Abstract
Cytochrome P450 (CYP) 3A4 is the most promiscuous of the human CYP enzymes and contributes to the metabolism of ≈50% of marketed drugs. It is also the isoform most often involved in unwanted drug–drug interactions. A better understanding of the molecular mechanisms governing CYP3A4–ligand interaction therefore would be of great importance to any drug discovery effort. Here, we present crystal structures of human CYP3A4 in complex with two well characterized drugs: ketoconazole and erythromycin. In contrast to previous reports, the protein undergoes dramatic conformational changes upon ligand binding with an increase in the active site volume by >80%. The structures represent two distinct open conformations of CYP3A4 because ketoconazole and erythromycin induce different types of coordinate shifts. The binding of two molecules of ketoconazole to the CYP3A4 active site and the clear indication of multiple binding modes for erythromycin has implications for the interpretation of the atypical kinetic data often displayed by CYP3A4. The extreme flexibility revealed by the present structures also challenges any attempt to apply computational design tools without the support of relevant experimental data.
Keywords: drug metabolism, structural flexibility, x-ray crystallography, inhibitor, substrate
Cytochrome P450 3A4 (CYP3A4) is the most abundant of the xenobiotic-metabolizing CYP isoforms, and interactions with CYP3A4 must always be taken into consideration during the development of new medications (1).
In recent years, a number of structures of mammalian CYP isoforms, including CYP2C5 (2), CYP2C9 (3, 4), CYP2C8 (5), CYP2B4 (6), CYP2A6 (7), and CYP3A4 (8, 9) have been solved. All of these structures were determined from modified versions of the protein where the N-terminal transmembrane helix was truncated and, in some cases, a number of mutations aimed at increasing solubility were introduced. The mammalian CYP structures all adopt the general CYP fold first described in 1987 when the structure of the bacterial P450 CYP101 was determined by x-ray crystallography (10).
The ligand-free structure of CYP3A4 was published in 2004 by two independent groups (8, 9). These structures are very similar (11). The most remarkable features are the short F and G helices (nomenclature adapted from Poulos et al.; ref. 10) and a large, highly ordered hydrophobic core of phenyl alanine residues above the active site (8, 9). CYP3A4 is known to metabolize large substrates such as bromocriptine (Mr 655 Da) and cyclosporine (Mr 1,203 Da). A number of studies also indicate that CYP3A4 displays ligand binding that does not follow Michaelis–Menten type kinetics, and it has been suggested that two or more ligand molecules can bind to the CYP3A4 active site simultaneously (12–15). In light of these observations, the volume of the active site in the published ligand-free structures is smaller than expected, comparable with the active-site cavity seen in CYP2C9 and CYP2C8, which led Williams et al. (8) to speculate that conformational changes may occur upon ligand binding. There are two CYP3A4–ligand complex structures published to date with metyrapone and progesterone (8). Metyrapone is bound via its pyridine nitrogen to the heme iron, in agreement with UV-visible absorption spectroscopy. Progesterone was not localized in the active site but was found in a peripheral site suggested to be an effector site. Surprisingly, the protein conformation in both complexes is very similar to the ligand-free structure (rmsd for all Cα atoms is 0.46 and 0.55 Å, respectively). However, both metyrapone and progesterone are small (Mr 226 and 314 Da, respectively) and may not require conformational changes in the protein. Erythromycin, a large CYP3A4 substrate (Mr 734 Da) was present during crystallization in the experiments described by Yano et al. (9). However, although addition of the ligand improved crystal quality, there was no evidence of bound erythromycin in the electron density (9).
The lack of conformational changes in response to ligand binding is in contrast with recently published work on rabbit CYP2B4 describing how different types of ligands induce remarkable conformational changes, demonstrated both by isothermal calorimetry and x-ray crystallography (6, 16, 17).
