Abstract
Toll-like receptors and other immune-signaling pathways play important roles as sensors of bacterial pattern molecules, such as peptidoglycan, lipoprotein, or teichoic acid, triggering innate host immune responses that prevent infection. Immune recognition of multiple bacterial products has been viewed as a safeguard against stealth infections; however, this hypothesis has never been tested for Staphylococcus aureus, a frequent human pathogen. By generating mutations that block the diacylglycerol modification of lipoprotein precursors, we show here that S. aureus variants lacking lipoproteins escape immune recognition and cause lethal infections with disseminated abscess formation, failing to elicit an adequate host response. Thus, lipoproteins appear to play distinct, nonredundant roles in pathogen recognition and host innate defense mechanisms against S. aureus infections.
Keywords: innate immunity, virulence
To establish a focus of infection and resultant bacteremia, Staphylococcus aureus overcomes physical protective barriers of the human body through invasion of soft tissues, surgical wounds, or medical devices (1). In the first hours after infection, staphylococci are cleared from the blood stream, in part by way of phagocytic killing but also by bacterial binding to host organ tissues (2). Staphylococci that escape killing replicate in infected tissues and generate proinflammatory responses mediated by the release of cytokines and chemokines from macrophages, neutrophils, and other immune cells (3, 4). The resulting massive invasion of immune cells to the site of infection is accompanied by central liquefaction necrosis and formation of peripheral fibrin walls in an effort to prevent microbial spread and allow for removal of necrotic tissue debris (5). When launched early and effectively, innate immune responses limit the establishment of infectious foci and thereby curb the severity of staphylococcal infections. These early events culminate in the activation of adaptive immune responses, during which T and B cells capable of specific antigen recognition lead to the eradication of staphylococci. Thus, the coordinated action of the innate and adaptive immune response is critical for efficient pathogen elimination.
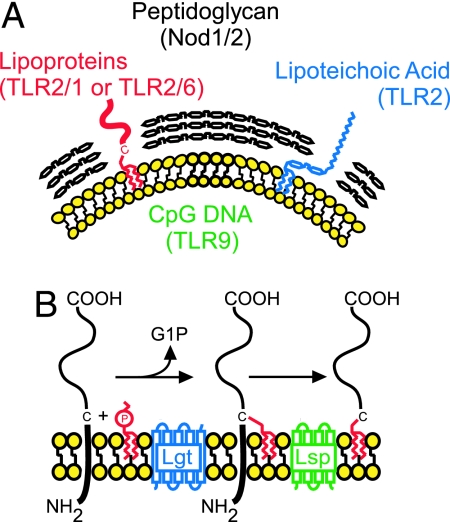
Binding of bacterial molecules called pathogen-associated molecule patterns (PAMPs) to dedicated Toll-like receptors (TLRs) or Nod proteins triggers specific signaling events and host responses to invading pathogens (6, 7). To date, a dozen different TLRs have been identified in mammals (8). TLR2 plays a critical role in host defense against S. aureus, because TLR2 knockout mice are highly susceptible to i.v. infection with staphylococci (9). Purified staphylococcal PAMPs activate immune signaling through TLR2 both in vivo and in cell culture (10–12) (Fig. 1A). Furthermore, bacterial lipoproteins also function as PAMPs, activating TLR2 signaling cascades (12–15). However, the contribution of individual PAMPs to host recognition of invading staphylococci by immune surveillance systems has not been studied.
Fig. 1.
Molecules of Gram-positive bacteria recognized by the innate immune system. (A) Schematic representation of proposed bacterial PAMPs to known TLRs and Nod1/2 (7, 8, 39). The plasma membrane and peptidoglycan are depicted in yellow and black, respectively. (B) Proposed pathway for lipoprotein maturation in S. aureus.
By generating mutations that block diacylglycerol attachment to lipoprotein precursors, we show that S. aureus variants bearing apolipoproteins escape immune recognition and cause disseminated abscess formation with increased lethality during infection. Furthermore, we show that immune cells do not infiltrate sites of infection carrying these mutant bacteria. Hence, it appears that acylation of lipoproteins is required for initiating and sustaining effective immune responses from infected hosts. Understanding the role of bacterial lipoproteins in mediating innate and adaptive immunity will be useful for the therapy of human S. aureus infections.
Results
Genetic Requirement of Staphylococcal PAMPs.
