Abstract
In this work, we provide evidence of a mechanism to reinforce the strength of an icosahedral virus by using its genomic DNA as a structural element. The mechanical properties of individual empty capsids and DNA-containing virions of the minute virus of mice are investigated by using atomic force microscopy. The stiffness of the empty capsid is found to be isotropic. Remarkably, the presence of the DNA inside the virion leads to an anisotropic reinforcement of the virus stiffness by ≈3%, 40%, and 140% along the fivefold, threefold, and twofold symmetry axes, respectively. A finite element model of the virus indicates that this anisotropic mechanical reinforcement is due to DNA stretches bound to 60 concavities of the capsid. These results, together with evidence of biologically relevant conformational rearrangements of the capsid around pores located at the fivefold symmetry axes, suggest that the bound DNA may reinforce the overall stiffness of the viral particle without canceling the conformational changes needed for its infectivity.
Keywords: capsid, virion, nanomechanics, finite element methods, atomic force microscopy
Investigation of the mechanical properties of biomolecular assemblies is important to understanding the relationship between physical structure and biological function (1) and for the application of biomaterials in the fabrication of molecular structures (2). Viruses are masterpieces of nanoengineering designed as replicating machines. In most infectious virus particles (virions), the protein shell (capsid) that encloses the nucleic acid genome reveals a minimalist architecture, based on the oligomerization of multiple copies of just one or a few types of structurally equivalent or quasiequivalent protein subunits (3, 4). However, even the most simple virion can accomplish many complex and sometimes conflicting functions during the infectious cycle (5). Virus capsids must be robust enough to protect the viral genome against physical–chemical assaults (6) but labile and/or flexible enough to release the infectious nucleic acid into a target cell (7, 8). Also, many virions accommodate a maximum amount of genetic information in the minimum space, as the nucleic acid is packed to crystal densities (9). To meet these and other stringent biological requirements, viral particles could have acquired outstanding mechanical properties, which are beginning to be revealed (10, 11). For example, it has been shown that on DNA packaging, the φ29 and λ bacteriophage capsids can withstand internal pressures as high as 60 (12) and 20 (13) bars, respectively. Several studies have provided insights into the forces involved in DNA ejection from or packaging in phage capsids (14–16). A recent study of φ29 empty capsids yielded a Young's modulus of 1.8 GPa (17), close to that of hard plastic. One of many important related aspects that have not been directly investigated yet is the influence of the enclosed genomic nucleic acid on the mechanical properties of the viral particle.
The parvovirus minute virus of mice (MVM) is among the smallest and structurally simplest viruses known. The parvovirus capsid is formed by 60 structurally equivalent subunits arranged in a simple (T = 1) icosahedral symmetry (18, 19). The atomic structures of the protein shell for both the empty capsid and the DNA-containing virion of MVM, as determined by x-ray crystallography, are very similar (19, 20). In addition, ≈25–30% of the single-stranded genomic DNA in the virion was crystallographically visualized as conformationally defined oligonucleotide stretches bound to 60 equivalent, small concavities located at symmetrical positions at the internal surface of the protein shell (19).
Results
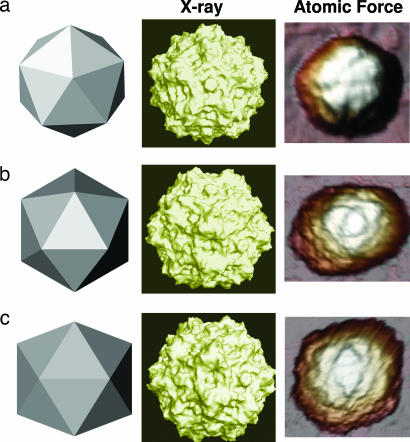
In this work, we perform atomic force microscopy (AFM) nanoindentation experiments to compare the mechanical properties of empty capsids and nucleic acid-containing virions of MVM. Viral particles were imaged under physiological conditions by AFM using “Jumping Mode” to accurately control the applied force (21). The agreement between the x-ray atomic model (19) and the AFM topographies (Fig. 1) was reflected by the same local topographic features being observed by both techniques, including the spikes located at the threefold axes, the narrow prominences located at the fivefold axes, and the grooves between those features. This agreement was not obvious because cryoelectron microscopy or crystallographic structural models reveal average features, whereas the AFM images show individual particles. We used those features to identify the orientation of the surface-immobilized icosahedral particles, i.e., along capsid fivefold (Fig. 1a), threefold (Fig. 1b), or twofold (Fig. 1c) symmetry axes. No differences in dimensions or topographic features between empty capsids and virions, or between individual viral particles, were found.
