Abstract
Meningitis caused by Escherichia coli K1 is a serious illness in neonates with neurological sequelae in up to 50% of survivors. A high degree of bacteremia is required for E. coli K1 to cross the blood–brain barrier, which suggests that the bacterium must evade the host defence mechanisms and survive in the bloodstream. We previously showed that outer membrane protein A (OmpA) of E. coli binds C4b-binding protein (C4bp), an inhibitor of complement activation via the classical pathway. Nevertheless, the exact mechanism by which E. coli K1 survives in serum remains elusive. Here, we demonstrate that log phase (LP) OmpA+E. coli K1 avoids serum bactericidal activity more effectively than postexponential phase bacteria. OmpA–E. coli cannot survive in serum grown to either phase. The increased serum resistance of LP OmpA+E. coli is the result of increased binding of C4bp, with a concomitant decrease in the deposition of C3b and the downstream complement proteins responsible for the formation of the membrane attack complex. C4bp bound to E. coli K1 acts as a cofactor to factor I in the cleavage of both C3b and C4b, which shuts down the ensuing complement cascade. Accordingly, a peptide corresponding to the complement control protein domain 3 of C4bp sequence, was able to compete with C4bp binding to OmpA and cause increased deposition of C3b. Thus, binding of C4bp appears to be responsible for survival of E. coli K1 in human serum.
Keywords: C4b-binding protein, complement, Escherichia coli K1, meningitis, outer membrane protein A
Introduction
Escherichia coli K1 is the leading cause of neonatal meningitis with mortality rates of the infected neonates as high as 40% and over half of the survivors suffering permanent neurological sequelae.1–3 Strains of E. coli K1 usually invade the blood of infants from the nasopharynx or gastrointestinal tract and then multiply in the blood to reach a certain threshold level of bacteremia before crossing the blood–brain barrier.2 To survive in the blood, E. coli must avoid the host's innate defence mechanisms such as the complement system and opsono-phagocytosis. Although, neonates are considered to be immunocompromised to some extent, the mechanism of E. coli serum resistance is still poorly understood. Interestingly, despite the presence of various serotypes of E. coli, only a small number of strains expressing K and O antigens are responsible for the majority of infections, with K1 and K5 being the predominantly found capsular antigens.4 Previous studies have demonstrated that increased production of K1 capsule dramatically influences the complement activation, opsonization and virulence of E. coli K1.5 However, Weiser and Gotschlich found that outer membrane protein A (OmpA) is necessary for serum survival as well, although the mechanism explaining this phenomenon is not defined.6 Other investigators have noted that only certain combinations of O-antigen with K-antigen are found repeatedly in E. coli K1 infections, which suggests that these bacterial surface structures act in association to evade the host's defence arsenal. Therefore, a consensus seems to be emerging that several mechanisms mediate serum resistance of Gram-negative pathogens such as E. coli K1.
The complement system is an important component of innate immunity in the host. Complement (C3) activation can proceed via three enzymatic cascades, the classical, the alternative, and the mannose-binding lectin (MBL) pathways.7 The classical complement pathway is initiated by the binding of C1q to the Fc portion of antibody–antigen complexes on the bacterial surface, while the alternative and lectin pathways are activated by cell surface components of bacteria. Activation of any of these three pathways leads to the opsonization of the target with the complement component C3b. Formation of C3b by a set of programmed enzymatic reactions proceeds to the assembly of a membrane attack complex (MAC) and bacteriolysis. In addition, the complement receptors present on phagocytes bind C3b and iC3b molecules on the bacteria to induce phagocytosis. However, it is not clear which of the three complement pathways play a major role in host defence against E. coli K1.
Several pathogenic bacteria have the capacity to avoid serum bactericidal activity by binding to serum inhibitors of the activated complement proteins C3b and C4b. Factor H (FH) is a host serum regulator of complement activation (RCA) and inhibits C3 activation in the alternative pathway. FH acts as a cofactor for factor I (FI) mediated cleavage of C3b leading to its inactivation and accelerates decay of alternative pathway C3-convertase (C3Bb, an enzymatic complex generating C3b) by displacing C3Bb. Neisseria gonorrhoeae has been shown to capture FH via bacterial sialic acid, which consequently decreased the deposition of C3b on the surface.8 Similarly, streptococci, e.g. Streptococcus pyogenes M6,9S. pyogenes M1810 and S. agalactiae Type 1,11, 12 have been shown to bind FH and FH-like proteins via surface proteins and to decrease the amount of C3 deposition on the bacterial surface.
C4b-binding protein (C4bp) is the predominant serum inhibitor of C3b activation via the classical pathway. C4bp is a cofactor in FI-mediated cleavage of C4b to C4d and increases the dissociation of the classical pathway C3 convertase.7 We showed previously that complement control protein (CCP) domain 3 (CCP3) of the α-chain of C4bp binds specifically to OmpA of E. coli K1 and that this interaction may play an important role in protection against serum killing.13 Our present study shows strong evidence that OmpA+E. coli grown to logarithmic phases (LP) bind greater amounts of C4bp and accordingly survive more efficiently in serum than those grown to postexponential phase (PEP). Also, the E. coli-bound C4bp exhibits FI cofactor activity, which leads to the degradation of C3b and C4b, thus regulating the activation of the classical complement pathway. We further demonstrate that such an interaction could be blocked by synthetic peptides directed to the CCP3 domain of C4bp α-chain.
Materials and methods
Bacterial strains, culture conditions and chemicals
All strains used in this study were derived from a cerebrospinal fluid clinical isolate of E. coli K1 strain RS218 (serotype O18:K1:H7) as described previously.14 E44 is a spontaneous rifampin-resistant mutant of RS218. A mutant lacking the entire ompA gene (E91) was generated from strain E44 in two steps.15 First, a tetracycline-resistance marker was mobilized from DME 558, which contains the marker neo-ompA, by P1 transduction to E. coli K12 strain BRE51 in which the ompA gene had been deleted. Then, a P1 lysate was used to transduce E. coli K1 strain E44. The tetracycline-resistant colonies were screened for lack of OmpA expression by Western blot analysis to identify mutant E91. Bacteria were grown in brain heart infusion broth (Difco, Detroit, MI) with appropriate antibiotics at the following concentrations: rifampin 100 μg/ml (E44); tetracycline 12·5 μg/ml (E91). Normal adult human serum (NHS) was prepared from blood obtained from healthy individuals with the approval of the Clinical Committee on Investigation of CHLA, pooled, divided into aliquots, stored at 70°, and used for experiments within 1 week. The NHS used in these studies was free of any antibody reactive to E. coli K1 as assessed by colony blot analysis. However, these strains did bind non-specific immunoglobulin G (IgG) as examined by immunofluorescence experiments using anti-human IgG coupled to fluorescein isothiocyanate (FITC). For some experiments, NHS was incubated either at 56° for 30 min to yield heat-inactivated serum or at 50° for 30 min to inactivate alternative complement pathway.
Antibodies and reagents
Antibodies to C1q, C3, C5, C9, FH, C4bp, C4b, C3b and FI were obtained from Calbiochem (San Diego, CA). Purified C3b, C4b and factor I proteins, polyclonal anti-C4 and monoclonal anti-C4d antibodies were obtained from Quidel Corp. (San Diego, CA). C8-deficient serum, antibodies to human IgG and IgM labelled with FITC, and all other chemicals were purchased from Sigma Chemical Co. (St Louis, MO). Purification of C4bp and FH were performed as described previously.16 Monoclonal antibodies to SfaS, a subunit of S-fimbria, were generously provided by Dr Hacker, University of Wurzburg, Germany.