To examine the contribution of staphylococcal PAMPs to immune recognition and disease pathogenesis, a collection of S. aureus bursa aurealis mutants (Phoenix library) (16) was examined for insertion mutants with defects in the biosynthesis of specific PAMPs. As expected, no transposon insertions in cell wall and lipoteichoic acid biosynthesis genes were identified, because these genes are required for staphylococcal growth (17). Lipoproteins are synthesized in the cytoplasm as precursors with an N-terminal signal peptide for secretion via the Sec pathway (18, 19). Lipoprotein diacylglycerol transferase (Lgt) catalyzes transfer of phosphatidylglycerol to the sulfhydryl moiety of a cysteine residue conserved in the signal peptides of all lipoprotein precursors (20, 21). The product of this reaction is then cleaved at the modified cysteine by lipoprotein (type II) signal peptidase (Lsp) (Fig. 1B) (22). In Gram-negative bacteria, the N-terminal cysteine residue of Braun’s murein lipoprotein (23) is modified by N-acyltransferase (Lnt), yielding mature N-acylated lipoprotein (20). Bursa aurealis insertions in lgt and lsp were identified in the Phoenix library; however, bioinformatic analysis revealed that lnt is not present in the genome of S. aureus. Thus, the amine of cysteine-diacylglycerol lipoprotein is likely not acylated in staphylococci.
Genetic Requirement for Processing of Staphylococcal Lipoproteins.
Wild-type S. aureus strain Newman grown in the presence of [3H]palmitate incorporated radiolabeled palmitate into lipoproteins, however an isogenic lgt variant did not (see Fig. 6, which is published as supporting information on the PNAS web site). This defect is not caused by reduced protein synthesis, because labeling with [35S]methionine revealed equal amounts of PrsA lipoprotein or its precursor in wild-type and lgt mutant staphylococci, respectively (see Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, cells were labeled with [35S]methionine for 1 min and PrsA was immunoprecipitated before and after a chase with nonradioactive methionine (see Fig. 8, which is published as supporting information on the PNAS web site). Wild-type bacteria synthesized a slower migrating precursor species (pro-PrsA) with an intact signal peptide that was converted to the mature form within 1 min. Signal peptide processing was blocked in lgt mutant staphylococci, showing accumulation of pro-PrsA. To determine whether bursa aurealis insertion in lgt affected both acylation (apo) and signal peptide cleavage, we compared lgt and lsp mutants. Pulse–chase labeling revealed that pro-PrsA processing did not occur in lsp mutants (Fig. 8, lsp), however this defect was restored upon expression of plasmid-encoded lsp (Fig. 8, lsp/pLsp). All biosynthetic defects were reversed upon transformation of lgt mutant staphylococci with the plasmid-encoded wild-type allele, indicating that the observed phenotypes are attributable to transposon insertion in the lgt gene (Fig. 8). Together, these data corroborate a model whereby Lgt-mediated acylation of prolipoprotein is a prerequisite for Lsp cleavage (20, 24). To examine the fate of apolipoproteins within cells, cultures were incubated for 5 min with [35S]methionine, and bacteria were converted to protoplasts before immunoprecipitation with specific antibodies (see Fig. 9, which is published as supporting information on the PNAS web site). Processing of PrsA and GmpC lipoprotein precursors was blocked in lgt mutants, however slower migrating variants of PrsA and GmpC were found associated with protoplasts (P) or cell wall fractions (CW). Protoplast association corroborates the notion that hydrophobic signal sequences were not removed by Lsp without diacylglycerol acylation as displayed in Fig. 8. The localization of newly synthesized cell wall anchored protein A (Spa) and secreted staphylococcal nuclease (Nuc) was not affected by the disruption of lgt, indicating that the mutant strain did not cause a general defect in protein secretion.
Contribution of Lipoprotein Processing to Bacterial Recognition by Immune Surveillance Systems.
