Abstract
The mb1 gene encodes the Ig-α signaling subunit of the B cell antigen receptor and is expressed exclusively in B cells beginning at the very early pro-B cell stage in the bone marrow. We examine here the efficacy of the mb1 gene as a host locus for cre recombinase expression in B cells. We show that by integrating a humanized cre recombinase into the mb1 locus we obtain extraordinarily efficient recombination of loxP sites in the B cell lineage. The results from a variety of reporter genes including the splicing factor SRp20 and the DNA methylase Dnmt1 suggest that mb1-cre is probably the best model so far described for pan-B cell-specific cre expression. The availability of a mouse line with efficient cre-mediated recombination at an early developmental stage in the B lineage provides an opportunity to study the role of various genes specifically in B cell development and function.
Keywords: Dnmt1, SRp20, loxP, enhanced yellow fluorescent protein, lymphocyte
The bacteriophage recombinase cre can efficiently delete DNA sequences that are flanked by loxP sites (floxed) even in eukaryotic cells (1). This feature has led to the frequent use of transgenic cre mice for the tissue-specific deletion or modification of floxed genes to access the function of a gene in a specific tissue (2).
Development of a B lymphocyte can be separated into several ordered steps encompassing commitment to the B lineage, somatic recombination and expression of its heavy chain and light chain Ig genes, and selection of the B cell antigen receptor repertoire (for reviews, see refs. 3–5). In the B cell system there are several transgenic mouse lines available that express cre in defined stages of B lymphocyte development. For example, CD19-cre mice (6) express cre from the pre-B cell stage on, whereas CD21-cre mice (7) express cre only in mature B cells. However, a cre transgenic mouse line with efficient cre-mediated deletion from the earliest pro-B cell stage was missing so far. We asked whether expression of the cre recombinase from the murine mb-1 locus would provide an even more efficient model for studying gene function specifically in B cell precursors. The mb1 gene encodes the Ig-α signaling subunit of the B cell antigen receptor (8, 9). It is strongly expressed in the B cell lineage beginning at the very early pro-B cell stage in the bone marrow and continues to be expressed in all later stages except plasma cells (10). The mb1-cre line was tested by intercrossing it to a floxed enhanced yellow fluorescent protein (EYFP) reporter mouse line. The analysis showed a very efficient and B cell-specific recombination. To further test the mb1-cre line, we bred it to several different lines bearing floxed genes, some of which are believed to be essential genes in all cell types. We show results for the splicing factor SRp20 and the DNA methylase Dnmt1. SRp20 belongs to a family of serine–arginine-rich proteins important for a variety of cellular functions surrounding mRNA including constitutive and alternative splicing, transport, translation, and degradation as well as genome stability (11). Deletion of the SRp20 gene in the mouse germ line blocks embryonic development at the morula-to-blastocyst transition (12), but its role in B cell development is not known. The DNA methyltransferase Dnmt1 is involved in transfer of the CpG methylation pattern from the parental to the daughter DNA strand during the S phase of the cell cycle (13). A reduction or loss of Dnmt1 activity has a drastic effect on cell function and, depending on the system, can lead to inhibition of DNA replication (14), T cell lymphoma (15), alterations in T cell development (16), and embryonic lethality (17).
The results presented here show that SRp20 and Dnmt1 are essential for B cell development and/or survival and that cre recombinase activity in the mb1-cre line is efficient and primarily restricted to the B lineage.
Results
Construction of a Targeted Mouse Line Expressing hCre from the Ig-α Locus.
A vector coding for a mammalian codon-optimized hCre (18) was designed to be inserted into the mb-1 WT locus (Fig. 1a). In the targeting vector, exons 2 and 3 of mb-1 were replaced by a hCre cDNA, which was fused at its 5′ end to the splice acceptor of exon 2 and at the 3′ end to a pA signal from SV40 (Fig. 1b). The construct also contains a modified form of exon 1 lacking the mb-1 ATG codon. Intron 1 was retained to provide splicing of the primary cre transcript and because it could contain transcriptional regulatory elements. A neo cDNA driven by the tk promoter and flanked by two flippase recombinase target (FRT) sites having the same orientation was introduced 3′ of hCre. Flanking short and long arms of DNA sequence homology derived from the mb-1 locus were also introduced (see Materials and Methods).
Fig. 1.
Targeting construct for the mb-1 locus with hCre recombinase. (a–c) mb-1 WT locus (a) and mb-1 locus (b) targeted by the mb1-cre before deletion of the neo cassette by the ACTB::Flpe mouse strain and after deletion of the neo cassette (c). The bar immediately upstream of the mb-1 promoter shows the approximate location of the probe used for the Southern blot analysis (shown in d) of genomic DNA isolated from WT and mb-1-targeted (mb1-hCre neo) ES cells. The endogenous mb-1 ATG in exon 1 was deleted. The FRT sites are represented by filled black arrowheads, and the tk neo cassette is represented by an open arrow. The mb-1 exons are shown as gray boxes numbered 1–5, and the EcoRI sites used for the Southern blot are labeled with R. The figure is not drawn to scale.