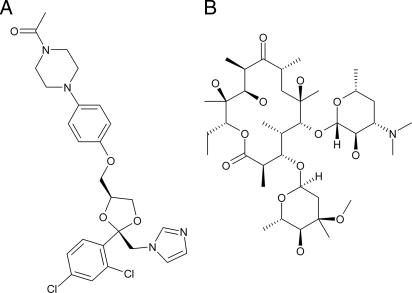
The aim of this work was to obtain structural information on CYP3A4 in complex with larger ligands and, thus, probe conformational changes. We hypothesized that ligand-induced conformational changes must occur also in CYP3A4 and, therefore, great effort was made to characterize the protein–ligand complex before crystallization. We choose to focus on ketoconazole and erythromycin (Fig. 1). The antifungal drug ketoconazole is a well characterized inhibitor of CYP3A4. Upon binding, it gives rise to a type II spectrum, indicative of direct heme binding, and complex formation thus can be followed spectrophotometrically during all stages in complex formation and crystallization. The macrolide antibiotic erythromycin was chosen because its properties in terms of heme coordination, size, and charge distribution are very different from ketoconazole, which would improve the probability for identification of a distinct set of interactions with CYP3A4. The resulting CYP3A4–ligand complex structures reveal a remarkable flexibility, which sheds light on the structural basis of ligand promiscuity, and have implications for efforts aiming to construct structure-based models for CYP3A4 inhibition and metabolism.
Fig. 1.
Chemical structures of ketoconazole (A) and erythromycin (B).
Results and Discussion
Initial studies using dynamic light scattering showed that addition of ketoconazole to purified N-terminally truncated CYP3A4 maintained in a buffer designed to stabilize the ligand-free enzyme (8, 9) caused protein aggregation (data not shown). By modifying the buffer composition, notably by lowering the salt concentration and by adding a polar solvent such as acetonitrile or dioxane, ligand-induced aggregation could be avoided. The preformed complexes then could be crystallized.
The structure of the CYP3A4–ketoconazole was determined to 2.8-Å resolution (see Table 1 and Methods). Upon binding of ketoconazole, the protein undergoes some dramatic conformational changes where the position of secondary structure elements are shifted relative to their positions in the structures of ligand-free CYP3A4. A superposition of the ketoconazole complex and the ligand-free structures yield a rmsd of 1.6 Å for all Cα atoms. The structural elements affected by the conformational changes include both those defining the active site and structural elements further away from the active site (Fig. 2A). The most prominent conformational changes are seen in helices F and G and the intervening loops. The hydrophobic cluster consisting of Phe-213, Phe-215, Phe-219, Phe-220, Phe-241, and Phe-304 seen in the ligand-free structures is broken up, and some of the hydrophobic side chains are exposed to the surrounding medium. The ligand-induced exposure of a hydrophobic patch on the protein surface provides a possible explanation for the need for a buffer with relatively low polarity. Residues 210–213, which lack secondary structure in the ligand-free structure, have adopted a helix-like shape that prolongs the F helix by one extra turn. An implication of the novel conformation of the F-F′ loop is that Arg-212, which is found in the active site in the ligand-free structures, is located on the surface of the protein in the ketoconazole complex. Similarly, Leu-211, which is located on the protein surface in the ligand-free structures, is inside the active site in the ketoconazole complex. This orientation is in line with the observation that a phenylalanine substitution of this residue affects homotropic cooperativity in testosterone binding (13).
Table 1.
Data collection and refinement statistics
| Ligand | Ketoconazole | Erythromycin |
|---|---|---|
| Data statistics | ||
| Space group | C2 | C2221 |
| Unit cell parameters | a = 229 Å, b = 66 Å,c = 146 Å, β=102.8° | a = 67.25 Å, b = 210.71 Å,c = 161.25 Å |
| Resolution range, Å | 30–2.8 (2.87–2.80)* | 30–2.75 |
| No. of reflections,total/unique | 195,505/52,634 | 218,119/30,289 |
| Redundancy | 3.7 (3.6) | 7.2 (7.4) |
| Data completeness, % | 98.8 (98.1) | 99.9 (100.0) |
| Average I/σI | 13.3 (5.5) | 16.4 (3.9) |
| Rmerge† | 0.85 (0.33) | 0.90 (0.52) |
| Refinement statistics | ||
| No. of molecules in asymmetric unit | 4 | 2 |
| rmsd values | ||
| Bond lengths, Å | 0.014 | 0.010 |
| Bond angles, ° | 1.60 | 1.36 |
| R factor‡ | 0.23 | 0.24 |
| Free R factor§ | 0.27 | 0.31 |
*Numbers within parentheses refer to the outer resolution shell.