C4bp peptides
Peptide sequences that represent fragments of CCP3 of C4bp were synthesized as described previously13 and used in the inhibition assays. The sequences of the peptides are as follows: CCP3-2: 141-EENFYAYGF-149 and CCP3-4: 156-DPRSFSLLGH-165. The bacteria were treated with each peptide (50 μm) for 1 hr on ice before incubating with serum. The peptides were present in the medium throughout the experiment.
Dot-blot analysis of C3b deposition on E. coli
Equal lables of bacteria were treated with various concentrations of either CCP2 or CCP4 peptide for 30 min on ice and then incubated with 40% NHS for varying periods at 37°. The bacteria were washed and an equal numbers of bacteria was loaded onto nitrocellulose. The lable of bacteria loaded from each sample was enumerated by plating the dilutions on blood agar. The nitrocellulose was then blocked with 5% bovine serum albumin in phosphate-buffered saline (PBS) for 1 hr followed by incubation with anti-C3 or anti-S-fimbria antibody for 1 hr at room temperature. The blot was washed three times with PBS and then incubated with peroxidase-conjugated secondary antibody. The blot was exposed to an X-ray film and the image was captured by scanning. The densities of the dots were quantified using ImageJ software (National Institutes of Health). The densities of the dots were normalized to the densities of the dots with anti-S-fimbria antibody and graphed.
Flow cytometry analysis
We used flow cytometry to determine the binding or deposition of complement proteins C1q, C3b, C5b and C9 on the surface of E. coli in NHS. OmpA+ and OmpA–E. coli K1 strains were grown for 6–8 hr to log phase (LP) or overnight (postexponential phase, PEP) in medium containing appropriate antibiotics. The cells were centrifuged and washed twice with Hanks' balanced salt solution (HBSS) at room temperature. The bacteria were re-suspended in HBSS and adjusted to 1 × 109 cells/ml at OD600. The bacterial suspension (100 μl) was incubated with NHS or C8-deficient serum (final concentration of 40%) for varying periods at 37°. The suspension was centrifuged at 12 000 g for 5 min and the pellet was re-suspended in PBS. To determine the bound components of the complement cascade, the bacteria were incubated with antibodies against C1q, C3, C5 and C9 (as a marker for the MAC) at room temperature for 30 min followed by three washings with PBS. The bacterial pellets were then incubated with FITC-conjugated secondary antibodies for 30 min at room temperature, washed and re-suspended in PBS. Flow cytometry analysis was carried out on a Becton-Dickinson Flow analyser at the University of Southern California, using Cell Quest software and approximately 10 000 gated events were recorded.
Serum bactericidal assays and Western blotting
Susceptibility of E. coli strains to complement-mediated killing was carried out using non-immune NHS or C8-deficient serum at a final concentration of 40%. The bacteria (105 cells) were suspended in HBSS or PBS along with serum and incubated at 37° for various time-points. The bacteria were then plated on blood agar in dilutions for quantitative determination. Serum bactericidal assays were also carried out using gelatin Veronal buffer (GVB) with either calcium and magnesium or EGTA. Routinely heat-inactivated NHS (56° for 30 min) was included as a control. In separate experiments, the E. coli strains were incubated with C4bp peptides on ice for 1 hr prior to the addition of NHS. The effect of these peptides on bactericidal survival was examined by the colony count method. For Western blot analyses, 109 colony-forming units (CFU) of OmpA+E. coli were incubated with NHS for varying periods as indicated at 37° and the bacteria were pelleted by centrifugation and re-suspended in PBS. The total lable of bacteria in the samples was estimated using OD600 readings during the final wash with PBS. In addition, a portion of the suspension was used to determine the bacterial count by plating the dilutions on the blood agar. A known lable of bacteria in PBS were then dissolved in Laemmli buffer containing β-mercaptoethanol, boiled, separated by 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), and transferred to nitrocellulose. The nitrocellulose membranes were blocked with 5% non-fat milk and then analysed by immunoblotting with antibodies to complement proteins.
C3b/C4b degradation assays
To examine cofactor activity of bacteria-bound C4bp, either OmpA+ or OmpA–E. coli (109 bacteria) were incubated with 200 nm purified C4bp for the indicated periods at 37°, the unbound C4bp was removed by centrifugation and the bacteria were washed with PBS three times. The bacteria coated with C4bp were incubated with either 500 nm C3b or 250 nm C4b and 60 nm of FI in PBS (final volume of the reaction was 250 μl). The samples were incubated for 1 hr at 37° and then centrifuged to remove the bacterial pellet. An aliquot (25 μl) of the supernatant was separated by SDS–PAGE, transferred to nitrocellulose, and then subjected to Western blotting with polyclonal anti-C3 antibody that recognizes all fragments of C3b. In some experiments, the bacteria were incubated with C8-deficient serum for varying periods. The bacteria were centrifuged, washed and treated with 100 mm hydroxylamine to release the bound C3b/iC3b fragments, boiled in the presence of SDS–PAGE buffer containing β-mercaptoethanol and separated by SDS–PAGE followed by immunoblotting with anti-C3 antibody. In addition, monoclonal anti-C4d antibody (Quidel Corp.), recognizing the alpha subunit and its product C4d, was used in the analysis of supernatants from C4b degradation assay.
K1 capsular polysaccharide determination
Cells were prepared as described by Vermeulen and colleagues5 while colorimetric assay was performed according to Warren17. Briefly, 1010 bacteria (as estimated by OD600) were harvested and washed once in PBS, pelleted and resuspended in 5 ml cold PBS for sonication. Twenty microlitres was removed for quantitative estimation of the bacteria and the remaining suspension was sonicated continuously at 50% power for 4 min on ice. Following sonication, 200 μl was mixed with 0·1 ml of the periodate solution (0·2 m in 9 m phosphoric acid). The tubes were shaken and allowed to stand at room temperature for 20 min. Then, 1 ml of sodium arsenite solution (10% in solution of 0·5 m sodium sulphate 0·1 N H2SO4) was added and shaken until the yellow-brown colour disappeared followed by the addition of 3 ml of thiobarbituric acid (0·6% in 0·5 m sodium sulphate). The tubes were shaken and heated vigorously in a boiling water bath for 15 min. Of this solution, 1 ml is transferred to another tube, which contains 1 ml of cyclohexanone. The tube was shaken twice and then centrifuged for 3 min at 1000 g The clear upper red-coloured cyclohexanone phase was collected and the absorbance was measured at 549 nm. The molecular extinction coefficient used was 57 000. The amount of N-acetylneuraminic acid present in a given sample is determined from the following equation: V × OD549/57 = 4·3 × OD549/57 = 0·075 × OD549, where V is the final volume of the test solution.
