseman and Elston revisited. Genet Epidemiol. 2000;19:1–17. doi: 10.1002/1098-2272(200007)19:1<1::AID-GEPI1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 43.Kelley MJ, Korczak JF, Sheridan E, Yang X, Goldstein AM, Parry DM. Familial chordoma, a tumor of notochordal remnants, is linked to chromosome 7q33. Am J Hum Genet. 2001;69:454–460. doi: 10.1086/321982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoedel KE, Martinez AJ, Mahoney TM, Contis L, Becich MJ. Chordomas: pathological features; ploidy and silver nucleolar organizing region analysis. A study of 36 cases. Acta Neuropathol. 1995;89:139–143. doi: 10.1007/BF00296357. [DOI] [PubMed] [Google Scholar]
- 45.Miozzo M, Dalpra L, Riva P, Volonta M, Macciardi F, Pericotti S, Tibiletti MG, Cerati M, Rohde K, Larizza L, Fuhrman Conti AM. A tumor suppressor locus in familial and sporadic chordoma maps to 1p36. Int J Cancer. 2000;87:68–72. [PubMed] [Google Scholar]
- 46.Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, Margalit H, Platzer M, Weiss A, Tsui LC, Rosenthal A, Kerem B. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashida S, Yamasaki K, Asada Y, Soeda E, Niikawa N, Kishino T. Construction of a physical and transcript map flanking the imprinted MEST/PEG1 region at 7q32. Genomics. 2000;66:221–225. doi: 10.1006/geno.2000.6206. [DOI] [PubMed] [Google Scholar]
- 48.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 49.Zenklusen JC, Hodges LC, LaCava M, Green ED, Conti CJ. Definitive functional evidence for a tumor suppressor gene on human chromosome 7q31.1 neighboring the Fra7G site. Oncogene. 2000;19:1729–1733. doi: 10.1038/sj.onc.1203488. [DOI] [PubMed] [Google Scholar]
- 50.Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, Richards RI. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 2000;60:1683–1689. [PubMed] [Google Scholar]
- 51.Paige AJ, Taylor KJ, Stewart A, Sgouros JG, Gabra H, Sellar GC, Smyth JF, Porteous DJ, Watson JE. A 700-kb physical map of a region of 16q23.2 homozygously deleted in multiple cancers and spanning the common fragile site FRA16D. Cancer Res. 2000;60:1690–1697. [PubMed] [Google Scholar]
- 52.Adams JC, Zhang L. cDNA cloning of human muskelin and localisation of the muskelin (MKLN1) gene to human chromosome 7q32 and mouse chromosome 6 B1/B2 by physical mapping and FISH. Cytogenet Cell Genet. 1999;87:19–21. doi: 10.1159/000015385. [DOI] [PubMed] [Google Scholar]
- 53.Adams JC, Seed B, Lawler J. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 1998;17:4964–4974. doi: 10.1093/emboj/17.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dameron KM, Volpert OV, Tainsky MA, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 55.Roberts DD. Regulation of tumor growth and metastasis by thrombospondin-1. FASEB J. 1996;10:1183–1191. [PubMed] [Google Scholar]
- 56.Mehta R, Kyshtoobayeva A, Kurosaki T, Small EJ, Kim H, Stroup R, McLaren CE, Li KT, Fruehauf JP. Independent association of angiogenesis index with outcome in prostate cancer. Clin Cancer Res. 2001;7:81–88. [PubMed] [Google Scholar]
- 57.Kershaw DB, Beck SG, Wharram BL, Wiggins JE, Goyal M, Thomas PE, Wiggins RC. Molecular cloning and characterization of human podocalyxin-like protein. Orthologous relationship to rabbit PCLP1 and rat podocalyxin. J Biol Chem. 1997;272:15708–15714. doi: 10.1074/jbc.272.25.15708. [DOI] [PubMed] [Google Scholar]
- 58.Schopperle WM, DeWolf WC. Human testicular germ cell tumor marker, gp200, identified as podocalyxin-like protein. J Urol. 2000;163:131. [Google Scholar]
- 59.Bork P, Doerks T, Springer TA, Snel B. Domains in plexins: links to integrins and transcription factors. Trends Biochem Sci. 1999;24:261–263. doi: 10.1016/s0968-0004(99)01416-4. [DOI] [PubMed] [Google Scholar]
- 60.Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 61.Christensen CR, Klingelhofer J, Tarabykina S, Hulgaard EF, Kramerov D, Lukanidin E. Transcription of a novel mouse semaphorin gene, M-semaH, correlates with the metastatic ability of mouse tumor cell lines. Cancer Res. 1998;58:1238–1244. [PubMed] [Google Scholar]
- 62.Tamagnone L, Comoglio PM. Control of invasive growth by hepatocyte growth factor (HGF) and related scatter factors. Cytokine Growth Factor Rev. 1997;8:129–142. doi: 10.1016/s1359-6101(97)00007-5. [DOI] [PubMed] [Google Scholar]