†Rmerge = Σ|I − 〈I〉|/ΣI.
‡R factor = Σ‖Fo| − k|Fc‖/Σ|Fo| where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
§The Free R factor was calculated as the R factor for 5% of the reflections that were not included in the refinement.
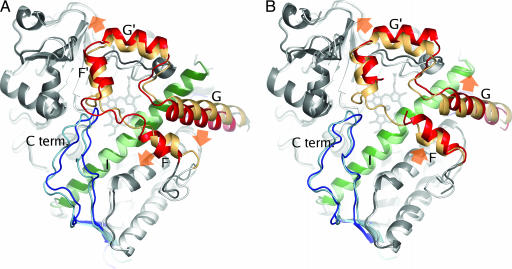
Fig. 2.
Overall structures of CYP3A4 in complex with ketoconazole (A) and erythromycin (B). Structures are shown in dark gray with color highlighting of helices F to G (residues 202–260) in red, helix I (residues 291–323) in green, and the C-terminal loop (residues 464–498) in blue. The complex structures are superimposed on the ligand-free structure (Protein Data Bank ID code 1TQN) shown in light colors. Orange arrows indicate the direction of coordinate shifts in the F-G region relative the ligand-free structure.
Other notable changes include a shift of up to 4 Å of the loop connecting the C-terminal β-sheets and a distortion of the I helix cleft centered around residue 307 (maximum Cα shift: 2 Å). Interestingly, a similar distortion also was observed in the structure of P450eryF in complex with ketoconazole (18).
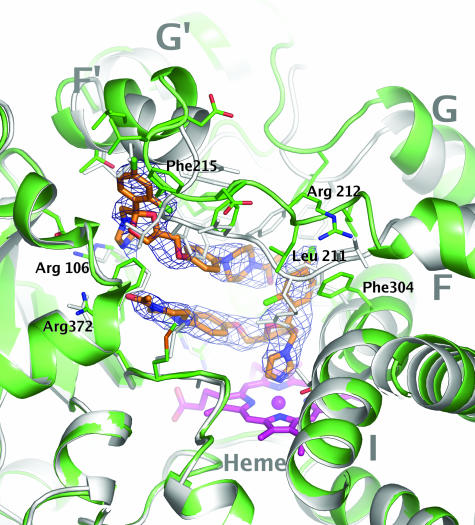
A ketoconazole molecule with its imidazole nitrogen bound to the heme iron could unambiguously be modeled into electron density in all four molecules in the asymmetric unit (Fig. 3). The ketoconazole keto group is located in a polar pocket lined by the side chains of Arg-372, Arg-106, and Glu-374. The interaction is stabilized further by hydrophobic interactions such as π-stacking involving the chlorobenzyl ring and the side chain of Phe-304. Surprisingly, a second molecule of ketoconazole was identified in the active site, stacked in an antiparallel orientation above the first ketoconazole molecule. The keto group is hydrogen bonded to the side chain of Ser-119. The head group, containing the imidazole and chlorobenzyl moieties, extends toward the surface of the protein. This region is poorly defined and could not be modeled in all molecules in the asymmetric unit. Simultaneous binding of multiple ligands has been suggested to be a mechanism responsible for atypical kinetics often displayed by CYP3A4 (13, 14, 20). The present structure provides structural evidence of simultaneous ligand binding in a mammalian P450 enzyme and an indication on how such drug–drug interactions, even between relatively large compounds, may occur. Although simultaneous binding of multiple ligands is thought to be a hallmark of CYP3A4, there are currently no reports in the literature on multiple binding events for ketoconazole. The hypothesis that the second binding site may be an artifact related to high concentrations of ketoconazole used in the crystallization experiments cannot be ruled out.
Fig. 3.
Ketoconazole binding to CYP3A4. Ketoconazole molecules are shown in orange stick representation. The heme group is shown in magenta. Secondary structure and stick representations of side chains within 4 Å from the ligands are shown in green. The C-terminal loop (residues 464–498) was omitted for clarity. A superposition of the ligand-free structure (Protein Data Bank ID code 1TQN) is shown in gray. The mesh represents a Fo − Fc difference map contoured at 4.5 σ calculated in the absence of ligands by using the program AutoBUSTER (19).