Results
Activation of complement system by LP OmpA+E. coli
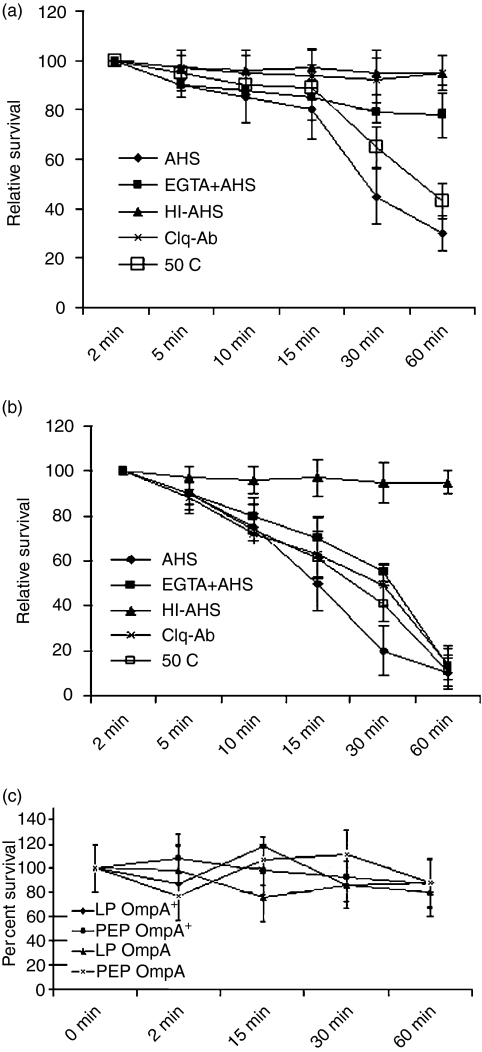
C3 of the complement system can be activated by bacterial surface components and such activation occurs via the classical, lectin and alternative pathways. Our previous studies have shown that despite similar binding of C1q, a strong activator of the classical pathway, from NHS to both OmpA+ and OmpA–E. coli, the survival of OmpA+E. coli in NHS was significantly greater than that of OmpA–E. coli.13 We subsequently identified that OmpA+E. coli binds C4bp, a regulator of complement activation via the classical pathway, indicating a prominent role of the classical pathway in killing E. coli K1. However, the contribution of the alternative pathway in serum killing of E. coli K1 was not assessed. Therefore, we examined the survival of both OmpA+ and OmpA–E. coli under conditions in which either classical or alternative pathway components were inactivated. More than 80% of OmpA+E. coli survived for up to 15 min in NHS (40%) and declined significantly to ∼30% after 1 hr of incubation (Fig. 1a). On the contrary, when the bacteria were incubated in heat-inactivated NHS they were not killed at any time point, indicating that intact complement is pivotal for serum bactericidal activity. Organisms incubated in NHS treated with EGTA, which inactivates both the classical and lectin pathways, had only 20% decrease in survival at 1 hr post-incubation. In addition, treatment of NHS with an anti-C1q antibody (polyclonal, monospecific to C1q) significantly inhibited serum bactericidal activity. In agreement with previous studies,18 pretreatment of NHS with anti-C1q antibody significantly prevented the binding of C1q to the bacteria as assessed by flow cytometry (data not shown). NHS, heated to 50°, which selectively inactivates the alternative pathway, showed very little difference on the OmpA+E. coli serum susceptibility as compared to untreated serum, yet again suggesting that the classical pathway is the major complement pathway that kills the majority of OmpA+E. coli. On the other hand, OmpA–E. coli was susceptible to NHS when either the classical complement pathway or alternative pathway was inactivated suggesting that the OmpA+ and the OmpA– strains had different patterns of survival (Fig. 1b). OmpA–E. coli survived only in heat-inactivated serum, suggesting that both classical and alternative complement pathways contribute to the killing of OmpA–E. coli. This stands in sharp contrast with the observation that mainly the classical pathway is responsible for the bactericidal activity against OmpA+E. coli. We also performed the survival assays in C8-deficient serum that precludes formation of the terminal membrane attack complex and lysis. Both the strains in their LP and PEP showed minimal killing even after 60 min incubation in C8-deficient serum as compared to normal NHS (Fig. 1c).
Figure 1.
Survival of Escherichia coli K1 strains in adult human sera. OmpA+ (a) and OmpA– (b)E. coli K1 strains grown overnight (postexponential) were subjected to serum bactericidal assays using adult human sera either untreated (NHS), heat-inactivated (HI-NHS), EGTA-treated (EGTA + NHS), incubated at 50° for 30 min, or preincubated with anti-C1q antibody. (c) In separate experiments, the bacteria were subjected to serum bactericidal assays using C8-deficient serum at a final concentration of 40%. The results are presented as means of at least three individual experiments and are expressed as relative survival with the survival of E. coli at 0 min being taken as 100%.
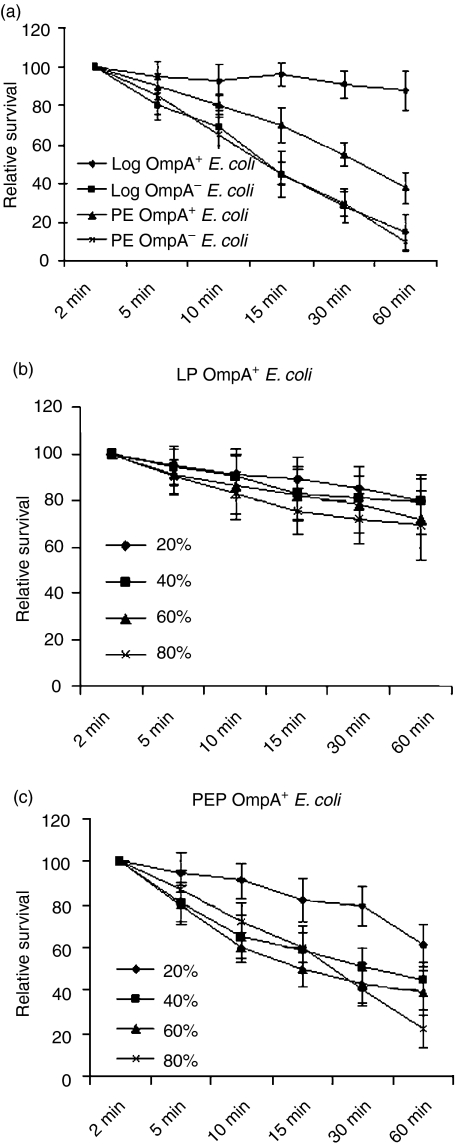
It has been demonstrated that growing conditions affect the surface structures in several bacterial pathogens.5 Therefore, we speculated that OmpA+E. coli were killed during the first hour of incubation in serum, possibly because they were grown overnight (PEP). To examine this possibility, we compared the logarithmic phase (LP) and PEP phase of both the strains of E. coli by subjecting them to serum bactericidal assays under similar conditions. As shown in Fig. 2(a), LP OmpA+E. coli continued to survive in serum without any signs of bactericidal susceptibility, suggesting that the LP OmpA+E. coli are more resistant to serum bactericidal activity when compared to PEP bacteria. In contrast, OmpA–E. coli in LP and PEP did not survive.
Figure 2.
Survival of LP and PEP OmpA+Escherichia coli K1 in various NHS concentrations. Serum bactericidal assays were performed on the bacteria grown to LP and PEP using NHS at a final concentration of 40% (a). Furthermore, OmpA+E. coli grown to LP and PEP were subjected to serum bactericidal activity with various serum concentrations for the indicated periods (b,c). The results are presented as means of at least three individual experiments and are expressed as relative survival with the survival of E. coli at 0 min set as 100%.