Fig. 1.
MVM particles as viewed along fivefold (a), threefold (b), and twofold (c) symmetry axes. (Left) Simplified cartoons. (Center) Molecular surface models derived from crystallographic data. The Protein Data Bank coordinates corresponding to the MVM virion (PDB ID code 1MVM; ref. 19) and the program Pymol (DeLano Scientific, San Carlos, CA) were used. (Right) Actual AFM images of individual MVM particles (image sizes: 60 × 60 nm). The MVM topographies appear laterally expanded because of the usual tip-sample dilation effects.
To measure the stiffness of the MVM particles, we carried out nanoindentations on individual intact particles in specific orientations. The recorded force-vs.-distance curves were fitted linearly to obtain the spring constant k along the direction of the applied force, assuming the virus and cantilever as two springs in a series (17). We obtained 194 force-vs.-distance curves by using 23 empty capsids (7, 11, and 5 particles oriented along the five-, three-, and twofold axes, respectively) (Fig. 2a). The spring constants obtained for the individual empty capsids in each orientation were represented in a histogram. Gaussian fits of the histograms yielded spring constants of 0.58 ± 0.13, 0.56 ± 0.15, and 0.58 ± 0.10 N/m along the five-, three-, and twofold symmetry axes, respectively (Fig. 2a). These results show that the spring constant of the MVM empty capsid is, within experimental error, isotropic, at least for the three symmetry orientations.
Fig. 2.
Comparison of the mechanical properties of MVM empty capsids (a) and virions (b). (Left) Shown are the crystallographic structures of the empty capsid (a) and DNA-filled virion (b) as space-filling models obtained by using the program RasMol (23). The models have been cut in half to show the particle interior. In the virion, the DNA stretches whose conformation was crystallographically solved are shown in green, coating in a periodic way 60 cavities of the capsid internal surface. (Right) The histograms obtained for empty capsids (a) and virions (b) are shown. They depict the stiffness (spring constant, k) values obtained for individual particles subjected to nano-indentation along fivefold (red), threefold (green), and twofold (blue) axes. (Insets) The k values from Gaussian fits obtained for empty capsids and virions along each symmetry axis. A Student t test for samples with an unequal variance reveals with >99% confidence that the three mean k values shown in b Inset do not overlap.
We then obtained 155 force-vs.-distance curves by using 28 DNA-containing virions (9, 11, and 8 particles oriented along the five-, three-, and twofold axes, respectively) (Fig. 2b). Gaussian fits of the histograms yielded spring constants of 0.6 ± 0.2, 0.8 ± 0.4, and 1.4 ± 0.5 N/m along the five-, three-, and twofold symmetry axes, respectively (Fig. 2b). A comparison of the spring constants of the empty capsid and the virion obtained in exactly the same conditions showed that the presence of the genomic DNA leads to an increase of the particle stiffness by 3%, 42%, and 140% when probed along five-, three-, and twofold symmetry axes, respectively.
We made a simple mechanical model of an icosahedron to understand how the genomic DNA can anisotropically affect the stiffness of the MVM particle. We used thin-shell finite element methods to construct a homogeneous icosahedral shell formed by 20 triangular faces of identical thickness, with an external diameter that approximately matches that of the MVM particle (25 nm). The applied load by the AFM tip was modeled as a point force. We also tested indentation with a parabolic tip (22) but decided that, because in the MVM particle the icosahedral faces are convex, for low indentations it is justifiable to consider the contact between tip and sample as a point load.