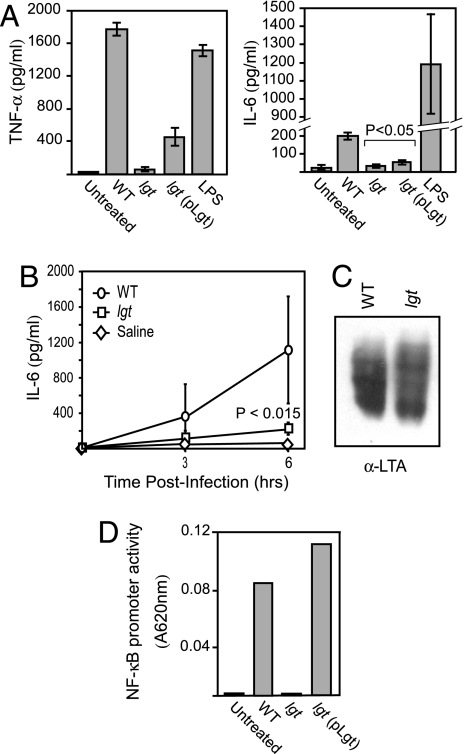
We asked whether host recognition of lgt mutants by elements of the innate immune system was affected. Incomplete Freund’s adjuvant-elicited mouse peritoneal macrophages were incubated with heat-killed staphylococci, and the production of proinflammatory cytokines was measured. lgt mutants induced significantly lower levels of TNF-α and IL-6 production than wild-type staphylococci (Fig. 2A). Complementation of this phenotype upon expression of wild-type lgt was reached within statistical significance (Fig. 2A, pLgt). The decrease in cytokine production was not due to macrophage cell death because no change in the number of viable cells was observed (data not shown). Similarly, the observed decrease in cytokine production was not secondary to alterations in the amount of lipoteichoic acid (LTA) or peptidoglycan, potent activators of the innate immune response (25–27). Wild-type and lgt mutant staphylococci appear to synthesize similar amounts of LTA, as determined by LTA extraction and immunoblotting (Fig. 2C). Release of IL-6 was measured in sera of infected animals for up to 6 h (Fig. 2B). Animals infected with lgt mutants failed to produce IL-6 compared with animals infected with wild-type staphylococci. These data are in agreement with a recent report showing that proinflammatory cytokine responses in various human cell lines are decreased upon incubation of lgt mutant compared with wild-type staphylococci (24). Because bacterial lipoproteins are known to engage TLR2, leading to cytokine production through NF-κB-dependent transcriptional activation, we used an NF-κB reporter assay system to assess the ability of wild-type and lgt mutant S. aureus to engage TLR2. Human 293 cell-stable transfectants, which express human TLR2, were transiently transfected with a reporter construct in which a NF-κB promoter drives expression of secreted alkaline phosphatase (SEAP). Cells were cocultured with wild-type or lgt mutant bacteria in medium containing colorimetric substrate for SEAP detection. As demonstrated in Fig. 2D, NF-κB activity was detected in 293TLR2 cells cocultured with wild-type S. aureus Newman. However, no activity was detected in 293TLR2 cells that remained uninfected or when these cells were cocultured with lgt mutant bacteria. Restoration of lgt expression by way of plasmid complementation (lgt/plgt) revealed levels of NF-κB induction similar to that induced by coculture with wild-type S. aureus. Importantly, no NF-κB inducible activity was present in parental 293 cells that lack TLR2 (data not shown). Together, these data strongly suggest that S. aureus-derived bacterial lipoproteins function through TLR2 in an NF-κB dependent fashion to induce the production of inflammatory cytokines.
Fig. 2.
Lack of recognition of lgt mutants by the innate immune system. (A) Incomplete Freund’s adjuvant-elicited peritoneal macrophages were harvested from C57BL/6 mice and treated with either heat-killed staphylococci (5 × 106 cfu/ml) or LPS (0.1 μg/ml) for 18 h or untreated. The production of TNF-α (Left) and IL-6 (Right) was measured by ELISA. LPS was used as a control. (B) Serum cytokine production in response to infection with live staphylococci in vivo. Fifteen mice infected with 5 × 106 cfu of bacteria were anesthetized, and three mice were terminally bled at 0, 3, and 6 h after infection, respectively. Sera were collected by centrifugation after clotting and assayed by ELISA for IL-6. (C) lgt mutants produce normal amounts of LTA. Saturated staphylococci cultures were disrupted by using a bead beater, and insoluble material was resuspended in 4% SDS and separated by SDS/PAGE, followed by immunoblot analysis using anti-LTA antibody. (D) 293 cells expressing TLR2 were transfected with a secreted alkaline phosphatase (SEAP) reporter plasmid under the control of an NF-κB inducible promoter. Coculture of these cells with wild-type (WT) S. aureus revealed induction of NF-κB promoter activity, whereas coculture with lgt mutant S. aureus failed to induce NF-κB promoter activity. Plasmid-encoded lgt (lgt/pLgt) restored TLR2-mediated NF-κB promoter activity.
Virulence of S. aureus lgt and lsp Mutants.