The CYP3A4–erythromycin complex crystallized in the C2221 space group and the structure was determined to 2.8 Å (see Table 1 for details).
The conformational changes induced by erythromycin were not as extensive as in the CYP3A4–ketoconazole complex; rmsd for all ordered Cα atoms was 1.2 Å when compared with the ligand-free structures. The most prominent differences are localized in the F-G region (Fig. 2B). The loop connecting helices F and F′ is poorly defined in the electron density and could not be modeled completely.
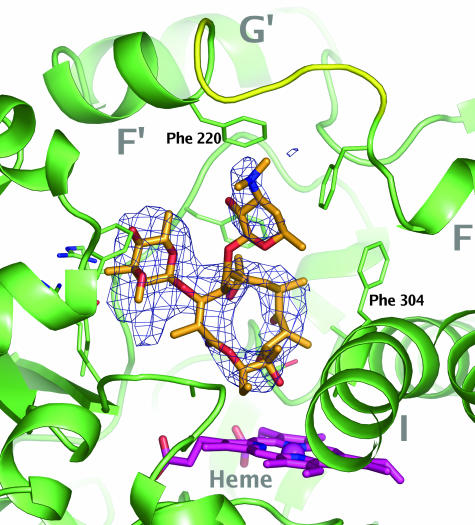
Electron density corresponding to the macrolide ring of erythromycin could be identified close to the heme group in one of the molecules in the asymmetric unit (Fig. 4). The quality of the data did not allow complete determination of the molecular configuration. Therefore, the macrolide cycle as determined by small molecule crystallography (21) (Cambridge Structural Database entry NAFTAF) was fitted as a rigid body into the density. The sugar moieties then were adjusted to match the residual density in the active site (Fig. 4). Because erythromycin is metabolized by demethylation of the d-desosamine group that is located 17 Å from the heme iron, it is clear that the model suggested by the x-ray data does not represent a productive binding mode. In contrast to the CYP3A4–ketoconazole interaction, the CYP3A4–erythromycin interaction is not characterized by high-quality interactions with the protein. In the final electron density maps, there was a substantial amount of residual density around the l-cladinose group, suggesting that other binding modes may exist. These data are in support of previous observations that a substrate may be localized inside the active site with no favored binding orientation (22). Although the two molecules in the asymmetric unit were very similar (rmsd for all ordered Cα atoms was 0.52 Å), the electron density was considerable poorer in molecule B than in molecule A, and it was not possible to model the ligand. The differences in the protein structures were focused mainly in the F′-G′ helices (residues 219–237), where the average differences in Cα positions is ≈1 Å in an optimal superposition of molecules A and B. This region is involved in crystal packing interactions in both molecules and the observed differences in protein–ligand interactions demonstrate how subtle variations in the external environment such as protein–protein interactions or membrane composition may modulate ligand binding.
Fig. 4.
Erythromycin binding to CYP3A4. Erythromycin is shown in orange stick representation. The heme is shown in magenta. Secondary structure and stick representations of side chains within 4 Å from the ligand are shown in green. A possible conformation for the disordered loop connecting helices F and F′ is shown in yellow. The mesh represents a Fo − Fc difference map contoured at 4.5 σ calculated in the absence of ligands by using the program AutoBUSTER (19).
Surprisingly, the nature of the coordinate shift relative the ligand-free structure in the F-G region is different in the two structures presented here. In an optimal overlay including all Cα atoms in the structurally invariant part of the protein (see Methods), the average shift of the Cα atoms included in the F-G region (residues 202–260) in the ketoconazole complex relative to the ligand-free structure (Protein Data Bank code 1TQN) is 3.1 Å. The corresponding shift for the erythromycin complex structure is 2.8 Å. However, although helices F′ and G′ shifts are similar, coordinate shifts of residues in helices F and G have opposite direction (Fig. 2). The average difference between the positions of Cα atoms for residues 202–260 in the ketoconazole and erythromycin complexes is 3.4 Å. The solvent accessible volume of the active site in the ligand-free structures was estimated to be ≈950 Å3 (calculations performed by using the LIGSITE algorithm; ref. 23). In the ketoconazole structure, the volume is increased to 1,650 Å3. In the erythromycin complex, the shift of the F helix has resulted in a large open cleft, and determination of the boundaries of the active site becomes somewhat arbitrary, but it is estimated that the volume is ≈2,000 Å3.