Our simplest hypothesis was that the DNA molecule, which in the MVM virion is known to be tethered to the protein shell (19) (Figs. 2b Left and 3), reinforces the particle by coating the entire capsid internal surface, thus increasing homogeneously the effective capsid wall thickness. We performed nanoindentation simulations on this model and obtained spring constant values as a function of the icosahedron wall thickness t (Fig. 4a). The relationship between wall thickness and spring constant was found to depend on the orientation of the icosahedron. Only at a wall thickness of ≈2 nm were the spring constant values obtained along the five-, three-, and twofold symmetry axes identical (Fig. 4a). This result is consistent with the isotropic spring constant experimentally obtained for the MVM empty capsid, for which the crystallographic data show a minimum wall thickness of ≈2 nm. At higher wall thicknesses, the spring constant along the different orientations increased in a nonisotropic manner. However, the relative rates along the three axes were not consistent with the experimentally observed anisotropic increase for the virion, whose spring constant along the twofold axis was higher than that along the threefold axis. Thus, the homogeneous wall model is consistent with the empty capsid results but not with those obtained with the full virion.
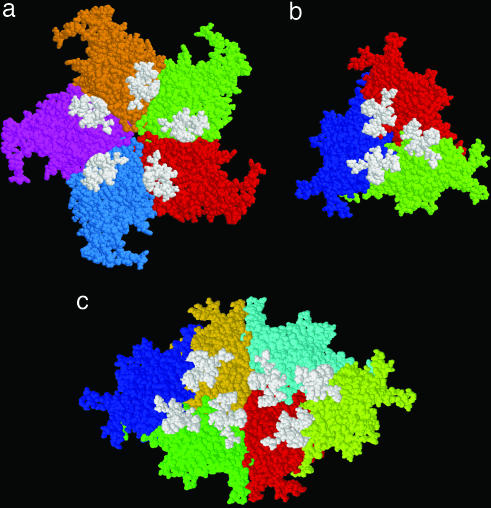
Fig. 3.
Space-filling representations of several symmetry-related MVM capsid subunits (in different colors) and crystallographically observed DNA stretches bound to those subunits (white). Views are from the particle interior and were obtained by using RasMol software (23). (a) Five protein subunits related by a fivefold symmetry axis. (b) Three subunits related by a threefold symmetry axis. (c) Six subunits related by a twofold symmetry axis. Distance measurements show that the center of gravity of each visible DNA stretch is located closer to the capsid twofold axis and further from the fivefold axis.
Fig. 4.
Finite element modeling. We used a rib size of 15 nm and a Young's modulus of 1.25 GPa. (a) The plot shows the calculated spring constant k along fivefold (red), threefold (green), and twofold (blue) symmetry axes as a function of the wall thickness t. (Inset) Homogeneous icosahedral model. (b) Five different reinforced icosahedral models with added circular patches of thickness tc positioned at different sites. Only 1 of the 20 faces of the icosahedron is shown for each model. Models 1 and 2 (circular patches at the three- or fivefold axis, respectively) did not predict the observed behavior. Models 3, 4, and 5 (circular patches positioned close to the twofold axes where the capsid-bound DNA is located) predicted a small increase of k along the fivefold axis and the largest one along the twofold axes. (c) Plot of the calculated spring constant k along fivefold (red), threefold (green), and twofold (blue) symmetry axes as a function of the added wall thickness tc in the circular regions depicted in gray in the reinforced icosahedral model used (Inset and model 4 in b), which are roughly coincident with the periodic locations where ordered DNA is bound to the capsid in the MVM virion.