Furthermore, to determine the influence of the amount of serum on E. coli K1 susceptibility, bacteria were incubated with various concentrations of NHS up to 60 min. As shown in Fig. 2(b) and 20% and 40% serum concentrations had no effect on LP OmpA+E. coli survival, whereas a slight increase in serum susceptibility was observed at 60% and 80% serum concentrations. Longer periods of incubation with NHS did not enhance serum bactericidal activity (data not shown). PEP OmpA+E. coli, on the other hand, survived well in 20% NHS when compared to concentrations beyond 40% in which 60–70% of bacteria were killed (Fig. 2c). In contrast, neither LP nor PEP OmpA–E. coli could survive in any given concentration of NHS (not shown, but similar to Fig. 1b). In addition, neither of the bacteria was killed in C8-deficient serum even at 80% concentration (data not shown). These results suggest that LP OmpA+E. coli resist serum bactericidal activity more efficiently when compared to PEP bacteria even at very high concentrations of serum.
Comparison of K1 capsular polysaccharide production and C4bp binding by LP and PEP E. coli
Vermeulen et al. found that E. coli grown to LP produces large amounts of K1 capsular polysaccharide that confer serum resistance on E. coli K1.5 Therefore, we examined whether increased survival of LP OmpA+E. coli K1 is the result of the presence of greater quantities of K1 capsule, when compared to LP OmpA–E. coli. The amount of K1 capsular polysaccharide on these bacteria was determined using a colorimetric assay.17
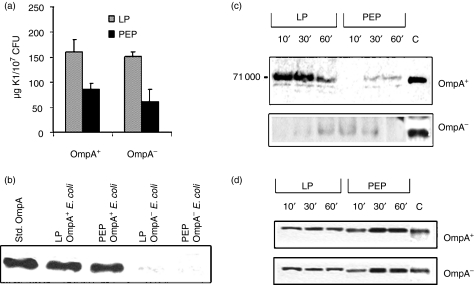
In the previous studies, the highest amount of capsule was observed on bacteria grown to a density of 105/ml in brain heart infusion broth. However, we were only able to perform the assay with detectable quantities of K1 capsule grown to a density of 107/ml. As shown in Fig. 3(a), LP E. coli, both OmpA+ and OmpA– contained twice the amount of K1 capsular polysaccharide when compared to PEP bacteria, indicating that greater survival of OmpA+E. coli when compared to OmpA–E. coli was not the result of the presence of more K1 capsular polysaccharide. We also determined K1 capsular polysaccharide content in the media of both phases of bacteria. The media of PEP bacteria contained ∼10 times more K1 capsular polysaccharide when compared to the content present in the media of LP bacteria, suggesting that the PEP E. coli K1 kept growing new K1 capsule while shedding it into the medium (data not shown). Since our studies have shown that OmpA expression confers serum resistance, we examined the quantity of OmpA expressed in these bacteria by Western blot analysis of outer membrane proteins using an anti-OmpA antibody. Interestingly, the results showed no significant difference in the quantities of OmpA between LP and PEP OmpA+E. coli (Fig. 3b). As expected, OmpA–E. coli did not show significant reactivity with the antibody.
Figure 3.
Assessment of K1 capsule and OmpA production, and the binding of C4bp and FH to LP OmpA+Escherichia coli. (a) Both OmpA+ and OmpA–E. coli grown to LP and PEP were used for estimation of K1 capsule as described in the Materials and methods section. Data represent at least three separate experiments and are expressed as μg of K1 capsule ± SD per 107 CFU. (b) Equal numbers of bacteria were suspended in SDS sample buffer containing β-mercaptoethanol and subjected to electrophoresis followed by Western blotting with an anti-OmpA antibody. Purified OmpA was included as a positive control. (c) and (d) Both LP and PEP OmpA+E. coli were incubated with 40% NHS for varying periods, the bacteria were collected by centrifugation, washed, dissolved in SDS buffer, and subjected to PAGE. After transferring the proteins to nitrocellulose, the blots were developed with anti-C4bp antibody (c) and anti-FH antibody (d). Purified C4bp and FH were loaded as positive controls on the gel.
Since the antibody against C4bp (Calbiochem) could not be used for flow cytometry analysis, Western blot analysis was carried out to examine the binding of C4bp to LP and PEP OmpA+E. coli. LP bacteria bound significantly greater amounts of C4bp after 10 min incubation with NHS, as compared to PEP E. coli (Fig. 3c). Interestingly, the amount of C4bp bound to OmpA+E. coli decreased by approximately 50% at 60 min postincubation. In contrast, OmpA–E. coli bound negligible quantities of C4bp, which is in agreement with our previous data.13 The differences in the binding of C4bp to LP and PEP OmpA+E. coli are not the result of unequal loading of the bacteria as equal lables of CFU were loaded in each lane after enumerating the colonies by plating the dilutions on the blood agar. Similar experiments were also carried out to examine the binding of the inhibitor of the alternative pathway, FH, to the surface of the bacteria and demonstrated that both LP and PEP OmpA+E. coli bound to similar amounts of FH (Fig. 3d). However, the binding of FH is unable to protect the OmpA–E. coli from serum killing, whereas increased binding of C4bp to OmpA+E. coli is able to increase the survival of the bacteria in NHS.
LP OmpA+E. coli inhibits the deposition of complement cascade beyond C3
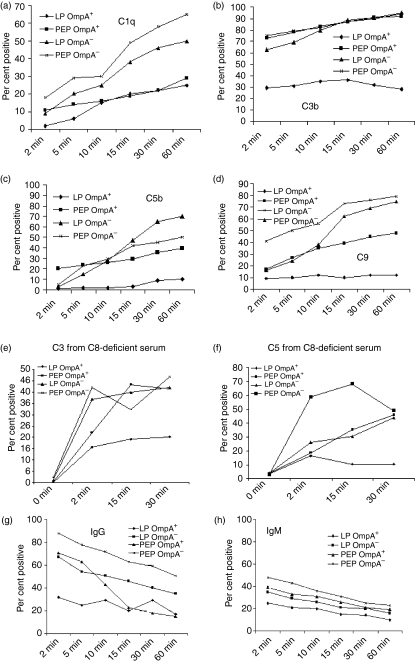
Having demonstrated that C4bp binds to OmpA+E. coli,13 we wished to identify the mechanism of serum resistance by E. coli K1. Flow cytometry was employed using antibodies against C1q, C3 (recognizes deposited C3b), C5 (recognizes deposited C5b), and C9 to verify any differences in the amounts of specific proteins of the complement cascade deposited on E. coli during the bactericidal reaction after incubation with 40% NHS. Cells were scored positive for complement binding if they were more fluorescent than cells treated without the primary antibody. Forward and side scatter analysis suggested cell membrane distortion upon serum exposure, however, this did not differ between OmpA+ and OmpA–E. coli. As shown in Fig. 4(a), OmpA+E. coli and OmpA–E. coli grown to LP bound equivalent amounts of C1q on their surface by 15 min postincubation. Interestingly, the deposition of C1q on LP OmpA–E. coli continued to increase up to 60 min, whereas C1q deposition on LP OmpA+E. coli stayed constant even at 60 min of incubation. The deposition of C3b on LP OmpA–E. coli was almost twice that of LP ompA+E. coli up to 15 min postincubation (Fig. 4b). This difference seemed to increase further as the time was increased to 60 min. In contrast, the deposition of C5b and C9 was significantly greater on LP OmpA–E. coli when compared to LP OmpA+E. coli (Fig. 4c,d). Interestingly, the depositions of C1q, C5b, and C9 on PEP OmpA+ and OmpA–E. coli were similar at all time-points. The deposition of C3b on PEP OmpA+E. coli was significantly lower than on OmpA–E. coli up to 10 min postincubation and increased by 60 min, although the binding was ∼20% lower. These data indicate that LP OmpA+E. coli resist serum killing more efficiently by inhibiting the deposition of C3b products and downstream complement proteins on its surface when compared to PEP OmpA+E. coli or the same bacteria lacking OmpA and grown to any phase. We also compared the deposition of C3b and C5b using C8-deficient sera to eliminate potential problems arising from the fact that some bacteria will be lysed in normal serum. The deposition of both C3b and C5b were similar on PEP OmpA+E. coli, and LP and PEP OmpA–E. coli (Fig. 4e,f). In agreement with the results of NHS, LP OmpA+E. coli significantly inhibited the deposition of both C3b and C5b.