BALB/c ES cells carrying the mb1-cre construct targeted to the mb-1 locus were generated by homologous recombination (Fig. 1d) and injected into blastocysts. Three chimeric mice were obtained, and one transmitted the mutation in the germ line. To obtain the final mb1-cre expression allele (Fig. 1c), the neo cassette was deleted by crossing the mice to the Flpe deleter strain (2).
mb1-cre Is More Efficient than CD19-cre in Deleting a Floxed Reporter in Early B Cells.
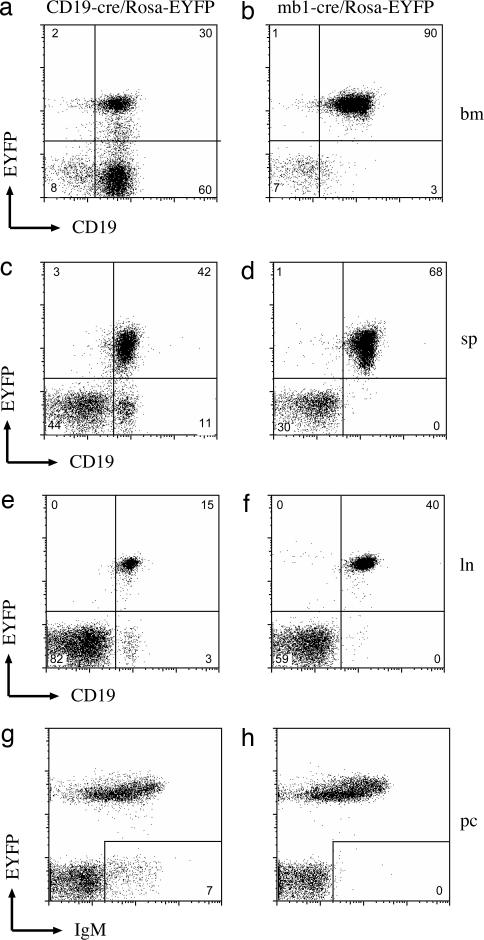
The mb1-cre mouse strain was first tested for the specificity of cre activity and for the efficiency of cre recombination in lymphoid organs by crossing it to the Rosa-floxed EYFP reporter mouse line (kindly provided by S. Srinivas, University of Oxford, Oxford, U.K.). This reporter mouse carries a modified Rosa locus containing a floxed phosphoglycerate kinase (PGK)-promoter-driven neo pA cassette upstream of a EYFP cDNA (19). Cre-mediated deletion of the PGK-neo pA cassette leads to EYFP expression. Rosa-floxed EYFP mice were also crossed with the previously published, B cell-specific, CD19-cre line (6) to compare cre recombination efficiencies in the mb1-cre and CD19-cre lines. The CD19-cre line contains a cre cDNA integrated into the B cell-specific CD19 gene. Flow cytometric analysis of cells derived from various tissues of the mb1-cre/Rosa-EYFP mice suggests that cre is primarily expressed in B cells (Fig. 2 and Fig. 7, which is published as supporting information on the PNAS web site). Very low levels of EYFP-positive T cells were detected in thymus (Fig. 7d), spleen (Figs. 2c and 7f), and lymph nodes (Fig. 2f). Virtually all B cells in the peritoneum were also EYFP-positive (Fig. 2h and data not shown). When the mb1-cre and CD19-cre lines are compared, recombination in the spleen is restricted to B cells for both intercrosses, but the efficiency in CD19-cre mice was ≈80% EYFP-positive B cells compared with 99% in the mb1-cre line (Fig. 2 c and d). The difference in the two cre lines was more pronounced when bone marrow cells were analyzed (Fig. 2 a and b). Here 33% of the B lineage cells in the lymphocyte gate were EYFP-positive with CD19-cre whereas the mb1-cre mice produced 97% EYFP-positive B lineage cells. This finding suggests an earlier and/or more efficient loxP site recombination in developing B cells of the mb1-cre mice.
Fig. 2.
mb-1-cre activity is detected in the B lineage. Cells from bone marrow (bm), spleen (sp), lymph nodes (ln), and the peritoneal cavity (pc) from CD19-cre/Rosa-EYFP mice (a, c, e, and g) or mb-1-cre/Rosa-EYFP mice (b, d, f, and h) were stained for CD19 or IgM and analyzed by FACS. The percentage of CD19-positive B cells that were also EYFP-positive is depicted in the upper right quadrant of each FACS plot. All plots show cells in the lymphocyte gate. The plots are representative of at least three mice analyzed for each genotype.
Efficient cre-Mediated Recombination Can Be Recapitulated in IL-7-Dependent Pre-B Cell Cultures.