Our second hypothesis was that the DNA-mediated anisotropic increase in the spring constant of the MVM virion is mainly due to the wedge-shaped DNA stretches (green patches in Fig. 2b Left) that are actually bound to concavities of the capsid internal surface, close to the twofold axes (19) (Fig. 3). These DNA stretches would increase the effective thickness of the capsid wall but only at specific locations (Figs. 2b Left and 3). To test this hypothesis, we modeled icosahedra with a wall thickness of 2 nm (to account for the isotropic stiffness of the empty capsid) and added reinforcements in the form of circular patches of increased thickness tc located at icosahedrally equivalent positions (Fig. 4b). We calculated the k values as a function of tc for five different models that differed in the positions of the patches. We found that the model that best fit the experiments was the one where the patches are approximately located at the positions where the capsid-bound DNA stretches are found in the virion, defining similar surface areas (Fig. 4b, model 4). This model predicted an anisotropic increase in stiffness that was highest along the twofold axis, somewhat lower along the threefold axis, and smallest along the fivefold axis (Fig. 4c). Slight repositioning of the circular patches (models 3 and 5) to account for the uncertainty of the exact locations of the irregular capsid–DNA interfaces, led to comparable results maintaining the model 4 anisotropy. In contrast, when we positioned the patches far from the locations where the DNA is actually bound to the capsid wall, i.e., near the twofold axis (Fig. 4b, models 1 and 2), the predictions were in complete disagreement with the experimental observations.
Discussion
Structure–function analyses increasingly support a view of the simpler virions as nanomachines optimized to carry out complex functions with a minimalist structure. For example, many experiments are repeatedly showing that the vast majority of point mutations made on the capsid of many simple viruses, including MVM, significantly reduce or abolish infectivity. Such mutations have been shown to impair cell receptor recognition, virus translocation, nucleic acid packaging, capsid conformational changes needed for infectivity, virion stability, etc. (for MVM, see refs. 24–30). Thus, only very specific structures and amino acid sequences of the capsid proteins allow the fulfillment of the many complex functions required for virus survival. In addition, the results presented here and previous evidence (4, 31, 32) show that the genomic nucleic acid of at least some small viruses not only passively carries information but also actively participates as a material component in the structure, properties, and/or function of the virion. In the case of flock house virus, the genomic RNA regulates the susceptibility of the particle to proteolysis (32). In the case of MVM, the genomic DNA contributes to the thermostability of the infectious virion through interactions between the capsid and the crystallographically visible DNA stretches (24). The present work provides direct evidence of the participation of the genomic nucleic acid in the anisotropic mechanical reinforcement of the virion.
A full understanding of how the nucleic acid molecule contributes to increase the stiffness of the MVM particle would require a complete structural description of the enclosed nucleic acid. This description is still unavailable for any icosahedral virus, because most of the viral nucleic acid molecule is asymmetrically arranged inside the symmetrical virion. However, evidence of substantial capsid–nucleic acid interactions (9, 31) and of a layer of partially ordered nucleic acid in close proximity to the inner capsid wall (33) is available for many viruses. Hence, our initial finite element icosahedral model was devised to simulate the effect of a global increase in wall thickness due to the DNA, but this model did not reproduce the experimental results. In fact, for parvoviruses like MVM, no experimental evidence supports any interaction between the DNA and the protein shell other than through the 60 crystallographically visible stretches (18–20). Interactions between some DNA phosphates and basic regions at the internal N-terminal segments of the parvovirus capsid proteins could potentially occur. However, these segments are loosely connected to the protein shell, and tethering of the DNA through these segments is not expected to reinforce the capsid wall. Thus, we modified our finite-element icosahedral model to mimic the effect of a local increase in wall thickness that was limited to those sites where the crystallographically visible DNA is bound (Figs. 2b Left, 3, and 4b, model 5). As described above, this model did remarkably match the experimentally observed stiffness anisotropy. These results indicate that the visible DNA stretches bound to 60 identical concavities of the capsid internal surface can mediate the anisotropic mechanical reinforcement of the MVM particle.
