Abstract
Enzymes are highly dynamic and tightly controlled systems. However, allosteric communication linked to catalytic turnover is poorly understood. We have performed an integrated approach to trap several catalytic intermediates in the α2-dimeric key enzyme of chlorophyll biosynthesis, glutamate-1-semialdehyde aminomutase. Our data reveal an active-site “gating loop,” which undergoes a dramatic conformational change during catalysis, that is simultaneously open in one subunit and closed in the other. This loop movement requires a β-sheet-to-α-helix transition to assume the closed conformation, thus facilitating transport of substrate toward, and concomitantly forming, an integral part of the active site. The accompanying intersubunit cross-talk, which controls negative cooperativity between the allosteric pair, was explored at the atomic level. The central elements of the communication triad are the cofactor bound to different catalytic intermediates, the interface helix, and the gating loop. Together, they form a molecular switch in which the cofactor acts as a central signal transmitter linking the subunit interface with the gating loop.
Keywords: negative cooperativity, x-ray crystallography, integrated approach, subunit communication, protein dynamics
Cofactors like heme, chlorophyll, coenzyme F430, and carotinoids are constructed from eight molecules of 5-aminolevulinate (ALA), which forms the building block for tetrapyrrols (1). In plants and bacteria, this compound is synthesized by the vitamin B6-dependent key enzyme glutamate-1-semialdehyde aminomutase (GSAM). GSAM catalyzes the isomerization of glutamate-1-semialdehyde (GSA) to ALA by an unusual intramolecular exchange of amino and oxo groups within the catalytic intermediate 4,5-diaminovalerate (DAVA) (2–4). Two unusual features are characteristic for this enzyme: (i) GSAM shows an asymmetric distribution of pyridoxamine 5′-phosphate (PMP) and pyridoxal 5′-phosphate (PLP) in solution, and (ii) the amine form of the cofactor is required for the initiation and the end of the catalytic cycle (Scheme 1, steps i, ii, vi, and vii), whereas the aldehyde form is needed for the intermediate steps (iii–v). To address the kinetic behavior of GSAM in solution, the process of reduction of the double PLP form by NaBH3CN has been monitored (4). The biphasic curve reveals that half of the enzyme is reacting rapidly and the other half slowly. This kinetic response is consistent with a negative cooperative behavior, in which the two enzyme subunits act asymmetrically, indicating intersubunit communication (4).
Scheme 1.
Proposed catalytic mechanism (2, 3). The reaction mechanism starts with the formation of a complex between PMP and GSA (i). After ketimine-5 formation (ii; KE-5), the double bond shifts and the external aldimine between PLP and 5′-DAVA is formed (iii; EA-5), followed by the formation of the internal aldimine between PLP and Lys-273 with reorientation of DAVA (iv; DAVA-IA). The second half of the reaction cycle (v–vii) is the backward reaction. Starting with the external aldimine between PLP and 4′-DAVA (v; EA-4), the ketimine-4 between PMP and ALA is established (vi; KE-4). Finally, ALA in the PMP form of GSAM can be released (vii). The catalytic intermediates marked with an asterisk were determined by x-ray crystallography.
Catalytic activity is tightly linked to the formation of transition states between enzyme and substrate to decrease the activation energy of the reaction (5, 6). This involves a coordinated process of substrate attraction and product release (7). Allosteric communication between distant sites is fundamental to enzyme function and often defines their biological role (8, 9). To establish coupling of these different functions, negative cooperative enzymes show a high degree of cross-talk between their allosteric components which can act as molecular switches connected via communicating elements (10). Systematic investigations of these enzymes are needed to get a consistent description of their dynamic properties (11).
In a number of recent solution NMR studies, the great importance of dynamics in allostery has been shown (12–14). Here, we present a combined approach using x-ray crystallography, single-crystal absorption microspectrophotometry, and site-directed mutagenesis to analyze the allosteric properties of GSAM. Trapping of several catalytic intermediates allowed a detailed structural insight into the cooperativity phenomena of GSAM and a prediction of the reaction trajectory.
Results
The PMP/PLP Form of GSAM: Overall Structure.