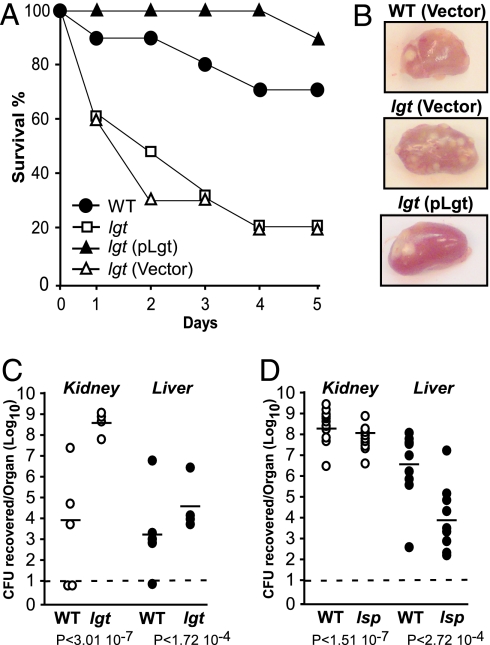
Wild-type S. aureus Newman or its isogenic lgt and lsp mutants (5 × 106 staphylococci) were administered i.v. via retro-orbital injection into mice. Disease progression was observed over 5 days, after which the animals were killed, and their internal organs were removed and inspected for abscess formation. Homogenized tissues were spread on agar medium and staphylococcal load within organ tissues enumerated by colony formation. In contrast to the sublethal infections of wild-type staphylococci that are eventually cleared by infected mice (Fig. 3A, filled circles), bursa aurealis insertion in lgt caused a rapid and pronounced mortality in infected animals over the experimental time course (Fig. 3A, open squares). This hypervirulent phenotype was not observed when lgt mutant strains carried the complementing pLgt plasmid (Fig. 3A, filled triangles). In contrast, mice infected with lgt mutant bacteria carrying only the vector control plasmid succumbed much more rapidly to infection (Fig. 3A, open triangles). Inspection of kidneys from animals infected with wild-type S. aureus revealed the presence of multiple raised, yellow lesions on the organ surface (Fig. 3B) that harbor collections of staphylococci and associated cellular debris. These lesions were more numerous in animals that had been infected with lgt mutant staphylococci. Animals infected with the complemented lgt mutant displayed gross pathology similar to that of the wild-type infected animals. Bacterial load in the organs of animals killed 48 h after infection was quantified. Data in Fig. 3C show that staphylococci lacking lgt proliferate to much higher numbers during infection compared with wild-type S. aureus (differences of 4.5 log in the kidneys and 1 log in the liver). Mice infected with lsp mutants did not develop acute, lethal disease (data not shown) and quantification of bacteria within infected organs (4 days after infection) demonstrated that this mutant displayed attenuated virulence with a severe defect in the ability to multiply in liver tissue (Fig. 3D). Previous studies demonstrated that lipoprotein processing by Lsp is required for the full virulence of Mycobacterium tuberculosis and Streptococcus pneumoniae in animal models of infection (28, 29). Additionally, a signature-tagged mutagenesis screen of S. aureus identified lsp as a factor contributing to virulence; however, the calculated LD50 of this lsp mutant in an animal model of i.p. infection was found to be similar to that of the wild-type parental strain (30). Thus, the increased virulence of lgt mutants appears to be solely caused by the lack of Lgt activity that results in a loss of lipoprotein acylation, not signal sequence removal.
Fig. 3.
Virulence of S. aureus lgt and lsp mutants. Six-week-old C57BL/6 mice were infected i.v. with 5 × 106 cfu for each strain. (A) Mice were monitored for the development of acute, lethal disease for 5 days upon infection of wild-type (WT) Newman, the lgt mutant strain, or lgt mutant carrying empty vector or complementing plasmid pLgt. (B) Kidneys were harvested from mice infected for 4 days with mutant strain lgt carrying the empty vector or infected for 5 days with WT Newman or complemented mutant strain lgt/pLgt. The kidneys were photographed to visualize the formation of abscesses. (C) In a separate experiment, animals infected with either WT or lgt mutant staphylococci and still alive 2 days after infection were killed. Kidneys and liver were removed and homogenized. Viable bacteria were counted after dilution and colony formation on tryptic soy agar. Statistical significance was examined with Student’s t test, and P values were recorded. The limit of detection (dashed line) was determined to be 100 cfu (102). (D) Dissemination to organs of WT or lsp mutant staphylococci 4 days after infection.
Growth of S. aureus lgt Mutant in Vivo.