Our simplified model was not meant to quantitatively match the observed increases of the spring constant along each of the different symmetry axes. The DNA stretches and corresponding binding sites in the capsid could be better described as DNA wedges filling capsid concavities. Local deformation/bending of these concavities could occur when the empty viral particle is pushed by the AFM tip. In the full virus, the cavities could be wedged by the DNA, impairing or distorting the bending at these positions. The different relative orientations of the DNA wedges with respect to the different symmetry axes (Fig. 3) could contribute to explain the anisotropy in the mechanical response of the virion against loading forces along those axes. Based on our simple mechanical model, it is appealing to speculate about the yet unknown distribution of the disordered DNA (≈70%). Because the experiments can be basically explained by assuming that only the visible DNA stretches contribute to the mechanical properties of the virions, it is plausible that the disordered DNA is distributed inside the capsid without strong interactions with the inner capsid wall.
To summarize, the experimental AFM results reveal an anisotropic mechanical reinforcement of the MVM particle mediated by the DNA. Furthermore, the finite element methods simulations demonstrate that capsid-bound DNA patches can explain the observed reinforcement.
Regarding possible biological implications of these observations, nature could have taken advantage of the presence of the nucleic acid molecule inside the virion to further reinforce the stiffness of the MVM particle by developing a periodic capsid–DNA interface through mutation and selection. A reinforcement of the MVM capsid could be needed for at least two reasons. First, we estimated the packing density of the nucleic acid inside the virion by calculating the values of two parameters, the packing efficiency ρ (defined in ref. 16) and Vm, which describes the internal capsid volume occupied per dalton of packed nucleic acid (9). Both values (≈35% and 1.7 Å3/Da, respectively) indicate that the packing density in MVM is very high, even surpassing that of single-stranded nucleic acids in molecular crystals and approaching that found for bacteriophage Φ29 (16), which is subjected to a high internal pressure due to the packed nucleic acid (12). Second, unlike other viruses, in MVM no substantial neutralization of the negative charges of the DNA phosphates may occur through interactions with basic capsid residues (24). Both the very high DNA packing density and potential mutual repulsions between closely packed phosphate groups may generate a substantial internal pressure. Binding of patches of the DNA itself to the internal capsid surface would provide the mechanical reinforcement needed to withstand such a pressure. A possible biological role for the anisotropic effect of the capsid-bound DNA can be also suggested. Entry and exit of the viral DNA during packaging and uncoating and extrusion of the N-terminal segments of some of the capsid protein subunits are needed for infectivity, and these translocation events probably occur through the pores located at the capsid fivefold axes (18–20, 24–27, 29, 34, 35). Some of these events are altered by mutation of residues located around the base or wall of the pores and may involve conformational rearrangements (18–20, 25–27, 29, 30, 34). Thus, completion of the biological cycle of MVM probably requires some capsid flexibility, at least around the pores (20, 26, 27, 29, 30, 34). The above evidence makes it tempting to propose that the anisotropic mechanical rigidity of the MVM virion, which is lower around the fivefold axis where the capsid pores are located, could have an adaptive biological role for allowing a maximum stiffness of the particle without canceling the conformational changes needed for virus infectivity.
Beyond the possible biological implications tentatively proposed, MVM provides an example of a 3D DNA network interacting with a protein shell to tailor its mechanical properties. Remarkably enough, during the last 10 years a sustained effort has been made to fabricate 3D networks of DNA (36), and only recently it was shown that DNA could indeed be used to construct rigid structures (2). MVM engineering solutions can be a source of inspiration to improve the present designs of these DNA artificial networks and other nano-objects, such as nanocontainers (37).
Materials and Methods
Production and Purification of MVM Empty Capsids and Virions.