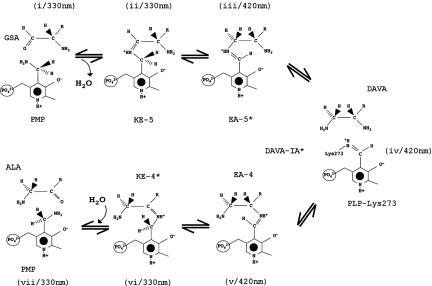
GSAM is a member of the α-family (subgroup II) of vitamin B6-dependent enzymes (4, 15). Each monomer of the compact homodimer (Mr of 92 kDa) contains 433 amino acid residues and is composed of three domains (Fig. 1A). The main PMP/PLP-binding domain, residues 70–326, contains a central seven-stranded β-sheet with one antiparallel and six parallel β-strands. Both flanking domains at the N and C termini are composed of a three-stranded antiparallel β-sheet surrounded by several α-helices on the outer surface. An outstanding feature of the structure is the large dimer contact area, which covers an area of ≈4,500 Å2 and is dominated by the interface helix. Both active sites of GSAM are located near this monomer–monomer interface and are composed of a number of crossover interactions. The phosphate groups of the cofactors are only 12 Å apart.
Fig. 1.
Crystal structure of GSAM in the PMP (KE-4)/PLP (DAVA-IA) form. (A) Overall stereo presentation of α2-dimeric GSAM. In subunit A, the N- and C-terminal domains as well as the cofactor binding domain are shown in different blue tones. Subunit B is shown in yellow. Cofactors and catalytic intermediates are highlighted. Both termini are denoted. The gating-loop regions disobeying local 2-fold symmetry and the interface helices (residues 121–138) are shown in blue (open) and red (closed), respectively. The gating loop is located at the dimer interface and extends toward the active site in the closed conformation. (B) Superposition of residues 150–183 in open (light blue/blue) and closed (yellow/red) conformation. Cofactors within the active sites are colored accordingly. The hinge element (Leu-158 and Ser-172) and Ser-163 are denoted. The β-hydroxy group of Ser-163 moves ≈22 Å.
Structural investigations of the PMP/PLP form of GSAM revealed an α2-dimeric enzyme showing deviations from the molecular twofold symmetry, which are presumably related to its function (Fig. 1; and see Table 1, which is published as supporting information on the PNAS web site). Its main characteristic is the “gating loop” (amino acid residues 150–183), a long loop covering the active-site pocket in the closed conformation (Fig. 1B). The location of this loop region suggests that it regulates access to the substrate-binding pocket. In the subunit containing PMP with the substrate bound in its ketimine-4 form (Scheme 1, step vi), the loop is open, thus allowing product release and substrate entry. In contrast, in the other subunit that contains PLP as internal aldimine and the intermediate DAVA (Scheme 1, step iv), the loop is closed, and access to the active site is obstructed by the short helical section of residues 164–168 (Fig. 1B).
The Active-Site Gating Loop in the Apoenzyme, and the Double PMP and Double PLP Forms of GSAM.
The movement of the gating loop can be defined as a concerted 90° rotation and 100° twisting of the plane between the loop helix (residues 164–168) and the hinge residues Leu-158 and Ser-172, respectively. The short helical segment, which marks the closed conformation, is disrupted in the open form, and only a short, twisted antiparallel β-sheet acts as a stabilizing element to allow spatial accommodation of the remaining loop residues.
To obtain a more precise analysis of the enzymatic reaction steps in context with the active-site gating-loop conformation, we determined the x-ray structures of the apoenzyme, the double PMP and double PLP forms of GSAM (see Fig. 4, which is published as supporting information on the PNAS web site). This was achieved by reconstituting the apoenzyme in such a way as to trap certain catalytic intermediates (see Materials and Methods). To exclude the influence of crystal contacts on loop conformations, we performed soaking experiments. For example, crystals of the double PLP form of GSAM were soaked with molar excess of DAVA (see Fig. 5, which is published as supporting information on the PNAS web site). A reorientation of the gating loop from closed to open conformation and vice versa can be observed also within the crystal (data not shown).
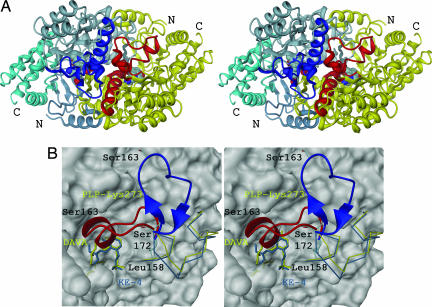
Different gating-loop conformations in GSAM can be correlated with the state of the cofactor and the respective catalytic intermediate in the active site. Besides a “disorder-to-order transition” effect of the cofactor/intermediate, the data allow an unambiguous assignment of the amine form to the open conformation. In the apo form of GSAM, the gating loop is disordered in both subunits, whereas the double PMP form is opened symmetrically with a DAVA molecule fixed (Fig. 2A). In contrast, in the double PLP form containing EA-5 (Scheme 1, step iii) in one monomer and DAVA-IA in the other (Scheme 1, step iv), only one gating loop is closed, whereas the other remains disordered.