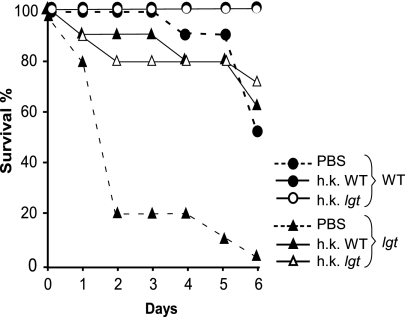
We wondered whether the proliferation of lgt mutant staphylococci could be the result of increased proliferation in the blood or increased resistance to phagocytosis. To distinguish between these possibilities, wild-type and lgt mutant bacteria were grown in the presence of fresh human whole blood, activated J774 murine macrophages, or freshly prepared human serum (see Fig. 10, which is published as supporting information on the PNAS web site). The results showed that lgt mutant bacteria proliferated much more slowly in the presence of blood or activated macrophages than S. aureus wild-type strain Newman but demonstrated similar proliferation in serum. These data suggest that the increased virulence observed in animals infected with the lgt mutant bacteria is not attributable to either enhanced proliferation or impaired clearance of the mutant bacteria. Furthermore, phagocytosis and macrophage killing of lgt mutants was not reduced compared with wild-type staphylococci, indicating that the mutant strain is not able to escape from phagocytic killing (data not shown). We sought to ascertain whether hypervirulence of lgt mutants may be caused by changes in exoprotein secretion or other traits associated with increased invasiveness. To examine this possibility, groups of 20 mice were inoculated i.p. either with buffer (PBS) or with 5 × 108 heat-killed bacteria of the wild-type or lgt mutant strains. This preinoculation with heat-killed wild-type or lgt mutant bacteria induced the generation of an IL-1 response in the host, detectable in peritoneal washes at 24 h (data not shown). Twenty-four hours after preinoculation, each group was divided into two groups of 10 mice, and cultures of lgt mutant or wild-type S. aureus Newman (5 × 106 staphylococci) were administered i.v. via retro-orbital injection. Disease progression was observed over 6 days (Fig. 4). Animals that were pretreated with the PBS alone developed acute, lethal disease upon i.v. challenge with the lgt mutant. However, pretreatment with heat-killed bacteria, either wild-type or mutant, resulted in protection against hypervirulence of lgt mutant or even killing by wild-type staphylococci. These results suggest that lgt mutants per se have not acquired a factor or trait that precipitates a hypervirulent state. Furthermore, heat-killed bacteria, administered in large quantity into the peritoneal cavity, do stimulate innate host defenses capable of containing infections caused by lgt mutants.
Fig. 4.
Pretreatment with heat-killed staphylococci protects animals from infection. Mice were pretreated with i.p. injection of either buffer (PBS) or heat-killed wild-type (WT) or lgt mutant bacteria 24 h before i.v. challenge with live (5 × 106) WT or lgt mutant bacteria. The percentage of survival after challenge was recorded over a 6-day time course.
Physiological Response to S. aureus lgt Mutant Infection.
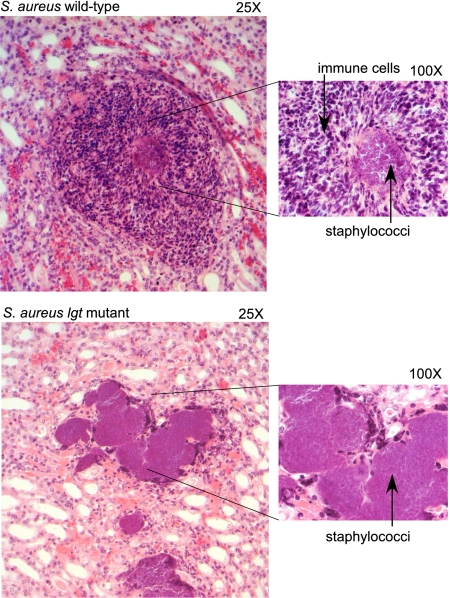
To probe the pathological consequence of infection, kidneys of animals infected with S. aureus wild-type Newman or lgt variants were removed 2 days after infection, formalin-fixed, and subsequently processed for microscopic evaluation of hematoxylin-eosin staining of thin sections (Fig. 5). As expected, infection with S. aureus strain Newman generated multiple small staphylococcal foci circumscribed by large numbers of infiltrating neutrophils. This zone of infection and inflammation is contained by a surrounding cuff of fibrin deposition. Surprisingly, physiologic host responses to infection with the S. aureus lgt mutant were abolished, exemplified by large collections of staphylococci in the kidney tissue with minimal neutrophil infiltration.