Purified empty capsids and infectious virions were obtained as described (28, 35) with some modifications. Mammalian NB324K cells were electroporated with a MVM (strain p) infectious recombinant plasmid originally engineered by P. Tattersall (Yale University Medical School, New Haven, CT) and coworkers (38) and obtained through J. M. Almendral (Centro de Biología Molecular, Madrid, Spain). The electroporated cells were diluted in DMEM plus 10% FCS, plated, and incubated at 37°C for 48 h. The cells were collected, resuspended in TE buffer (50 mM Tris·HCl, pH 7.5/1 mM EDTA) containing 0.2% SDS, and sonicated, and the cell extract was clarified by centrifugation. The supernatant was deposited on a layer containing 20% sucrose in TE buffer supplemented with 0.2% SDS and centrifuged for 21.5 h at 16,000 rpm in a SW40 rotor at 10°C (Beckman, Fullerton, CA). The sediment was thoroughly resuspended in TE buffer containing 0.2% Sarkosyl. The suspension was centrifuged in a cesium chloride gradient for 29.5 h at 50,000 rpm in a TFT 75.13 rotor (Kontron, Zurich, Switzerland) at 10°C. The empty capsids and virions were separated by taking advantage of their different buoyant densities (1.363 and 1.373, respectively). The bands corresponding to empty capsids and virions were clearly separated, as determined by assaying the hemagglutination activity of fractions from the gradient as described (35). The fractions of interest were extensively dialyzed against PBS. To completely exclude the possibility of any cross-contamination of virions and capsids, only the central fractions of well resolved peaks were used. As further controls, the presence of viral DNA in the fraction corresponding to virions and its absence in the fraction corresponding to capsids were assessed as described (24). In addition, the fraction corresponding to virions was layered on a cesium chloride gradient, recentrifuged as above, and extensively dialyzed again. As expected, no particles were detected in the fractions with a density corresponding to empty capsids. The purity and integrity of empty capsids and virions were assessed by transmission electron microscopy, and their concentration was estimated by UV-spectrophotometry.
AFM of Viral Particles.
Stocks of purified empty capsids (1 μM) and virions (0.3 μM) in PBS buffer (pH 7.2) were used. For AFM experiments, the stocks were diluted 20 times. A single drop (20 μl) of diluted stock was deposited on a silanized glass surface (17). The drop was left on the surface for 30 min and then rinsed twice with 20 μl of PBS. This surface treatment ensures proper virus adsorption. The tip was also prewetted with 20 μl of PBS. The AFM (Nanotec Electrónica, Madrid, Spain) was operated in Jumping Mode (21) in liquid. We used two different rectangular cantilevers, RC800PSA and BL-RC150VB-HW (Olympus, Tokyo, Japan), with spring constants of 0.05 ± 0.01 and 0.03 ± 0.01 N/m, respectively. The maximum normal force during AFM imaging was always ≈100 pN. AFM images were processed by using WSxM software (www.nanotec.es).
To determine the stiffness of empty capsids and virions, once individual particles were located on the surface, the lateral piezo scan was stopped when the tip was on top of the equatorial area of the particle. Then, force-vs.-distance curves were obtained by elongating the z-piezo until the tip established mechanical contact with the virus particle, and a nanoindentation was performed. Although in AFM experiments the Hertz model is widely used (39, 40), it does not apply when indenting shells. The observed linear behavior is expected from thin-shell mechanics, which predicts a linear elastic response for the indentation of a homogeneous spherical shell up to an indentation on the order of the shell thickness (41). To avoid particle damage, the maximum applied force was limited to 0.9 nN with typical indentations of ≈2 nm. We observed that after a few contact events the force-vs.-distance curve exhibited marked steps (42), which corresponded to an irreversible modification of the virus particle. In this case, we moved to another particle. In total, 51 viral particles were probed, and 349 force-vs.-distance curves were obtained, averaging ≈7 force-vs.-distance curves per virus. The force-vs.-distance curves were processed assuming the cantilever and the virus to be two springs in a series obtaining the stiffness (spring constant, k) of the virus particle along the direction of the applied force (17, 42).
Finite Element Modeling.