Figure 4.
Deposition of complement proteins on Escherichia coli K1. The bacteria grown to LP (a,c) and PEP (b,d) were incubated with 40% NHS for the periods indicated. The bacteria were then collected by centrifugation and subjected to flow cytometry with anti-antibodies detecting C1q, C3b, C5b, or C9. The experiments were repeated at least three times with similar results. In separate experiments, both LP and PEP bacteria were incubated with C8-deficient serum for varying periods and examined by flow cytometry for the deposition of C3b (e)and C5b (f). In similar flow cytometry analysis, the bacteria were also examined for the binding of IgG and IgM antibodies from NHS using FITC-conjugated anti-IgG or anti-IgM antibodies (g,h). The data represent means of per cent of positive cells in population in the positive gate ± standard deviation from four different experiments.
Although both PEP OmpA+ and OmpA–E. coli showed similar binding of C1q by flow cytometry analysis, LP OmpA+E. coli bound significantly lower amounts of C1q when compared to LP OmpA–E. coli. Neisseria gonorrhoeae, which binds IgG in addition to C4bp, also regulates the levels of IgM binding to the surface to avoid complement activation.19 Therefore, the binding of antibodies (IgG and IgM) present in non-immune sera to LP bacteria was examined by flow cytometry analysis. Even non-immune sera contain natural IgM antibodies that recognize bacterial lipid antigens and apparently there is a pool of cross-reacting IgG antibodies. As shown in Fig. 4(g), LP OmpA+E. coli bound 50% less IgG as compared to LP OmpA–E. coli at all time points. The binding of IgG was also 50% greater to PEP OmpA+E. coli when compared to LP OmpA+E. coli until 5 min postincubation with the difference in binding disappearing by 60 min. In contrast, the binding of IgM is only slightly greater to OmpA–E. coli compared to that of IgM bound to OmpA+E. coli in both growth phases. Extended incubation did not change the amount of IgM bound to the bacteria (data not shown), suggesting that OmpA+E. coli, in addition to C4bp binding, may also influence the binding of IgG to bacterial surface, and that both factors may contribute to the final outcome, which is the decreased bacterial killing.
C4bp bound to E. coli K1 exhibits cofactor activity against C3b and C4b
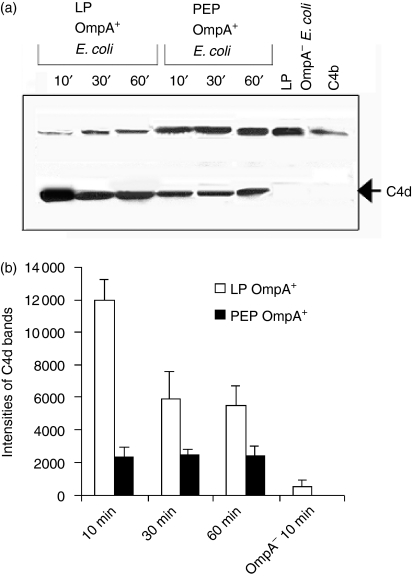
C4bp regulates the classical pathway activity by serving as a cofactor in the inactivation of C4b in the presence of FI, and yields the fragments C4d and C4c.7 The anti-C4d antibody recognizing both the intact α-chain of C4b and its cleaved product C4d was used to analyse the cofactor activity of C4bp bound to the cells. Briefly, OmpA+E. coli grown to LP and PEP were incubated with pure C4bp, unbound C4bp was removed and then pure C4b and Factor I were added. Incubation proceeded for the indicated periods, followed by washing and pelleting down of the bacteria. Western blotting of the proteins in the supernatant with anti-C4d antibodies showed a band of ∼45 000 MW, indicating that LP OmpA+E. coli incubated with pure C4bp cleaved C4b more efficiently as compared to PEP OmpA+E. coli(Fig. 5a). In contrast, the LP OmpA–E. coli showed no such cleavage of added C4b up to 60 min incubation (only 10 min data-point is shown). Densitometric analysis of bands corresponding to C4d and normalization to equal lables of bacteria loaded in each lane indicated that LP OmpA+E. coli generated two or three times more C4d than PEP OmpA+E. coli (Fig. 5b).
Figure 5.
Degradation of C4b by C4bp bound to OmpA+Escherichia coli. (a)Either OmpA+ or OmpA–E. coli grown to LP and PEP were incubated with NHS or purified C4bp for various periods at 37°. The bacteria were then collected by centrifugation, washed, and incubated with C4b and FI for 1 hr at 37°. The bacteria were then pelleted and a portion (30 μl) of the supernatant was dissolved in SDS buffer, and subjected to immunoblotting with anti-C4d antibody (recognizes α-chain of C4b and C4d). The experiments were carried out at least three times with similar results. (b)The bands of C4d on the blot were scanned, the intensities of the bands were normalized to number of bacteria loaded, and plotted on graphs.
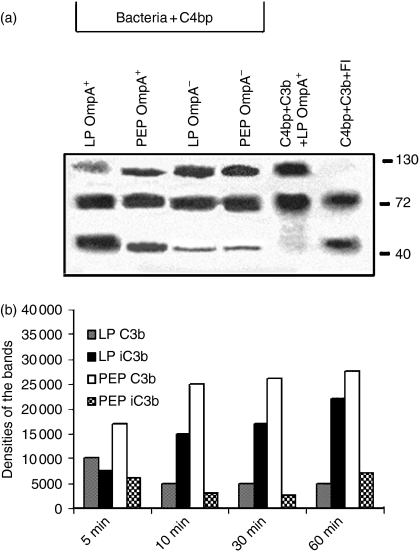
To assess the ability of the bound C4bp as a cofactor in the cleavage of C3b, both LP and PEP OmpA+E. coli were incubated with purified C4bp, washed, centrifuged and the bacterial pellet was further incubated with C3b and FI for the indicated periods. The bacteria were centrifuged, washed and an aliquot of the supernatant (20 μl) was then subjected to Western blotting with anti-C3 antibody. As shown in Fig. 6(a), LP OmpA+E. coli enhanced the cleavage of C3b (presence of 43 000–46 000 MW degradation product) when compared to other bacteria. No cleavage was observed in the absence of FI, indicating that the bacteria by themselves could not cause the degradation of C3b. Interestingly, OmpA–E. coli also showed some effect on the cleavage of C3b in both growth phases, which could be the result of the cofactor activity of C4bp bound to these bacteria in minute quantities as we showed previously.13 In addition, OmpA+ and OmpA–E. coli were treated with C8-deficient serum (to avoid lysis of the bacteria) for varying periods and the bound C3b fragments were released with hydroxylamine. Analysis of C3b fragments by Western blotting with anti-C3 antibody showed that the generation of iC3b on OmpA+E. coli was less when compared to PEP OmpA+E. coli (Fig. 6b). However, the amount of C3b bound to LP OmpA+E. coli was notably lower than the C3b on the surface of PEP OmpA+E. coli. These results suggest that C4bp bound to E. coli K1 acts as a cofactor to FI to inactivate both C3b and C4b; thus it may further provide protection against opsono-phagocytosis.