Fig. 5.
Pathological substrate of infection caused by S. aureus wild-type and lgt mutant strains. Kidneys of 6-week-old C57BL/6 mice infected with 5 × 106 cfu S. aureus Newman (wild-type) or its isogenic lgt variant were analyzed 2 days after infection. Formalin-fixed tissues were embedded, sectioned, stained with hematoxylin/eosin, and viewed at ×25 (Left) and ×100 (Right) magnification.
Discussion
S. aureus is a physiological commensal of the human skin and nares (31). Breaches in local defense, such as a skin cut or hair follicle trauma, provide this pathogen with an opportunity to gain access to deeper tissues. S. aureus is capable of causing infections of any organ tissue. These infections may culminate in life-threatening bacteremia. Despite medical advances, the frequency of both community- and hospital-acquired S. aureus infections has increased steadily, and the treatment of these infections is becoming even more difficult with the emergence of antibiotic-resistant strains. This increased emergence of antibiotic resistance necessitates the identification of novel therapies that are capable of interfering with the virulence of multidrug-resistant strains of S. aureus.
The establishment of staphylococcal abscesses with liquefaction necrosis represents the sum of all pathogenetic events implemented by the activity of virulence factors, bacterial molecules sampled by the host and the corresponding host responses (32–34). The innate immune system plays an integral role in determining the outcome of the infection. Virulence studies using knockout mice have shown that both TLR2 and the signaling molecule MyD88 play a critical role in the innate immune response to staphylococcal infection (9), suggesting that TLR2-recognition of one or more staphylococcal PAMPs results in signaling through this adaptor molecule.
We have examined the contribution of staphylococcal lipoproteins during infection by targeting the only two genes surmised to be involved in this process, lgt and lsp (Fig. 1B). Taken together, our experiments reveal that lgt mutants that lack diacylglycerol-modified lipoproteins, but not lsp variants that accumulate uncleaved modified lipoproteins, escape detection by host innate immune surveillance systems. This result is surprising, because bacteria elaborate many different pattern molecules (peptidoglycan, teichoic acid, N-formyl methionine), each of which was hitherto thought sufficient to activate innate immune responses. In concert with our data, the recent finding that interleukin-1 receptor/MyD88 (but not TLR2) signaling pathways are essential for neutrophil recruitment and host responses to staphylococcal infection (35) highlights the complexity of the host/pathogen interaction. Staphylococcal diacylglycerol lipoproteins are therefore not only required for bacterial transport reactions (36) but are also essential in triggering the host innate response to infection. Because early innate responses to an invading pathogen provide a critical template for adaptive immune responses, staphylococcal lipoproteins assume a central role in defense against this pathogen. These results suggest further that immunomodulatory therapies with a diacylglycerol lipoprotein may be useful for treatment or prevention of human infections caused by S. aureus.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Escherichia coli and S. aureus were grown in Luria–Bertani broth and tryptic soy broth, respectively, at 37°C. Chloramphenicol and erythromycin were used at 10 mg/liter, and ampicillin was used at 100 mg/liter. lgt, lsp, nuc, spa, and gmpC mutants were obtained from the Phoenix (ΦNΞ) library (16). Each Phoenix isolate is a derivative of the clinical isolate Newman (16, 37). All bursa aurealis insertions were transduced into wild-type S. aureus Newman by using bacteriophage φ85. Additional alleles were generated by replacing the lsp and lgt coding region in strain Newman with the ermC cassette by allelic exchange as described in ref. 38. The pLgt complementation plasmid was generated by cloning the hprK promoter (275 bp upstream of the hprK lgt yvoF yvcD translational start site) upstream of the lgt coding region in E. coli–S. aureus shuttle vector pOS1. pLsp was generated by cloning lsp downstream of the hprK promoter in pOS1.
Macrophage Assays.
Three days after i.p. injection with incomplete Freund’s adjuvant, peritoneal cavities of 6- to 8-week-old C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) were washed with cold, serum-free Hanks’ balanced salt solution. Cells were plated in triplicate at a density of 2 × 106 cells per well by using 24-well dishes and serum-free RPMI medium 1640. After 2 h of incubation at 37°C in an atmosphere with 5% CO2, plates were carefully washed three times with prewarmed, serum-free medium to remove nonadherent cells and fresh RPMI medium 1640 containing 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, 100 units/ml streptomycin, and 50 μM 2-mercaptoethanol. Macrophage cultures were treated with 5 × 106 cfu/ml washed, heat-killed staphylococci or 0.1 μg/ml LPS (Sigma-Aldrich, St. Louis, MO) in RPMI medium 1640 containing 10% FBS. Macrophage culture supernatants were collected 18 h after the addition of proteins and analyzed by ELISA for concentration of IL-6 (BD Biosciences, San Jose, CA) and TNF-α (R&D Systems, Minneapolis, MN) according to the manufacturer’s recommendations.