Fig. 2.
Transition of the gating loop between opened, closed, and reopened conformation. (A) View of the opened gating loop from the crystal structure of the double PMP form of GSAM (2). Fixation of DAVA with its 4′-amino group suggests that the gating loop offers a channeling mechanism to transport substrate into the active-site pocket. The backbone helix is suggested to be the anchoring point for GluTR (26). (B) Active-site pocket in the closed gating-loop conformation. Shown is a superposition of intermediate step iii/preparation 3 (stick-and-ball mode and water as red spheres) and step iv/preparation 4 (in yellow). In the external aldimine, the carboxy group fixation of the intermediate is mediated by three water molecules (W1–W3) and Ser-29. In the DAVA-IA state, all hydrogen bonds between the catalytic intermediate and the water molecules are disrupted and W3 is replaced by the carboxy group of DAVA. (C) Active-site pocket in the reopened and disordered gating-loop conformation. Shown is a superposition of step vi/preparation 4 (stick-and-ball mode and water as red spheres) and step iv/preparation 3 (in yellow). Electrostatic interactions of the catalytic intermediate with Tyr-301* and Ser-163 are absent. In KE-4 and EA-5, Glu-406 reveals multiple conformations and the carboxy group of ketimine-4 interacts again with waters W2 and W3. Dotted lines in black and yellow indicate hydrogen bonds in the EA-5/KE-4 and the DAVA-IA states, respectively. Residues marked with an asterisk depict the other subunit.
The Gating Loop in the Closed Conformation: Proposed Reaction Trajectory.
Closing of the loop creates a very shallow active-site pocket of ≈600 Å3, which seems to be necessary for the “entropy trap” (Fig. 2B) (16, 17). The closed gating loop occludes the active-site cleft from the solvent during catalysis and prevents the release of catalytic intermediates. In the EA-5 state (Scheme 1, step iii), Ser-163 is involved in a complex network of hydrogen bonds with Tyr-301* from the adjacent subunit and the carboxy group of the catalytic intermediate (Fig. 2B). The most critical step in the reaction mechanism is the reorientation of the intermediate DAVA after disconnection from the cofactor (Scheme 1, step iv). To facilitate such a concerted reaction, DAVA has to be shifted longitudinally and rotated axially to bring the 5′-amino and subsequently the 4′-amino group in proper position to the internal aldimine. Based on our findings, we propose a reaction trajectory that involves a shuttle-movement of DAVA driven by electrostatic interactions of both, the N terminus with Glu-406 and the C-terminal moiety with Ser-29 and Arg-32, respectively (Fig. 2 B and C). The process is accompanied by rotations of the catalytic intermediate in distinct 120° steps. This rotation is completed by van der Waals contacts of Val-31 and Trp-67 with the aliphatic part of the intermediate, together with varying repulsive forces of the dipole moment caused by reorientation of the short gating-loop helix (18). Val-31 in the substrate-binding loop (residues 29–32) moves ≈1.0 Å to allow repositioning of DAVA relative to the Schiff base linkage of the internal aldimine. In the DAVA-IA state, Ser-163, although forming only a water-mediated hydrogen bond with Tyr-301*, contributes significantly to the helical conformation of the closed gating loop. Consequently, the disrupted network of hydrogen bonds between Ser-163 and the catalytic intermediate may result in the reopening of the loop (Scheme 1, step vi) and product release (Fig. 2C).
The Communication Triad and Intersubunit Signaling.
The course of reduction of GSAM by borohydrid in solution is biphasic, consistent with the existence of an asymmetric dimer in which both subunits react dependent on each other (4). If both subunits of GSAM assume two distinct and complementary conformations, and if they oscillate from opened to closed forms, they have to do this in a coordinated way, raising the question as to how both subunits cross-talk to each other.