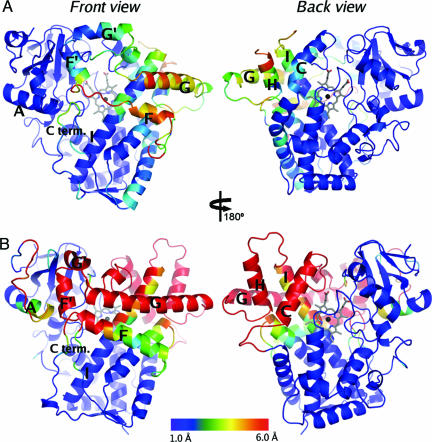
Despite the large increase in active site volume upon ligand binding, the conformational differences observed in CYP3A4 are less pronounced than those seen in the structures of CYP2B4 (6, 16, 17) (Fig. 5). However, the conformational differences between the available CYP2B4 structures seem to be, at least in part, related to crystal contacts involving large parts of the active-site cavity CYP2B4 (6, 17) rather than ligand interactions. These structures are thought to represent membrane-bound conformations with open channels connecting the hydrophobic core of the membrane with the active site. The conformational changes in CYP3A4 largely map into three of the five plastic regions (PR), which were identified from the CYP2B4 structures (17), suggesting that the existence of defined plastic regions may be a common feature for membrane-bound P450 enzymes. The largest conformational changes are seen in the region corresponding to PR4. In CYP2B4, this region comprises the C-terminal half of the F helix, helices F′, G′, G, H, and half of helix I together with the intervening loops. In CYP3A4, the observed structural plasticity in PR4 also extends toward the N-terminal side, connecting it with PR3, which contains the C-terminal part of helix E. Pronounced structural plasticity also is observed in PR5, which includes the C-terminal loop (residues 475–485). In contrast, there is very little flexibility observed in the regions corresponding to PR1 (the loop preceding helix A and most of helix A) and PR2 (helix B′, B′/C loop, and helix C). For CYP2B4, shifts of helix C were suggested to be central to conformational transitions of PR2, PR3, and PR4 (17), but the observations of structural flexibility in CYP3A4 suggest that these transitions can be independent of the C helix conformation.
Fig. 5.
Comparison of observed structural flexibility in CYP3A4 (A) and CYP2B4 (B). For each isoform, the secondary structure of the most compact conformation (Protein Data Bank ID codes 1TQN for CYP3A4 and 1SUO for CYP2B4) is shown colored according to the maximum observed difference in Cα position in an optimal superposition of all available structures (Protein Data Bank ID codes 1TQN, 2J0C, and 2J0D for CYP3A4 and 1PO5, 1SUO, and 2BDM for CYP2B4). Maximum observed Cα differences >6 Å are shown in red. Each isoform is shown in two orientations related by a 180° rotation about a vertical axis in the plane of the paper. Heme groups are shown in stick representation.
The ligand-induced conformational changes reported here demonstrate the flexibility of CYP3A4 and give some insights into how diverse substrates and inhibitors can be accommodated in the active site. The ability of the enzyme to accommodate multiple ligands simultaneously further adds to the promiscuity because the active site shape and charge distribution may be modulated by a second ligand. It is highly unlikely that the CYP3A4 conformational space has been covered by the present structures alone, and additional CYP3A4–ligand complexes will be required to complete the picture. An implication of the structural flexibility in CYP3A4 is that the development of general methods for prediction of binding modes and site of metabolism based on protein structures will be very challenging. The degree of success is likely to vary depending on the compound series of interest. Nevertheless, recent work within AstraZeneca demonstrates that the protein conformation seen in the ketoconazole structure performed well in structure-based site-of-metabolism predictions for many CYP3A4 substrates (24).