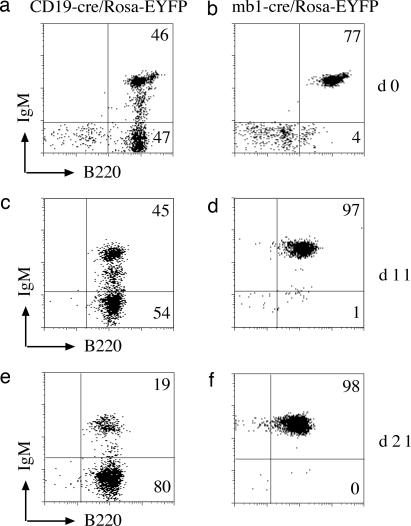
The comparison of bone marrow-derived lymphocytes from CD19-cre and mb1-cre mice suggested that the mb1-cre was considerably more active than CD19-cre at early stages of B cell development. To examine this more closely, total bone marrow cells from CD19-cre/Rosa-EYFP and mb1-cre/Rosa-EYFP mice were cultured in vitro with IL-7. Under these conditions, an almost pure population of proliferating pre-B cells was obtained within a few days of culturing (20). Flow cytometric analysis of bone marrow-derived mb1-cre/Rosa-EYFP pre-B cells cultured up to 21 days with IL-7 showed almost 100% EYFP-positive cells with mb1-cre throughout the cultivation (Fig. 3 b, d, and f). For CD19-cre, 50% of the pre-B cells were positive for EYFP at day 0 (Fig. 3a), and this decreased to 19% after 21 days of cultivation (Fig. 3 c and e). These data demonstrate that, with mb1-cre but not CD19-cre, a homogeneous population of pre-B cells carrying a recombined gene can be generated.
Fig. 3.
Comparison of CD19-cre and mb1-cre activities in IL-7-dependent, bone marrow-derived pre-B cell cultures. Bone marrow-derived pre-B cells from CD19-cre/Rosa EYFP (a, c, and e) or mb1-cre/RosaEYFP (b, d, and f) mice were cultured for the indicated times in the presence of IL-7 and then analyzed by FACS. Cells were stained with anti-B220 antibodies. Similar results were obtained for two mice for each genotype.
Using the mb1-cre Line to Study B Cell Development.
The efficiency of the mb1-cre encouraged us to use this line to study the effect of deleting several floxed loci on B cell development. Data will be presented here for two genes, the splicing factor SRp20 and the DNA methyltransferase Dnmt1. SRp20 belongs to a family of serine–arginine-rich splicing factors shown to modify the splicing efficiency of some but not all splice sites. Mouse embryos deficient in SRp20 do not develop beyond the morula stage (12).
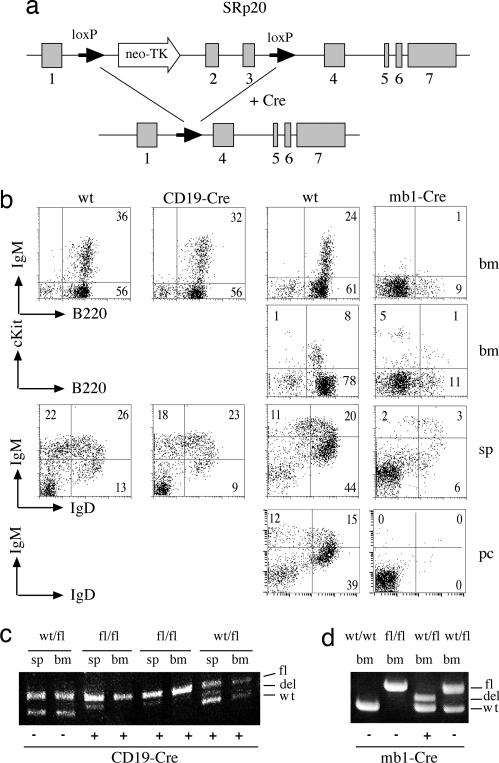
To test whether the splicing factor SRp20 is essential for B cell development or function, the mb1-cre line was intercrossed with a mouse line carrying a floxed version of SRp20 (Fig. 4a) (12). For comparison, the SRp20 line was also crossed to the CD19-cre line. FACS analysis of spleen, thymus, and bone marrow showed drastic reductions in the number of B cells in the mb1-cre background and only mild effects with CD19-cre (Fig. 4b). A strong reduction in B cell precursors was already apparent in c-kit-positive pro-B cells in mb1-cre/SRp20 bone marrow (Fig. 4b). PCR analysis of genomic DNA extracted from sorted B cells derived from spleen and bone marrow of CD19-cre/SRp20 mice indicates that only a small proportion of the splenocytes carry a deletion in the SRp20 gene whereas no deletion was detected in the bone marrow (Fig. 4c). This is true even for heterozygous (WT/fl) mice where selection for nondeleted alleles is not expected. In contrast, mb1-cre mice heterozygous for the floxed SRp20 allele show a complete loss of the floxed allele already in bone marrow B cells (Fig. 4d), again indicating an early and very efficient cre activity in the mb1-cre line. The drastic reduction of immature and mature B cells in the mb1-cre/SRp20 mice indicates that SRp20 is essential for pre-B cell survival and/or differentiation. Analysis of the peritoneal B cell population indicated that B1 cells are also drastically affected by mb1-cre-mediated SRp20 deletion (Fig. 4b).