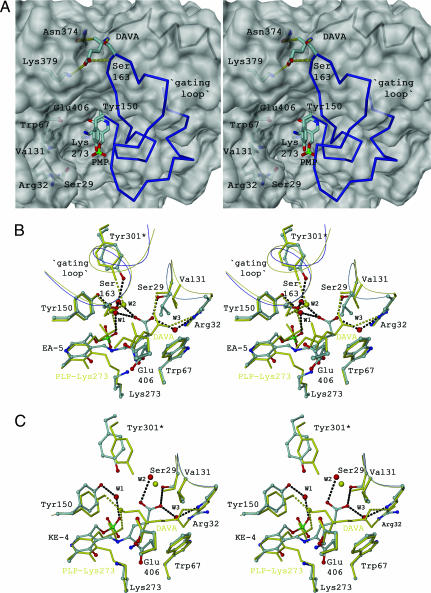
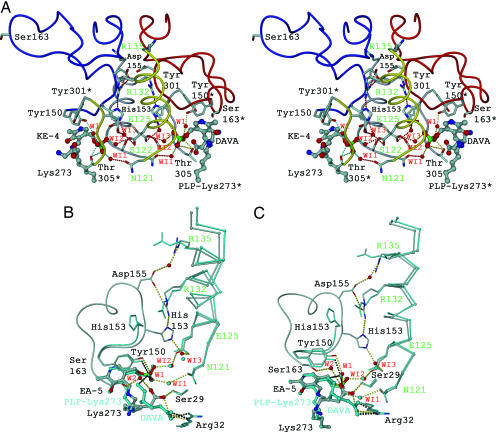
It has been shown that allosteric communication in proteins is characterized by evolutionarily conserved structural networks of amino acid interactions (19, 20). A multiple sequence alignment of GSAM reveals that only a small subset of residues forms physically connected networks that link distant functional sites in the tertiary structure (see Fig. 6, which is published as supporting information on the PNAS web site). Remarkably, each of the four clusters of invariant residues is involved in substrate binding, cofactor fixation, formation of the interface helix, or recruiting of the gating loop (Fig. 3A). The physical integration of this network is particularly striking given that it comprises ≈50% of the whole intersubunit interface. A key element is the interface helix, which at its N-terminal end is involved in electrostatic crossover interactions (residues Ser-122 and Glu-125* and vice versa) and fixation of the cofactor phosphate group via the positive dipole moment (18). At the C-terminal end of the helix, Arg-132* is involved in asymmetric crossover interactions with gating-loop residues of the adjacent chain, His-153 and Asp-155, respectively (Fig. 3A). Arg-135 is directed from the opposite site to the short antiparallel α-helical stretch, which marks the only contact between both gating-loop regions.
Fig. 3.
The communication triad and intersubunit cross-talk. (A) Invariant amino acid residues within the PMP/PLP form of GSAM (also see Fig. 5). Residues of the interface helix involved in crossover interactions are highlighted in green and denoted in single-letter code. All waters shown are conserved among the GSAM structures. The diminished minicore of the cofactor is completed by a network of water molecules providing contacts to intermediate and active-site residues Tyr-150 (W1) as well as intersubunit contacts in the second layer (WI1–WI3). (B and C) Cross-talk in the double PLP form of GSAM (Scheme 1, superposition of steps iii and iv) in two different orientations. The EA-5 state is shown in stick-and-ball mode, whereas DAVA-IA is highlighted in cyan. Interactions are marked in dotted lines. Beginning from residue Ala-154, the gating loop of the subunit containing DAVA-IA is disordered.
Significant structural changes accompanying different catalytic steps affect the interaction between cofactor and intermediate. Such changes are subsequently communicated to the interface helix and the gating loop (Fig. 3A). These three elements define a communication triad, which acts as a molecular switch element determining the catalytic state of the allosteric pair. The cofactor is connected across the interface with the gating loop forming a network of packing interactions. In effect, reorientation of DAVA (Scheme 1, step iv) causes a change in the electrostatic balance by which the catalytic intermediate is fixed within the active-site pocket (Fig. 3 B and C). During the propeller-like rotation, the 5′-amino group of DAVA is moved into hydrogen-bonding distance with a catalytic water molecule (W1), which mediates the interaction with Tyr-150 and the phosphate group of the cofactor. The two long interface helices oriented in parallel, which can be considered as transition elements between cofactors and gating-loop regions, transmit this signal via rigid-body displacement (in average 0.67 Å in Cα-positions between the KE-4/DAVA-IA and EA-5/DAVA-IA state). Local bending of the helices (rms deviation of >1.0 Å) causes changes in key contact points between cofactor, interface helix, and gating loop (Fig. 3 B and C). As a consequence, His-153 undergoes a dramatic spatial reorientation and becomes oriented inside the hinge region of the gating loop by losing both fix points, namely Arg-132* and water-mediated Glu-125*, respectively. Induced by a domino effect, both invariable arginines, Arg-132* and Arg-135, change their spatial orientation, which leads to a disruption of the salt bridge Arg-132*–Asp-155 and a reorientation of Arg-135.