Figure 6.
C4bp bound to OmpA+Escherichia coli exhibits cofactor activity to FI in cleavage of C3b. (a)Either OmpA+ or OmpA–E. coli grown to LP and PEP were incubated with purified C4bp for 30 min at 37°. The bacteria were then collected by centrifugation, washed, and incubated with C3b, and FI for 1 hr at 37°. The bacteria were then pelleted and a portion of the supernatant was dissolved in SDS buffer, and subjected to immunoblotting with anti-C3 antibody (which recognizes α- and β-chains and the cleavage products). The experiments were carried out at least twice with similar results. (b)In separate experiments, LP and PEP OmpA+E. coli were incubated with 40% C8-deficient serum for various periods, centrifuged, washed, the bacterial pellets were treated with hydroxylamine and mixed with SDS sample buffer containing β-mercaptoethanol. The proteins were then subjected to Western blotting with anti-C3 antibody, the bands on the blot were scanned and their intensities were normalized to total bacteria added and plotted on graphs.
Peptide that represents the C4bp binding region to OmpA enhances C3b deposition
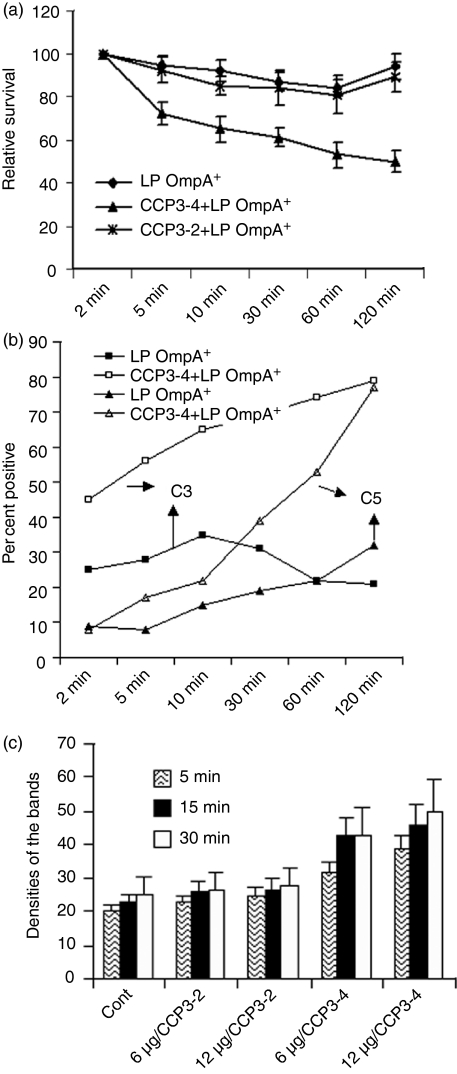
Our previous studies have shown that the binding of C4bp to OmpA was significantly blocked by incubating the bacteria with a peptide that represents the region of C4bp involved in the binding of OmpA.13 Therefore, we used this peptide to examine whether blocking of C4bp binding to OmpA increases serum bactericidal activity against LP OmpA+E. coli. The bacteria were incubated with the peptide, designated as CCP3-4 for 1 hr on ice prior to adding the NHS. Another peptide, CCP3-2, derived from the same domain of C4bp but unable to block the interaction, was used as a control. As shown in Fig. 7(a), treating the bacteria with CCP3-4 increased the susceptibility of LP bacteria to NHS bactericidal activity by up to 50% when compared to either untreated or treated with CCP3-2 peptide. Further increase in the peptide concentration did not result in increased killing of the bacteria. The bacteria were also analysed by flow cytometry for C3b and C5b deposition at various time-points (Fig. 7b). The deposition of both C3b and C5b on LP OmpA+E. coli surface was enhanced by 20–30% in the presence of CCP3-4 peptide, whereas the same bacteria pretreated with CCP3-2 did not show such increased deposition.
Figure 7.
CCP3-4 peptide treatment of LP OmpA+Escherichia coli increases the deposition of C3b and the serum sensitivity of the bacteria. (a)LP OmpA+E. coli were incubated with either a control peptide (CCP3-2) or a blocking peptide (CCP3-4) for 1 hr on ice and then subjected to serum bactericidal activity. The data represent means ± standard deviation from at least three separate experiments. (b)In separate experiments the bacteria treated with CCP3-4 or CCP3-2 were incubated with 40% NHS for various periods, centrifuged to collect the bacteria, washed and subjected to flow cytometry analysis using anti-C3 and anti-C5 antibodies. The bacteria incubated without primary antibody were used as a negative control. The data represent means of percentage of positive cells in the positive gate ± SD from three different experiments. (c)LP OmpA+E. coli were treated with or without peptides as described in (a) and equal number of bacteria were adsorbed onto a nitrocellulose filter. After drying, dot blot analysis was carried out using either anti-C3 or anti-S-fimbria antibody. The blots were exposed on an X-ray film, the film was scanned, the densities of the dots were determined, and plotted on graphs. The experiments were performed at least three times and the data represent mean ± SD.
In addition, for quantitative determination of C3b deposition on the bacteria treated with the peptides, dot blot assays were performed. As shown in Fig. 7(c), the deposition of C3b on LP OmpA+E. coli increased gradually with increased concentration of CCP3-4, whereas CCP3-2 showed no effect on the deposition. At a concentration of 12 μg, CCP3-4 peptide treatment increased the deposition of C3b by 50%, suggesting that the inhibition of C4bp binding by CCP3-4 peptide enhances the C3b deposition, probably for subsequent killing of the bacteria. Furthermore, LP OmpA+E. coli, which was preincubated with CCP3-4 followed by NHS, revealed no significant cleavage of C4b to C4d in contrast to CCP3-2-treated bacteria (data not shown). These results suggest that increased killing of CCP3-4-treated LP OmpA+E. coli is the result of increased deposition of C3b and other downstream complement proteins.