We used the program FEMLAB 3.1i (Comsol, Zoetermeer, The Netherlands). Icosahedral shell models were composed of 20 triangular faces, all with 15-nm sides. The icosahedron diameter was 24.3 nm when measured between parallel ribs. For Young's modulus, linearly scaling with the icosahedron spring constant, we used 1.25 GPa to obtain values for the spring constant close to experimental results. The models were made from thin shells, where compression in the normal direction of the plate is ignored and buckling does not occur. Icosahedra were supported by a nondeformable virtual surface and indented by a point force.
Acknowledgments
We thank C. F. Schmidt for discussion and expertise in the mechanical modeling, P. Tattersall and J. M. Almendral for providing the infectious MVM plasmid, and F. Moreno for discussion and critically reading the manuscript. This work was supported by Comunidad de Madrid (CM) Grants S-0505/MAT/0303 (to P.J.d.P., J.G.-H., M.G.M., and P.A.S.), GR-MAT-0254 (to P.J.d.P. and P.A.S.), Ministerio de Educación y Ciencia Grant BIO2003-04445 (to M.G.M.), and MAT-2004-05589-C02-01 (to J.G.-H., P.J.d.P., and C.C.) and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular. M.G.M. is an associate member of the Centro de Biocomputación y Física de los Sistemas Complejos, Zaragoza, Spain.
Abbreviations
- AFM
atomic force microscopy/microscope
- MVM
minute virus of mice.
Note Added in Proof.
Recently, two articles about mechanical properties of viruses were published (43, 44). In ref. 43, the elasticity of the cowpea chlorotic mottle virus is investigated as a function of mutations. In ref. 44, the effect of maturation on murine leukemia virus particle stiffness is reported.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. J.W. is a guest editor invited by the Editorial Board.
References
- 1.Hamm CE, Merkel R, Springer O, Jurkojc P, Maier C, Prechtel K, Smetacek V. Nature. 2003;421:841–843. doi: 10.1038/nature01416. [DOI] [PubMed] [Google Scholar]
- 2.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 3.Rossmann MG, Johnson JE. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JE, Speir JA. In: Encyclopedia of Virology. Granoff A, Webster R, editors. London: Academic; 1999. pp. 1946–1956. [Google Scholar]
- 5.Chiu W, Garcea RL, Burnett RM. Structural Biology of Viruses. Oxford: Oxford Univ Press; 1997. [Google Scholar]
- 6.Moody MF. J Mol Biol. 1999;293:401–433. doi: 10.1006/jmbi.1999.3011. [DOI] [PubMed] [Google Scholar]
- 7.Rossmann MG, Greve JM, Kolatkar PR, Olson NH, Smith TJ, MCKinlay MA, Rueckert RR. In: Structural Biology of Viruses. Chiu W, Garcea RL, Burnett RM, editors. Oxford: Oxford Univ Press; 1997. pp. 105–133. [Google Scholar]
- 8.Chow M, Basavappa R, Hogle JM. In: Structural Biology of Viruses. Chiu W, Garcea RL, Burnett RM, editors. Oxford: Oxford Univ Press; 1997. pp. 157–186. [Google Scholar]
- 9.Johnson JE, Rueckert RR. In: Structural Biology of Viruses. Chiu W, Garcea RL, Burnett RM, editors. Oxford: Oxford Univ Press; 1997. pp. 269–287. [Google Scholar]
- 10.Zandi R, Reguera D, Bruinsma RF, Gelbart WM, Rudnick J. Proc Natl Acad Sci USA. 2004;101:15556–15560. doi: 10.1073/pnas.0405844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruinsma RF, Gelbart WM, Reguera D, Rudnick J, Zandi R. Phys Rev Lett. 2003;90:248101. doi: 10.1103/PhysRevLett.90.248101. [DOI] [PubMed] [Google Scholar]