Immunoblot Analysis of LTA.
Saturated staphylococci cultures grown in tryptic soy broth for 12 h were disrupted by using a bead beater, and insoluble material was recovered by centrifugation at 16,000 × g, boiled in 4% SDS for 30 min to disrupt membranes, separated by SDS/PAGE, and analyzed by immunoblot using LTA-specific monoclonal antibodies (HyCult BioTechnology, Uden, The Netherlands).
NF-κB Reporter Assay.
A total of 293 parental cells (293null) and 293 cells expressing the TLR2 receptor (293TLR2C.6; Invivogen, San Diego, CA) were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS, l-glutamine, blasticidin, and normocin according to the manufacturer’s protocol. On day 0, cells were counted and plated at a density of 1 × 106 cells per well in six-well plates with 3 ml of medium lacking antimicrobial supplements. On day 1, cells were transiently transfected with 5 μg of pNiFty2-secreted alkaline phosphatase (SEAP) plasmid DNA (Invivogen) by using Lipofectamine 2000 (15 μl of lipofectamine mixed with DNA; Invitrogen). On day 2, medium from transfected cells was aspirated and replaced with 1 ml HEK-Blue detection medium (Invivogen). Overnight cultures of staphylococci were diluted 1:100 into fresh medium and grown to OD660 0.5 (≈2 × 108 cfu/ml). Staphylococci were sedimented by centrifugation, washed, and suspended in PBS, and 1 × 107 cfu in 20 μl suspension were added to each well of transfected 293 cells, followed by an 18-h incubation. Medium was removed from the wells. Cells, staphylococci, and debris were sedimented by centrifugation at 13,000 × g for 1 min, and absorbance of supernatant was measured at OD620 as a measure of NF-κΒ promoter activity.
Virulence Studies.
S. aureus strains were grown at 37°C overnight in tryptic soy broth, diluted 100-fold in fresh broth, and incubated at 37°C until OD660 of 0.5. Cells were washed, diluted, and suspended in PBS, and 100 μl of bacterial suspension was injected i.v. into 6-week-old female C57BL/6 mice. Viable staphylococci were enumerated by colony formation on tryptic soy agar to quantify the infection dose (≈5 × 106 cfu). At the indicated time points, i.e., days after challenge, mice were killed by CO2 asphyxiation. Spleen, kidneys, and liver were removed, and organs were homogenized in 1 ml of 1% Triton X-100 in PBS. Dilutions of the homogenates were plated on agar for enumeration of viable staphylococci. Statistical data analysis was performed with Student’s t test by using the software Analyze-it (Analyze-it Software, Leeds, U.K.). For histology, kidneys of infected animals were placed in 10% neutral-buffered formalin. Fixed tissues were embedded in paraffin, sectioned, mounted on slides, and stained with hematoxylin and eosin. For complementation experiments, mice were administered chloramphenicol in their drinking water (0.5 mg/ml) 24 h before infection and until the end of the experiment. For protection experiments, heat-killed bacteria (5 × 108 cfu) were inoculated into groups of 20 6-week-old C57BL/6 mice i.p., and 24 h later the animals were challenged with live bacteria in suspension injected i.v. The time to death was recorded over 6 days.
Supporting Information.
For details regarding protein labeling and pulse–chase analysis, fractionation of radiolabeled staphylococci, and bacterial proliferation in the presence of human blood, cultured macrophages, or human serum, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank K. DeBord for technical assistance and C.-R. Wang, O. Schneewind, and members of our laboratory for discussion. J.B.W. is a National Institute of Child Health and Human Development (NICHD) Fellow of the Pediatric Scientist Development Program (NICHD Grant Award K12-HD00850). W.A.W. was supported by Molecular and Cellular Biology Training Grant T32GM007183 awarded by the National Institutes of Health/National Institute of General Medical Sciences to the University of Chicago. This work was supported by U.S. Public Health Service Grant AI055838 (to D.M.).