The present structures of CYP3A4 also may have implications for the interpretation of kinetic data on drug metabolism. There are two main hypotheses to explain non-Michaelis–Menten kinetics and drug–drug interactions. One model involves multiple ligand-binding sites and kinetic models including two or even three sites have been described (13, 14, 20). The other model relies on the existence of kinetically distinct conformers (25, 26) and is supported by presteady-state kinetics (25) as well as high-pressure spectroscopic experiments (15). Although it is not possible to draw any conclusions regarding the existence of multiple conformational states in the absence of ligands, the present structures show that CYP3A4 can adopt multiple ligand-bound protein conformations and accommodate simultaneous ligand binding even of relatively large compounds. Based on the available data, none of these hypotheses can be excluded and it suggests, rather, that both mechanisms are available to the enzyme and may even be in play simultaneously. This adaptability is likely to be an explanation for the extreme chemical diversity in CYP3A4 substrates.
Methods
Protein Expression and Purification.
A plasmid encoding human CYP3A4 was obtained from the LINK scheme (University of Dundee, Dundee, U.K.) and the coding sequence, modified according to Williams et al. (8), was subcloned into the expression vector pCWori. CYP3A4 was produced in Escherichia coli XL1 blue cells grown at 25°C. The cells were collected 50 h after induction and disrupted in a high salt lysis buffer by passage once through a high-pressure homogenizer. CYP3A4 was purified by affinity chromatography with Ni-NTA resin (Qiagen, Valencia, CA), desalted, and then applied to a cation exchange column (HiTrap CM Sepharose; Amersham Pharmacia Biotech, Uppsala, Sweden) in the presence of 10 mM CHAPS.
Complex Formation and Crystallization.
Different biochemical methods, e.g., dynamic light scattering and analytical ultracentrifugation, were used to identify a number of modifications to the buffer previously used in crystallization of the ligand-free CYP3A4 (8, 9) needed to avoid protein aggregation upon ligand addition. Crystals were obtained by the hanging-drop vapor diffusion method with protein at a concentration of 20–27 mg/ml in a solution containing 50 mM potassium phosphate (pH 7.2), 0.2 mM potassium chloride, 20% (vol/vol) glycerol, 2 mM DTT, and 1 mM EDTA. Ketoconazole or erythromycin dissolved in acetonitrile was added to the protein to yield a final compound:protein ratio of ≈10:1. Any undissolved compound was removed by centrifugation before setting up crystallization trials. Crystals formed over a period of 1–4 days at 20°C against a well solution containing 0.1 M Hepes (pH 7–7.5), 0.2 M lithium sulfate, 0.8–1.0 M lithium chloride, and 30% PEG 4000.
Data Collection and Structure Determination.
Crystals were quickly passed through a crystallization solution supplemented with 20% glycerol and flash-cooled in liquid nitrogen. Data were collected at beamline ID14 EH4 at a wavelength of 0.9760 Å. The data were processed by using MOSFLM (27), scaled, and further reduced by using the CCP4 suite of programs (28). The initial structure of the CYP3A4–ketoconazole complex was solved by molecular replacement using the ligand-free form of CYP3A4 (Protein Data Bank ID code 1W0E) as a search model. Four molecules were found by using the program Molrep (28). In all four molecules, portions of the model, including helices F to G, did not fit the electron density. Map improvement by water addition and atoms update by using ARP/wARP (29) and REFMAC5 (30) followed by fourfold noncrystallographic symmetry averaging resulted in clearly interpretable electron density maps. Model rebuilding was performed within O (31), and refinement was performed by using REFMAC5. Tight noncrystallographic symmetry restraints were used in the initial rounds of refinement but were gradually released as the model improved. The CYP3A4–erythromycin structure was solved by molecular replacement using the ligand-free form of CYP3A4 (Protein Data Bank ID code 1W0E) as a search model. Subsequent model building and refinement was done by using O and REFMAC5. For statistics for the final models, see Table 1.
Structure Analysis.