Fig. 4.
mb1-cre/SRp20 shows a stronger B cell phenotype than CD19-cre/SRp20. (a) cre-mediated recombination of the targeted SRp20 locus (upper line) results in deletion of SRp20 exons 2 and 3. The loxP sites in introns 1 and 3 are indicated with solid arrows. (b) Single-cell suspensions from bone marrow (bm), spleen (sp), and peritoneal cavity (pc) were stained for B220, cKit, IgM, and IgD. The numbers in the plots indicate the percentages of total cells in the lymphocyte gate falling in each quadrant or region. For one mouse, a low number of B1 B cells was detected in the peritoneal cavity. DNA was isolated from sorted B cells derived from bone marrow, and spleen was PCR-amplified with primers specific for the floxed SRp20 locus. Bands corresponding to the floxed (fl), deleted (del), or WT alleles were resolved on agarose gels. Mice heterozygous (WT/fl), homozygous (fl/fl), or WT (wt/wt) for the floxed SRp20 gene either with (+) or without (−) the CD19-cre (c) or mb1-cre (d) gene were analyzed. The plots are representative of five mice analyzed for each genotype.
The Dnmt1 gene is thought to be the major methyltransferase maintaining DNA methylation in somatic cells. The murine Dnmt1 locus has been floxed (21) in such a way that induction of cre recombinase results in the deletion of exons 4 and 5 (Fig. 5a). As a consequence of the deletion, splicing of exon 3 to exon 6 causes a frame shift. The resulting, presumably nonfunctional, peptide would contain the first 75 of the 1,621 aa of the mature protein. Flow cytometric analysis of bone marrow and spleen cells derived from these mice reveals a complete block in B cell development already in the bone marrow (Fig. 5b Center). This block appears to be similar to that observed in Ig-α-deficient mice (Fig. 5b Right) (22). However, the reduced number of CD19+ cells seen in Dnmt1-deficient mice (the relative proportions of CD19+ cells in the lymphocyte gate for WT, mb1-cre/Dnmt1, and Ig-α knockout were 74%, 10%, and 62%, respectively) suggests that Dnmt1 may be important for survival and accumulation of developmentally blocked pro-B cells.
Fig. 5.
Deletion of Dnmt1 by mb1-cre results in a dramatic block in B cell development. (a) Schematic of the floxed Dnmt1 locus. Cre-mediated recombination deletes exons 4 and 5 of the Dnmt1 gene and a flanking hygromycin cassette introduced during construction of the floxed Dnmt1 strain. (b) B cells derived from the bone marrow (bm), peritoneal cavity (pc), and spleen (sp) of WT (Left), mb1-cre/Dnmt1 (Center), or mb1-deficient (Right) mice were analyzed by flow cytometry after staining for CD19, B220, cKit, IgM, and IgD. The plots are representative of at least three mice analyzed for each genotype.
For both mb1-cre/SRp20 and mb1-cre/Dnmt1 mice, attempts to culture B cell precursors from bone marrow in the presence of IL-7 failed (data not shown), probably because of the early and efficient deletion of the essential SRp20 and Dnmt1 genes, which may affect cell survival, proliferation, and/or the IL-7 responsiveness of mutant B cell precursors.
Tissue Specificity of the mb1-cre-Mediated Recombination.
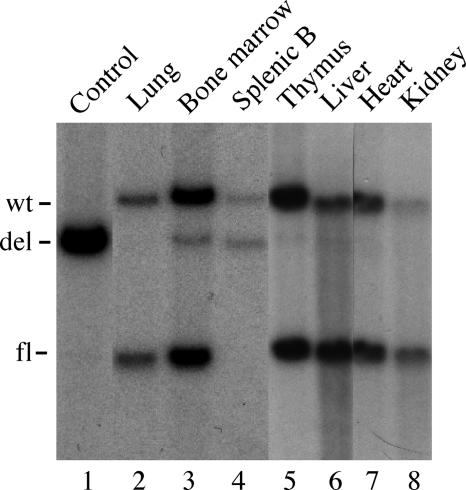
The results of the experiments described here suggest that cre-mediated recombination in the mb1-cre line is primarily restricted to the B cell lineage (see Discussion for a summary). To directly address this question, a Southern blot was performed by using DNA isolated from various organs of mb1-cre/Dnmt1 mice (Fig. 6). As expected, mice heterozygotes for both the floxed Dnmt1 locus and the mb1-cre allele (double heterozygotes) show recombination of the Dnmt1 locus in genomic DNA isolated from total bone marrow (Fig. 6, lane 3) and from sorted splenic B cells (Fig. 6, lane 4). As could be expected from the FACS results presented in Fig. 5, analysis of sorted (CD19+) B cells from the spleen showed a complete absence of the unrearranged Dnmt1 allele in double heterozygote mice (Fig. 6, lane 4). This finding again underscores the high efficiency of mb1-cre-mediated recombination in the B lineage. A weak recombined band can sometimes also be seen in thymus, liver, and kidney. Although we suspect that most of this signal is due to contaminating B cells in the organ, we cannot rule out a low level of recombination in other tissues. We have sometimes seen a small number of EYFP-positive T cells by FACS in mb1-cre/Rosa-EYFP mice and a rearranged Dnmt1 allele by PCR in kidney DNA of homozygous mb1-cre/Dnmt1 mice. We believe such ectopic recombination must be minor because we have no evidence for a phenotype in any other cell type than the B lineage in both test lines for the apparently essential SRp20 and Dnmt1 genes. We also have no evidence that expression of the hCre recombinase from the mb1 locus in the B lineage is detrimental to B cell development or survival because the number of cells in the B subfractions in spleen and bone marrow is unchanged compared with WT littermates (Table 1).