Abbreviations
- LTA
lipoteichoic acid
- PAMP
pathogen-associated molecule pattern
- TLR
Toll-like receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lowy FD. New Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Thakker M, Park J-S, Carey V, Lee JC. Infect Immun. 1998;66:5183–5189. doi: 10.1128/iai.66.11.5183-5189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas CA, Li Y, Kodama T, Suzuki H, Silverstein SC, El Khoury J. J Exp Med. 2000;191:147–155. doi: 10.1084/jem.191.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson IM, Mazmanian SK, Schneewind O, Bremell T, Tarkowski A. Microb Infect. 2003;5:775–780. doi: 10.1016/s1286-4579(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson P, Lindberg M, Haraldsson I, Wadstrom T. Infect Immun. 1985;49:765–769. doi: 10.1128/iai.49.3.765-769.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medzhitov R, Preston-Hurlburt P, Janeway CAJ. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 7.Medzhitov R. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 8.Takeda K, Kaisho T, Akira S. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi O, Hoshino K, Akira S. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 11.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 13.Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 14.Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, Maitland M, Norgard MV, Plevy SE, Smale ST, et al. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 15.Hirschfeld M, Kirschning CJ, Schwandner R, Wesche H, Weis JH, Wooten RM, Weis JJ. J Immunol. 1999;163:2382–2386. [PubMed] [Google Scholar]
- 16.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. Proc Natl Acad Sci USA. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuhaus FC, Baddiley J. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inouye S, Wang S, Sekizawa J, Halegoua S, Inouye M. Proc Natl Acad Sci USA. 1977;74:1004–1008. doi: 10.1073/pnas.74.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Heijne G. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 20.Tokunaga M, Tokunaga H, Wu HC. Proc Natl Acad Sci USA. 1982;79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan K, Gupta SD, Sankaran K, Schmid MB, Wu HC. J Biol Chem. 1993;268:16544–16550. [PubMed] [Google Scholar]
- 22.Choi DS, Yamada H, Mizuno T, Mizushima S. J Biol Chem. 1986;261:8953–8957. [PubMed] [Google Scholar]
- 23.Braun V, Bosch V. Eur J Biochem. 1972;28:51–69. doi: 10.1111/j.1432-1033.1972.tb01883.x. [DOI] [PubMed] [Google Scholar]
- 24.Stoll H, Dengjel J, Nerz C, Gotz F. Infect Immun. 2005;73:2411–2423. doi: 10.1128/IAI.73.4.2411-2423.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morath S, Stadelmaier A, Geyer A, Schmidt RR, Hartung T. J Exp Med. 2002;195:1635–1640. doi: 10.1084/jem.20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deininger S, Stadelmaier A, von Aulock S, Morath S, Schmidt RR, Hartung T. J Immunol. 2003;170:4134–4138. doi: 10.4049/jimmunol.170.8.4134. [DOI] [PubMed] [Google Scholar]
- 27.Inohara N, Chamaillard M, McDonald C, Nunez G. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 28.Petit CM, Brown JR, Ingraham K, Bryant AP, Holmes DJ. FEMS Microbiol Lett. 2001;200:229–233. doi: 10.1111/j.1574-6968.2001.tb10720.x. [DOI] [PubMed] [Google Scholar]
- 29.Sander P, Rezwan M, Walker B, Rampini SK, Kroppenstedt RM, Ehlers S, Keller C, Keeble JR, Hagemeier M, Colston MJ, et al. Mol Microbiol. 2004;52:1543–1552. doi: 10.1111/j.1365-2958.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 30.Mei JM, Nourbakhsh F, Ford CW, Holden DW. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 31.Archer GL, Climo MW. N Engl J Med. 2001;344:55–56. doi: 10.1056/NEJM200101043440110. [DOI] [PubMed] [Google Scholar]
- 32.Dinges MM, Orwin PM, Schlievert PM. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick RP. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 34.Archer GL. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 35.Miller LS, O'Connell RM, Gutierrez MA, Pietras EM, Shahangian A, Gross CE, Thirumala A, Cheung AL, Cheng G, Modlin RL. Immunity. 2006;24:79–91. doi: 10.1016/j.immuni.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Skaar EP, Humayun M, DeBord KL, Schneewind O. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 37.Duthie ES, Lorenz LL. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 38.Bae T, Schneewind O. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Travassos LH, Girardin SE, Philpott DJ, Blanot D, Nahori M-A, Werts C, Boneca IG. EMBO Rep. 2004;5:1000–1006. doi: 10.1038/sj.embor.7400248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.