- 12.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 13.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Proc Natl Acad Sci USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odijk T. Biophys J. 1998;75:1223–1227. doi: 10.1016/S0006-3495(98)74041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purohit PK, Kondev J, Phillips R. Proc Natl Acad Sci USA. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanovska IL, Pablo PJC, Ibarra B, Sgalari G, MacKintosh FC, Carrascosa JL, Schmidt CF, Wuite GJL. Proc Natl Acad Sci USA. 2004;101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsao J, Chapman MS, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith TJ, Rossmann MG, Compans RW, Parrish CR. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 19.Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. Struct Folding Des. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 20.Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, Agbandje-McKenna M. J Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Herrero F, de Pablo PJ, Fernandez-Sanchez R, Colchero J, Gomez-Herrero J, Baro AM. Appl Phys Lett. 2002;81:2620–2622. [Google Scholar]
- 22.Schaap I, Carrasco C, de Pablo PJ, MacKintosh FC, Schmidt CF. Biophys J. 2006;91:1521–1531. doi: 10.1529/biophysj.105.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayle RA, Milner-White EJ. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 24.Reguera J, Grueso E, Carreira A, Sanchez-Martinez C, Almendral JM, Mateu MG. J Biol Chem. 2005;280:17969–17977. doi: 10.1074/jbc.M500867200. [DOI] [PubMed] [Google Scholar]
- 25.Maroto B, Valle N, Saffrich R, Almendral JM. J Virol. 2004;78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reguera J, Carreira A, Riolobos L, Almendral JM, Mateu MG. Proc Natl Acad Sci USA. 2004;101:2724–2729. doi: 10.1073/pnas.0307748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farr GA, Cotmore SF, Tattersall P. J Virol. 2006;80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombardo E, Ramirez JC, Agbandje-McKenna M, Almendral JM. J Virol. 2000;74:3804–3814. doi: 10.1128/jvi.74.8.3804-3814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valle N, Riolobos L, Almendral JM. In: Parvoviruses. Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. London: Edward Arnold; 2005. pp. 291–304. [Google Scholar]
- 30.Carreira A, Mateu MG. J Mol Biol. 2006;360:1081–1093. doi: 10.1016/j.jmb.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Casjens S. In: Structural Biology of Viruses. Chiu W, Garcea RL, Burnett RM, editors. Oxford: Oxford Univ Press; 1997. pp. 3–37. [Google Scholar]
- 32.Bothner B, Schneemann A, Marshall D, Reddy V, Johnson JE, Siuzdak G. Nat Struct Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 33.Baker TS, Olson NH, Fuller SD. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotmore SF, D'Abramo AM, Ticknor CM, Tattersall P. Virology. 1999;254:169–181. doi: 10.1006/viro.1998.9520. [DOI] [PubMed] [Google Scholar]
- 35.Hernando E, Llamas-Saiz AL, Foces-Foces C, McKenna R, Portman I, Agbandje-McKenna M, Almendral JM. Virology. 2000;267:299–309. doi: 10.1006/viro.1999.0123. [DOI] [PubMed] [Google Scholar]
- 36.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 37.Douglas T, Young M. Nature. 1998;393:152–155. [Google Scholar]
- 38.Gardiner EM, Tattersall P. J Virol. 1988;62:2605–2613. doi: 10.1128/jvi.62.8.2605-2613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson KL. Contact Mechanics. Cambridge, UK: Cambridge Univ Press; 2001. [Google Scholar]
- 40.A-Hassan E, Heinz WF, Antonik MD, D'Costa NP, Nageswaran S, Schoenenberger CA, Hoh JH. Biophys J. 1998;74:1564–1578. doi: 10.1016/S0006-3495(98)77868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau LD, Lifshitz EM. Theory of Elasticity. New York: Pergamon; 1986. [Google Scholar]
- 42.de Pablo PJ, Schaap IAT, MacKintosh FC, Schmidt CF. Phys Rev Lett. 2003;91:98101. doi: 10.1103/PhysRevLett.91.098101. [DOI] [PubMed] [Google Scholar]
- 43.Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJL, Schmidt CF. Proc Natl Acad Sci USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kol N, Gladnikoff M, Barlam D, Shneck RZ, Rein A, Rousso I. Biophys J. 2006;91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]