Fig. 6.
Tissue specificity of mb1-cre recombination in floxed Dnmt1 mice. Recombination of the floxed Dnmt1 locus was accessed by Southern blotting using a probe derived from intron 2 of the Dnmt1 gene (see Materials and Methods). Genomic DNA from heart, kidney, thymus, liver, lung, bone marrow, and sorted splenic B cells was prepared from mice heterozygous for both the floxed Dnmt1 allele (WT/fl) and the targeted mb1-cre allele (WT/mb1-cre). As a control, genomic DNA derived from in vitro cultured bone marrow-derived pro/pre-B cells from mb1-cre/Dnmt1(fl/fl) bcl2 transgenic mice was also analyzed (lane 1).
Table 1.
B cell population sizes during development in mb1-cre mice
| Mice | Total | CD19+ |
Transitional 1 | Marginal zone | Follicular | Immature | Recirculating | Pro/pre | |
|---|---|---|---|---|---|---|---|---|---|
| No. | % | ||||||||
| Spleen | |||||||||
| WT | 79.5 ± 14.8 | 28.4 ± 7.5 | 35.5 ± 5.2 | 2.3 ± 1.1 | 0.4 ± 0.3 | 25.5 ± 5.8 | — | — | — |
| mb1-Cre | 96.3 ± 23 | 31.5 ± 8.5 | 32.6 ± 1.9 | 2.3 ± 0.8 | 0.5 ± 0.4 | 28.0 ± 7.2 | — | — | — |
| Bone marrow | |||||||||
| WT | 23.5 ± 2.7 | 4.6 ± 0.9 | 19.8 ± 4.6 | — | — | — | 1.5 ± 0.3 | 0.3 ± 0.2 | 0.4 ± 0.1 |
| mb1-Cre | 21.5 ± 2.1 | 4.0 ± 0.9 | 18.5 ± 3.0 | — | — | — | 1.5 ± 0.4 | 0.2 ± 0 | 0.3 ± 0.1 |
All cell numbers are ×106. All B cell populations were based on a lymphocyte gate. Cells were derived from heterozygote and WT littermates on a BALB/c background with four mice for each genotype. Transitional 1 B cells: CD19+, IgM-high/intermediate, IgD-low, CD23-negative, and CD21-low. Marginal zone B cells: CD19+, IgM-high, CD21+, and CD23-negative. Follicular B cells: CD19+, CD21+, and CD23+. Immature B cells: B220-intermediate and IgM+. Recirculating B cells: B220-high and IgM-intermediate. Pro/pre-B cells: B220+ and IgM-negative. —, not applicable.
Discussion
In this study we introduce the mb1-cre mouse strain as a unique cre transgenic line for efficient B cell-specific deletion of floxed genes. Because the mb-1 gene is expressed very early in B cell development, even before VDJ recombination at the IgH locus begins, the mb1-cre transgenic line enables efficient cre recombination in bone marrow and in ex vivo pre-B cell cultures. In agreement with this, B cell development was partially blocked at the pre-B cell stage when mb1-cre mice were crossed to mice bearing a floxed SRp20 gene and completely blocked at the pro-B cell stage when crossed to mice bearing a floxed Dnmt1 gene. Previous studies showed that lack of a functional SRp20 or Dnmt1 gene is lethal in developing mouse embryos (12, 17). Consequently, the question of whether these genes are essential for B cell development or function could not be asked by using constitutive knockout lines. The drastic block in B cell development seen in the bone marrow and spleen of mb1-cre/SRp20 and mb1-cre/Dnmt1 lines demonstrates that both genes are essential for B cell development and/or survival. Because both of these genes are ubiquitously expressed, it is also reasonable to suppose that loss of their function would be lethal not only in B cells but also in most, if not all, other cell types. However, no other abnormalities were observed in the mb1-cre/SRp20 or mb1-cre/Dnmt1 mice, strongly suggesting that cre expression in mb1-cre mice is B cell-specific. The results showing that no B1 B cells develop in mb1-cre/SRp20 and mb1-cre/Dmnt1 mice suggest that SRp20 and Dmnt1 are absolutely required for the development of both B1 and B2 subsets.