Discussion
Many pathogenic micro-organisms avoid host defence mechanisms and successfully survive in the host. Since the complement system is the first line of defence and a major arm of innate immunity, microbes have evolved strategies to regulate complement at various steps of the cascade. Escherichia coli K1, a causal agent of meningitis in neonates that primarily disseminates into the central nervous system by haematogenous spread, must avoid serum bactericidal activity to survive and establish infection.2 Numerous factors seem to affect the resistance of a given micro-organism including the presence of outer membrane proteins such as M proteins,9 O-antigens of LPS such as O7 and O18, and sialic acid-containing capsules.4,20 In addition, there are now several lines of evidence indicating that cooperation takes place between these surface-bound elements. For example, it has been noted for several decades that of all the possible combinations of O and K antigens available, O18:K1 is predominant in pathogenic E. coli K1 strains that cause neonatal meningitis.20 Our recent studies showed that E. coli binds to C4bp via OmpA, thereby potentially manipulating the classical pathway of complement and avoiding killing by serum. In addition, these studies showed that although FH purified from serum binds to both OmpA+ and OmpA–E. coli it does not effect the OmpA–E. coli survival in serum, further substantiating the role of C4bp in protecting E. coli K1 from serum bactericidal activity.
In the present study, we demonstrate that OmpA+E. coli grown to LP resist serum bactericidal activity more efficiently than the same bacteria grown to PEP and OmpA–E. coli grown to both phases. The survival efficiency of LP OmpA+E. coli is the result of its ability to decrease and delay C3b deposition and subsequent MAC formation and lysis. Vermeulen et al. have previously shown that growth conditions affect the production of the capsules of bacteria.5 In agreement with this observation, we have also found that LP E. coli produces more K1 capsular polysaccharide as compared to the PEP bacteria. However, the survival of LP OmpA–E. coli did not improve despite increased capsule production, indicating that OmpA but not the capsule is the major factor contributing to the serum resistance of E. coli K1. Although the expression of OmpA in LP and PEP OmpA+E. coli is similar, C4bp binding was more significant to log phase (LP) bacteria. It is possible that the presence of more K1 capsular polysaccharide in LP OmpA+E. coli stabilizes the highly mobile external loops of OmpA, which may lead to better interaction with C4bp. Alternatively, O-antigens, which are known to be regulated by growth conditions, may also be responsible for increased binding of C4bp and this possibility remains to be examined.
Interestingly, LP OmpA+E. coli bound at least 50% less of IgG when compared to PEP bacteria, which correlated well with higher binding of C1q to PEP OmpA+E. coli early in incubation. In contrast, the binding of IgM to LP OmpA+E. coli was only 10% lower than that of PEP bacteria. This initial regulation of antibody binding may be one of the factors responsible for unimpeded survival of LP OmpA+E. coli when compared to PEP E. coli. During this period, the bound C4bp aids cleavage of C4b and C3b and regulates the complement cascade, which provides additional protection against serum bactericidal activity. A lable of pathogens known to date to bind C4bp are, Neisseria gonorrhoeae, group A streptococcus, Bordetella pertussis, Moraxella catarrhalis and Candida albicans.19,21–25 In all the above cases bound C4bp serves as the FI cofactor. In the case of streptococci, binding of C4bp has been shown to be the major mechanism for serum resistance.21,26 Of note, C4bp bound to FHA of B. pertussis does not seem to significantly protect from bactericidal activity of serum.24
Several bacteria survive complement attack by avoiding alternative complement pathway activation. It is known that sialic acid enhances the binding of FH to any cell surface, which promotes inactivation of C3b by FI.27 Sialic acid, present as a terminal sugar on the lipo-oligosaccharides of N. gonorrhoeae, may be responsible for the binding of FH, which leads to the deposition of lower amounts of C3b and iC3b on the bacterial surface to decrease complement attack. In other cases, such as S. pneumoniae, there are specific membrane proteins capturing FH.28,29 Some pathogens, such as Streptococcus pyogenes, bind both C4bp and FH, which provides protection from all complement pathways. We do not know yet what surface component of E coli K1 binds FH, but we presented conclusive evidence that the binding of FH does not contribute to serum survival of the bacteria. In addition, the OmpA–E. coli did not survive in the presence of EGTA, which entirely blocks the classical pathway. Therefore, the bound FH does not protect OmpA–E. coli from bactericidal activity even in the presence of alternative pathway alone. At this point it is not clear why the alternative pathway does not deposit more C3b on the surface of OmpA+E. coli, when compared to OmpA–E. coli. One possible explanation is that the amount of C3b deposited on E. coli by the classical pathway is below the threshold levels required to activate the alternative pathway. The resistance to serum killing of haemophilus and streptococcus may be similar to that of E. coli, as some these strains bind FH but this does not seem to account for serum resistance.29,30
By preventing activation of the complement cascade beyond C4b because of the binding of C4bp, OmpA protects the bacterium from the effects of complement activation such as opsonization by C3b and iC3b, and from direct killing of bacteria by lysis because of MAC formation. Although antibody opsonization may be very effective for promoting phagocytosis, our previous studies have shown that the sole presence of antibody is not sufficient for killing of the bacteria by phagocytes.31 These studies have also revealed that OmpA binds directly to phagocytes in the presence of serum, suggesting that the binding site for C4bp on OmpA could be different from the binding site for the receptor on phagocytes. Entry of E. coli into macrophages by mechanism(s) other than that of OmpA-mediated bacteria renders the bacteria susceptible for killing, indicating that targeting OmpA may be a valuable approach for increasing serum bactericidal activity as well as phagocytic activity. An interesting finding of this study was the decrease in associated complement components in the presence of a peptide that corresponds to the C4bp-binding region for OmpA. We have previously shown the CCP3-4 is a potent inhibitor of C4bp-binding to E. coli and also enhances killing of bacteria by serum, suggesting that CCP3-4 blocked the C4bp-mediated inactivation of complement. Inhibition of C4b cleavage by OmpA+E. coli pretreated with CCP3-4 and the increased deposition of C3b confirm this concept. Therefore targeting this region on C4bp could be a valuable therapeutic approach to increase the serum bactericidal activity.
In summary, our studies show that LP E. coli K1 is more resistant to complement attack than that of PEP E. coli. This is because of the binding of greater amounts of C4bp to the LP OmpA+E. coli, which exhibits cofactor activity to FI and thereby inactivates C3b and C4b. As a result of the deposition of low quantities of C3b and C4b, the bacteria reduce the amplification of the complement cascade. Growth of E. coli K1 during initial periods in the blood may reflect LP growth conditions and may be required for successful dissemination into the central nervous system. Neutralization of this resistance would prove useful in the prevention and/or treatment of neonatal meningitis.
Acknowledgments
This work was supported by NIH grants HD 41525 and AI40567 (N.V.P) and by the Swedish Research Council (A.B.).