The different conformations of CYP3A4 and CYP2B4 were analyzed by using the program ESCET (32). The largest identified structurally invariant region was used to generate an optimal superposition of structures. The conformationally invariant region for CYP3A4 comprises residues 30–107, 118–131, 138–145, 166–171, 224–228, 312–477, and 487–496, which corresponds to 65% of all ordered residues. The largest conformationally invariant part of CYP2B4 comprises 58% of the protein, including residues 28–37, 58–70, 73–99, 300–473, and 481–492.
Acknowledgments
We thank Bert Larsson for assistance with large-scale fermentation; Philip R. Mallinder for producing the DNA construct; Cristian Johansson for help with crystallization of the CYP3A4–erythromycin complex; and Philip R. Mallinder, Lovisa Afzelius, Tommy B. Andersson, and Tomas Lundqvist for useful discussions and critical reading of the manuscript.
Abbreviations
- CYP
cytochrome P450
- PR
plastic regions
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. J.H.D. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors for CYP3A4 in complex with ketoconazole and erythromycin have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2J0C and 2J0D, respectively).
See Commentary on page 13565.
References
- 1.Gibbs MA, Hosea NA. Clin Pharmacokinet. 2003;42:969–984. doi: 10.2165/00003088-200342110-00003. [DOI] [PubMed] [Google Scholar]
- 2.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 3.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 4.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. J Biol Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 5.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 6.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 8.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 9.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 10.Poulos TL, Finzel BC, Howard AJ. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 11.Scott EE, Halpert JR. Trends Biochem Sci. 2005;30:5–7. doi: 10.1016/j.tibs.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Atkins WM. Annu Rev Pharmacol Toxicol. 2005;45:291–310. doi: 10.1146/annurev.pharmtox.45.120403.100004. [DOI] [PubMed] [Google Scholar]
- 13.Harlow GR, Halpert JR. Proc Natl Acad Sci USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenworthy KE, Clarke SE, Andrews J, Houston JB. Drug Metab Dispos. 2001;29:1644–1651. [PubMed] [Google Scholar]
- 15.Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Biochem Biophys Res Commun. 2003;312:121–30. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 16.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. J Biol Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 18.Cupp-Vickery JR, Garcia C, Hofacre A, McGee-Estrada K. J Mol Biol. 2001;311:101–110. doi: 10.1006/jmbi.2001.4803. [DOI] [PubMed] [Google Scholar]
- 19.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Acta Crystallogr D. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 20.He YA, Roussel F, Halpert JR. Arch Biochem Biophys. 2003;409:92–101. doi: 10.1016/s0003-9861(02)00484-8. [DOI] [PubMed] [Google Scholar]
- 21.Stephenson GA, Stowell JG, Toma PH, Pfeiffer RR, Byrn SR. J Pharm Sci. 1997;86:1239–1244. doi: 10.1021/js9701667. [DOI] [PubMed] [Google Scholar]
- 22.Cameron MD, Wen B, Allen KE, Roberts AG, Schuman JT, Campbell AP, Kunze KL, Nelson SD. Biochemistry. 2005;44:14143–14151. doi: 10.1021/bi051689t. [DOI] [PubMed] [Google Scholar]
- 23.Hendlich M, Rippmann F, Barnickel G. J Mol Graphics. 1997;15:359–363. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 24.Afzelius L, Hasselgren Arnby C, Broo A, Carlsson L, Isaksson C, Jurva U, Kjellander B, Kolmodin K, Nilsson K, Raubacher F, Weidolf L. Drug Metab Rev, 2006 doi: 10.1080/03602530600969374. in press. [DOI] [PubMed] [Google Scholar]
- 25.Koley AP, Buters JT, Robinson RC, Markowitz A, Friedman FK. J Biol Chem. 1997;272:3149–3152. doi: 10.1074/jbc.272.6.3149. [DOI] [PubMed] [Google Scholar]
- 26.Atkins WM, Wang RW, Lu AY. Chem Res Toxicol. 2001;14:338–347. doi: 10.1021/tx0002132. [DOI] [PubMed] [Google Scholar]
- 27.Leslie AG. Acta Crystallogr D. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project No 4 Acta Crystallogr D. 1994;50:750–763. [Google Scholar]
- 29.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Schneider TR. Acta Crystallogr D. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]