Using the EYFP reporter line, we observed a low frequency (<1%) of EYFP-positive T cells in the thymus, spleen, and lymph nodes (Figs. 2 and 7 and unpublished observations). We believe that there are three conceivable explanations for this low frequency of EYFP-positive T cells. First, because it is not known to what extent the mb-1 gene locus is transcriptionally active in uncommitted lymphoid precursors, the rare EYFP-positive cells could be derived from cre recombination in a common lymphocyte progenitor that could give rise to both B and T lineages. Second, the mb1 gene locus may be active (and thus cre recombinase would be expressed) in some T cells either by stochastic transcription of the WT gene or as a consequence of the modifications made to produce the knockin cre. Specifically, exons 2 and 3 of the mb1 gene, which were deleted during the creation of the mb1-cre line, may contain regulatory motifs that influence the tissue-specific expression of this gene. Third, it is also possible that a small number of pro-B cells redifferentiate to the T cell lineage after activating the mb-1 gene and expressing cre (23, 24). Because T cell development in the crosses to the SRp20 and Dnmt1 mice is normal (data not shown) and because it is known that T cell development is completely blocked in the absence of Dnmt1 (16), the level of cre-mediated recombination in T cells of mb1-cre/Dnmt1 mice is probably minimal. We have very infrequently observed a recombined EYFP reporter being transmitted in the germ line of mb1-cre/EYFP mice, presumably the result of rare cases where cre was expressed in germ cells. The frequency of such “ectopic” recombination may also vary depending on the floxed locus.
There are several reports showing that the proportion of recombined cells in CD19-cre mice is higher in later stages of B cell development, and this is probably because the CD19 promoter is more active in mature B cells than in immature B cells. FACS analysis shows increased CD19 expression during B cell development (25), and cre-recombinase expression in CD19-cre mice was shown to increase with B cell maturation (26). The CD19-cre mouse strain has been successfully used to inactivate floxed Pax-5, Blimp-1, and IkB kinase genes in later stages of B cell development (27–29). When the CD19-cre was combined with an apparently nonessential floxed gene, the frequencies of recombined B cell precursors in the bone marrow were in the 75–80% range (6, 29) or 33% in our hands (Fig. 2). The level of recombination in B cells increased to 80–98% in the spleen (29). These efficiencies change drastically when the floxed gene is essential for B cell development (Figs. 4 and 5 and unpublished results). In this case, the 20–60% unrecombined precursor B cells in the bone marrow are sufficient to generate almost normal levels of peripheral B cells that lack the recombination (Fig. 4b). The more dramatic difference in efficiency between CD19-cre and mb1-cre seen in the bone marrow would be consistent with the fact that B cells in the bone marrow are enriched for early stages of B cell development. The observation that some B cells in the CD19-cre/EYFP line express less EYFP compared with the mb1-cre/EYFP line (Figs. 2, 3, and 7) presumably reflects cells that have not had time to accumulate high levels of EYFP because the recombination of the reporter locus occurred shortly before analysis. This low EYFP expression may also be attributed to the different expression levels of CD19 at the different B cell stages and/or the fact that the cre cDNA in the CD19-cre allele is not the humanized form. The humanized cre has been reported to be more efficiently expressed in mouse cells (18).
Of particular interest is the finding that only a fraction of pre-B cells in IL-7-dependent cultures from CD19-cre/EYFP mice were positive for EYFP, indicating inefficient cre activity, which is in sharp contrast to the mb1-cre/EYFP mice showing cre-mediated recombination in virtually all pre-B cells (Fig. 3). Because most of the cells in the IL-7-dependent cultures of CD19-cre/EYFP pre-B cells are CD19-positive, the CD19 promoter (presumably on both alleles) must be transcriptionally active, and thus cre recombinase should be expressed. Also, accessibility reasons cannot explain the differences in recombination efficiency between CD19-cre and mb1-cre because the Rosa-EYFP gene locus is recombined in both cases. One possible explanation may be the above-mentioned low expression levels of CD19 in early B cell stages. These resulting low levels of cre in IL-7-dependent pre-B cultures may not be sufficient to allow efficient recombination. In summary, the mb1-cre mouse strain is a valuable tool for early and efficient cre-mediated B cell-specific recombination in vivo and in ex vivo cultured pre-B cells.
Materials and Methods
Generation of Targeting Vector and Targeted ES Cell Clones.
The short (2.1-kb) and long (9.6-kb) homology arms for the targeting construct were isolated from mouse mb-1 genomic clones derived from the BALB/c strain (30) (kindly provided by N. Sakaguchi, Kumamoto University, Kumamoto, Japan). Exons 2 and 3 of the mb-1 gene were replaced by a cDNA encoding a mammalian codon-optimized cre recombinase (hCre) followed by a SV40 poly(A) signal. The hCre cDNA was derived from the pBluehCre plasmid kindly provided by R. Sprengel (Max Planck Institute for Medical Research, Heidelberg, Germany). In addition, mb-1 exon 1 was truncated to remove the ATG codon whereas intron 1, including the splice donor and acceptor sites, was retained without modifications. A neo cDNA cassette under the tk promoter and flanked by two FRT sites having the same orientation was introduced 3′ of hCre.