Abbreviations
- C3b
activated form of C3
- C4bp
C4b-binding protein
- FH
factor H
- FI
factor I
- iC3b
inactivated C3b
- LP
logarithmic phase
- MAC
membrane attack complex
- NHS
normal human serum
- OmpA
outer membrane protein A
- PEP
post exponential phase
- RCA
regulator of complement activation
References
- 1.Dawson KG, Emerson JC, Burns JL. Fifteen years of experience with bacterial meningitis. Pediatr Infec Dis J. 1999;18:816–22. doi: 10.1097/00006454-199909000-00014. [DOI] [PubMed] [Google Scholar]
- 2.Deitzman DE, Fischer GW, Schoenknecht FD. Neonatal Escherichia coli septicemia: bacterial counts in blood. J Pediatr. 1974;85:128–30. doi: 10.1016/s0022-3476(74)80308-2. [DOI] [PubMed] [Google Scholar]
- 3.Grimwood K, Anderson P, Tan L, Nolan T. Twelve-year outcomes following bacterial meningitis: further evidence for persisting effects. Arch Dis Child. 2000;83:1111–16. doi: 10.1136/adc.83.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jann K, Jann B. Polysaccharide antigens of Escherichia coli. Rev Infect Dis. 1987;5:S517–26. doi: 10.1093/clinids/9.supplement_5.s517. [DOI] [PubMed] [Google Scholar]
- 5.Vermeulen C, Cross A, Byrne WR, Zollinger W. Quantitative relationship between capsular content and killing of K1-encapsulated Escherichia coli. Infect Immun. 1988;56:2723–30. doi: 10.1128/iai.56.10.2723-2730.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiser JN, Gotschlich EC. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun. 1991;57:2252–8. doi: 10.1128/iai.59.7.2252-2258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sim RB, Dodds AW. The Complement System: an introduction. In: Dodds AW, Sim RB, editors. Complement: a Practical Approach. Oxford: Oxford University Press; 1997. pp. 1–17. [Google Scholar]
- 8.Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, Rice PA. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med. 1998;187:743–52. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thern A, Stenberg L, Dahlback B, Lindahl G. Ig-binding surface proteins of Streptococcus pyogenes also bind human C4b-binding protein (C4 BP), a regulatory component of the complement system. J Immunol. 1995;154:375. [PubMed] [Google Scholar]
- 10.Johnsson E, Thern A, Dahlback B, Heden LO, Wikstrom M, Lindahl G. Human C4 BP binds to the hypervariable N-terminal region of many members in the streptococcal M protein family. Adv Exp Med Biol. 1997;418:505–10. doi: 10.1007/978-1-4899-1825-3_120. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Caballero D, Alberti S, Vivanco F, Sanchez-Corral P, Rodriguez de Cordoba S. Assessment of the interaction of human complement regulatory proteins with group A Streptococcus. Identification of a high-affinity group A Streptococcus binding site in FHL-1. Eur J Immunol. 2000;30:1243–53. doi: 10.1002/(SICI)1521-4141(200004)30:4<1243::AID-IMMU1243>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 12.Areschoug T, Stalhammar-Carlemalm M, Karlsson I, Lindahl G. Streptococcal beta protein has separate binding sites for human factor H and IgA-Fc. J Biol Chem. 2002;277:12642–8. doi: 10.1074/jbc.M112072200. [DOI] [PubMed] [Google Scholar]
- 13.Prasadarao NV, Blom AM, Villoutreix BO, Linsangan LC. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J Immunol. 2002;169:6352–60. doi: 10.4049/jimmunol.169.11.6352. [DOI] [PubMed] [Google Scholar]
- 14.Prasadarao NV, Wass CA, Weiser JN, Stins MF, Huang SH, Kim KS. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:143–51. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bremer E, Silhavy TJ, Maldener M, Cole T. Isolation and characterization of mutants deleted for the sulA-ompA region of the E. coli K12 chromosome. FEMS Microbiol Lett. 1986;33:173–8. [Google Scholar]
- 16.Blom AM, Kask L, Dahlbäck B. CCP1-4 of the C4b-binding protein α-chain are required for factor I mediated cleavage of C3b. Mol Immunol. 2003;39:547–56. doi: 10.1016/s0161-5890(02)00213-4. [DOI] [PubMed] [Google Scholar]
- 17.Warren L. The thiobarbituric assay of sialic acid. J Biol Chem. 1959;234:1971–5. [PubMed] [Google Scholar]
- 18.Roos A, Bouwman LH, Munoz J, et al. Functional characterization of the lectin pathway of complement in human serum. Mol Immunol. 2003;39:655–68. doi: 10.1016/s0161-5890(02)00254-7. [DOI] [PubMed] [Google Scholar]
- 19.Ram S, Cullinane M, Blom AM, et al. Binding of C4b-binding protein to porin: a molecular mechanism of serum resistance of Neisseria gonorrhoeae. J Exp Med. 2001;193:281–95. doi: 10.1084/jem.193.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pluschke G, Mercer A, Kusecek B, Pohl A, Achtman M. Induction of bacteremia in newborn rats by Escherichia coli K1 is correlated with only certain O (lipopolysaccharide) antigen types. Infect Immun. 1983;39:599–608. doi: 10.1128/iai.39.2.599-608.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berggard K, Johnsson E, Morfeldt E, Persson J, Stalhammar-Carlemalm M, Lindahl G. Binding of human C4 BP to the hypervariable region of M protein: a molecular mechanism of phagocytosis resistance in Streptococcus pyogenes. Mol Microbiol. 2001;42:539–51. doi: 10.1046/j.1365-2958.2001.02664.x. [DOI] [PubMed] [Google Scholar]
- 22.Nordstrom T, Blom AM, Forsgren A, Riesbeck K. The emerging pathogen Moraxella catarrhalis interacts with complement inhibitor C4b binding protein through ubiquitous surface proteins A1 and A2. J Immunol. 2004;173:4598–606. doi: 10.4049/jimmunol.173.7.4598. [DOI] [PubMed] [Google Scholar]
- 23.Jarva H, Ram S, Vogel U, Blom AM, Meri S. Binding of complement inhibitor to serogroup B Neisseria meningitides. J Immunol. 2005;174:6299–307. doi: 10.4049/jimmunol.174.10.6299. [DOI] [PubMed] [Google Scholar]
- 24.Berggard K, Johnsson E, Mooi FR, Lindahl G. Bordetella pertussis binds the human complement regulator C4 BP. role of filamentous hemagglutinin. Infect Immun. 1997;65:3638–43. doi: 10.1128/iai.65.9.3638-3643.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meri T, Blom AM, Hartmann A, Lenk D, Meri S, Zipfel PF. The hyphal and yeast forms of Candida albicans bind the complement regulator C4b-binding protein. Infect Immun. 2004;72:6633–41. doi: 10.1128/IAI.72.11.6633-6641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlsson F, Berggard K, Stalhammar-Carlemalm M, Lindahl G. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med. 2003;198:1057–68. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meri S, Pangburn MK. Discrimination between activators and non-activators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci USA. 1990;87:3982–6. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarva H, Janulczyk R, Hellwage J, Zipfel PF, Bjorck L, Meri S. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded Hic protein that binds to short consensus repeats 8–11 of factor H. J Immunol. 2002;168:1886–94. doi: 10.4049/jimmunol.168.4.1886. [DOI] [PubMed] [Google Scholar]
- 29.Janulczyk R, Iannelli F, Sjoholm AG, Pozzi G, Bjorck L. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem. 2000;275:37257–63. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 30.Kotarsky H, Gustafsson M, Svensson HG, Zipfel PF, Truedsson L, Sjobring U. Group A streptococcal phagocytosis resistance is independent of complement factor H and factor H-like protein 1 binding. Mol Microbiol. 2001;41:817–26. doi: 10.1046/j.1365-2958.2001.02496.x. [DOI] [PubMed] [Google Scholar]
- 31.Sukumaran SK, Shimada H, Prasadarao NV. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect Immun. 2003;71:5951–61. doi: 10.1128/IAI.71.10.5951-5961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]