ES cells containing the hCre integrated into the mb-1 locus (mb1-cre) were produced by electroporating 1 × 107 BALB/c ES cells (31) in 900 μl of transfection buffer (20 mM Hepes, pH 7.0/137 mM NaCl/5 mM KCl/0.7 mM Na2HPO4/6 mM glucose/0.1 mM 2-mercaptoethanol) with 60 μg of linearized vector at 240 V and 475 μF. ES cells were cultured in complete DMEM selection medium (10% FCS, l-glutamine, sodium pyruvate, and penicillin/streptomycin) containing G418 (320 μg/ml). After 12 days, 220 ES cell colonies were screened by Southern blot, and two clones gave the expected bands on the targeted allele. One clone was injected into C57BL/6J blastocysts at the transgene facility of the Max Planck Institute of Immunobiology. Three chimeric mice were obtained, and one of these transmitted the targeted mb-1 locus to subsequent generations. The neo cassette was deleted by crossing the resulting mice to the Flpe deleter strain (2). The resulting mb1-cre line (BALB/c × C57BL/6 F1) was backcrossed to C57BL/6 and BALB/c mice, and the results presented here are from experiments performed on mice backcrossed for at least four generations.
The sources of various floxed reporter or test lines crossed to the mb1-cre line were as follows: SRp20 (12), Dnmt1 (21), R26R YFP (19), CD19-cre (6), and Flpe deleter (2).
Mice used throughout these experiments were 6–8 weeks old. All mice were maintained in a barrier mouse facility at the animal facility (Max Planck Institute of Immunobiology). All animal studies were approved by the German Animal Rights Office.
Southern Blot and PCR Analysis.
To characterize modifications of the mb-1 locus, a 170-bp genomic mb-1 fragment located 2 kb 5′ of the mb-1 promoter was amplified by PCR using the following oligonucleotides: mb1extprobe sense, 5′-TGTGAAGTCATAACTTCTTTGG-3′; mb1extprobe antisense, 5′-AGCAAACCAAACCAAGGCTCAGTGC-3′. This fragment was used as an external probe to discriminate between WT (6.4 kb) and the mb1-cre-targeted allele (7.2 kb) when hybridized to EcoRI-digested genomic DNA. Homozygous mice were identified by the lack of B lymphocytes in peripheral blood caused by the absence of Ig-α.
Genomic DNA isolated from sorted cells or tail biopsies was used for PCR genotyping as described previously (12). For mb1-cre detection, a hCre PCR was used with the primers hCre dir (5′-CCCTGTGGATGCCACCTC-3′) and hCre rev (5′-GTCCTGGCATCTGTCAGAG-3′). The conditions were 30 cycles of 94°C for 45 sec, 58°C for 60 sec, and 72°C for 1 min, resulting in a 450-bp product. For the SRp20 gene, the primers Xi1 (5′-TTGATTGCGACAGGACTTT-3′) and X16X3 (5′-GATTACCGCAGGAGGAGT-3′) were used as forward primers for the deleted and floxed SRp20 gene, respectively. The primer Xi3R (5′-AGAACGGATGATTGGGAA-3′) was used as a common reverse primer for both the deleted and floxed alleles. The PCR conditions were as follows: 31 cycles of 50 sec at 94°C, 20 sec at 56°C, and 50 sec at 72°C. The 531- and 664-bp products correspond to the deleted and floxed alleles, respectively. Southern blot analysis of deletions in the Dnmt1 locus was performed on SpeI-digested genomic DNA isolated from various organs. The probe used was a 767-bp fragment derived from intron 2, and it was generated by PCR using primers dnmt1sF (5′-AGGTAGTCTAGGTGCCCTG-3′) and dnmt1sR (5′-CAGCCTCCAGAATGTGTATC-3′).
Preparation of Cell Suspension from Lymphoid Organs.
Femurs were flushed with DMEM to extract cells, and spleens were minced through a nylon mesh cell strainer (Falcon; BD, Heidelberg, Germany) to obtain a single-cell suspension in DMEM/10% FCS. Erythrocytes were depleted by incubating cell preparations from bone marrow, spleen, and lymph nodes in lysis buffer (150 mM NH4Cl/10 mM KHCO3) for 2 min on ice. Mice were bled from the tail vein in the presence of heparin (Liquemin; Roche Diagnostics, Mannheim, Germany), and peripheral blood lymphocytes were purified after lysing the erythrocytes. Mouse peritoneal cells were isolated with 5 ml of PBS buffer. Splenic B cells were purified by staining single-cell suspensions with B220-phycoerythrin (PE) and sorting on a MoFlo device (Dako, Glostrup, Denmark). The purity was always >95% B cells as tested by FACS analysis.
Antibodies for FACS Analysis.
For each sample, 2 × 105 cells were incubated with various combinations of antibodies as indicated in the figure legends. Staining was done for 20 min on ice. The antibodies used for lymphocyte staining were anti-mouse IgM-PE clone, anti-mouse CD5-Biotin (clone 53-7.3;), anti-mouse B220-PE or peridinin–chlorophyll–protein complex (PerCP) (clone RA3-6B2), anti-mouse CD19-PE or PerCP-cy5.5 (clone 1D3), anti-mouse IgD-Biotin or IgD-PE (clone 11-26), anti-mouse CD43-PE (clone S7), anti-mouse c-Kit-PE (clone ACK 45), anti-mouse CD21/CD35-biotin (clone 7G6), and anti-mouse CD23 PE (clone B3B4) (all from BD). Anti-mouse IgM-cy5 was from Jackson ImmunoResearch Laboratories (West Grove, PA). Goat anti-mouse IgM-PE and anti-mouse IgD-PE clone (clone 11-26) were from Southern Biotechnology Associates (Birmingham, AL). Biotinylated Abs were detected by streptavidin-PerCP (BD). Propidium iodide (Sigma–Aldrich, Hamburg, Germany) was added to the samples immediately before FACS analysis to detect dead cells.
Four-color flow cytometry was performed on a FACS or FACSCalibur flow cytometer (BD), and 50,000–100,000 events were collected by sample. Flow cytometric profiles were analyzed by using CELLQuest (BD) and FlowJo (Tree Star, Ashland, OR) software.
Supplementary Material
Acknowledgments
We thank Dr. Sankar Srinivas for the Rosa26-EYFP mice, Dr. Rudolf Jaenisch (Whitehead Institute for Biomedical Research, Cambridge, MA) for the Dnmt1 mouse line, Dr. Susan Dymecki (Harvard Medical School, Boston, MA) for the flpe line, Dr. Rolf Sprengel for hCre cDNA, Dr. Klaus Rajewsky (Harvard Medical School) for the CD19-cre line, and Dr. Benoit Kanzler (Max Planck Institute of Immunobiology) for ES cell injections and the production of transgenic mice. We also thank Catrin Eschbach and Ingrid Fidler for valuable technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB620.
Abbreviations
- EYFP
enhanced yellow fluorescent protein
- FRT
flippase recombinase target
- PE
phycoerythrin
- PGK
phosphoglycerate kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ghosh K, Van Duyne GD. Methods. 2002;28:374–383. doi: 10.1016/s1046-2023(02)00244-x. [DOI] [PubMed] [Google Scholar]
- 2.Branda CS, Dymecki SM. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 3.Matthias P, Rolink AG. Nat Rev Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 4.Melchers F. Nat Rev Immunol. 2005;5:571–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 5.Nemazee D. Annu Rev Immunol. 2000;18:19–51. doi: 10.1146/annurev.immunol.18.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rickert RC, Roes J, Rajewsky K. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Reth M, Wienands J. Annu Rev Immunol. 1997;15:453–479. doi: 10.1146/annurev.immunol.15.1.453. [DOI] [PubMed] [Google Scholar]
- 9.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Sakaguchi N, Kashiwamura S-i, Kimoto M, Thalmann P, Melchers F. EMBO J. 1988;7:3457–3464. doi: 10.1002/j.1460-2075.1988.tb03220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveley BR. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jumaa H, Wei G, Nielsen PJ. Curr Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 14.Knox JD, Araujo FD, Bigey P, Slack AD, Price GB, Zannis-Hadjopoulos M, Szyf M. J Biol Chem. 2000;275:17986–17990. doi: 10.1074/jbc.C900894199. [DOI] [PubMed] [Google Scholar]
- 15.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 16.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melogosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 17.Li E, Bestor TH, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 18.Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 19.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemming A, Brummer T, Reth M, Jumaa H. Nat Immunol. 2003;4:38–43. doi: 10.1038/ni862. [DOI] [PubMed] [Google Scholar]
- 21.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 22.Pelanda R, Braun U, Hobeika E, Nussenzweig MC, Reth M. J Immunol. 2002;169:865–872. doi: 10.4049/jimmunol.169.2.865. [DOI] [PubMed] [Google Scholar]
- 23.Rolink A, Nutt S, Melchers F, Busslinger M. Nature. 1999;401:603–606. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 24.Taghon TN, David E-S, Zuniga-Pflucker JC, Rothenberg EV. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoham T, Rajapaksa R, Boucheix C, Rubinstein E, Poe JC, Tedder TF, Levy S. J Immunol. 2003;171:4062–4072. doi: 10.4049/jimmunol.171.8.4062. [DOI] [PubMed] [Google Scholar]
- 26.Schwenk F, Sauer B, Kukoc N, Hoess R, Müller W, Kocks C, Kühn R, Rajewsky K. J Immunol Methods. 1997;207:203–212. doi: 10.1016/s0022-1759(97)00116-6. [DOI] [PubMed] [Google Scholar]
- 27.Horcher M, Souabni A, Busslinger M. Immunity. 2001;14:779–790. doi: 10.1016/s1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro-Shelef M, Lin K-I, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 29.Pasparakis M, Schmidt-Supprian M, Rajewsky K. J Exp Med. 2002;196:743–752. doi: 10.1084/jem.20020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashiwamura S, Koyama T, Matsuo T, Steinmetz M, Kimoto M, Sakaguchi N. J Immunol. 1990;145:337–343. [PubMed] [Google Scholar]
- 31.Noben-Trauth N, Kohler G, Burki K, Ledermann B. Transgenic Res. 1996;5:487–491. doi: 10.1007/BF01980214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.