Discussion
Structural and Functional Complexity of the Active-Site Gating Loop.
By performing a combination of single-crystal microspectrophotometry and x-ray crystallography, our results show that binding of the cofactor by GSAM causes adjustments of the active-site gating loop. In contrast to Hennig et al. (4), the present work shows two separate gating-loop conformations in GSAM as a function of the chemical modification of the cofactor. Therefore, the cross-talk between cofactor binding and loop segment serves as a prerequisite for substrate channeling and subsequent catalysis. A similar function of flexible-loop regions has been shown for several other enzymes, including tyrosyl-tRNA synthetase, triosephosphate isomerase, and DOPA-decarboxylase (21–23). In all cases, several amino acids within the loop center are highly conserved and essential for catalytic activity by placing them in close proximity to the active-site pocket (24). Structural investigations of the human branched-chain α-keto acid dehydrogenases (hBCKD) suggest the concerted regulatory mechanism through thiamin diphosphate binding to be a general principle that might also apply to other cofactor-dependent enzymes (25).
The gating loop undergoes a dramatic conformational change during the opening and closing shift (Fig. 1). As shown in the double PMP form (Fig. 2A), a DAVA molecule is fixed between Ser-163, Asn-375, and the backbone helix (residues 190–202), suggesting a substrate channeling upon loop closing. This finding is supported by the proposed model of interaction between the V-shaped glutamyl-tRNA reductase (GluTR) and GSAM (26). GluTR reduces activated glutamine to glutamate-1-semialdehyde, which is known as an extremely reactive α-amino aldehyde with a strong tendency to polymerize. In the suggested ternary complex, the large void of GluTR is occupied by GSAM, allowing efficient penetration of the transient aldehyde GSA. The importance of Ser-163 for the catalytic mechanism has been revealed by site-directed mutagenesis (27). The observed specific activity together with a diminished affinity to the enamine mechanism-based inhibitor (4-amino-5-fluoropentanoic acid), however, is suggestive of a more complex function of this single gating-loop residue (see above). According to the induced fit theory (28), the open form of GSAM binds the substrate, whereas the catalysis can only proceed in the closed form, before the enzyme reopens to release the product. The contribution of the loop to the reaction was analyzed by a deletion mutant (Δ159–172) of GSAM (29). Kinetic studies revealed that removal of the gating loop not only increases the dissociation constant for DAVA (100-fold), but also lowers the catalytic efficiency by a factor 30 (kcat/KM = 2.1 mM−1·s−1).
Intersubunit Communication.
In the Koshland–Nemethy–Folmer model of negative cooperativity, the protein in the absence of ligand is symmetric, with binding sites of equal affinity (30). This is confirmed by the double PMP form of GSAM, revealing an enzyme with both gating-loop regions in the open conformation and symmetrical connections in key point interactions mediated by Arg-132. As ligands bind successively, the protein loses its symmetry and the relative affinities for subsequent ligands are changed. Closing of one subunit causes the simultaneous opening of the other, so that GSAM exists in two complementary conformations and switches between open and closed forms. Evidence to support such a cooperative catalytic mechanism in GSAM and the hypothesis of a cross-talk between protomers comes from (i) its biphasic kinetic behavior and (ii) the fact that unless preparations of GSAM are deliberately converted into either the double PMP or the double PLP form, the enzyme in solution invariably contains both forms of the cofactor (2). The asymmetric activity cycles suggested by this behavior allow for kinetically and structurally separated reactions. This model is underlined by the kinetic behavior of the enzyme with both subunits in the PLP form revealing a significantly decreased GSA turnover (31). GSAM resists conversion into the double PLP form during the normal activity, because this would render the enzyme essentially inactive.
The design of several point mutations within the communication triad was guided by comparisons of individual mean square displacements of the nonhydrogen atoms derived from Debye–Waller factors of individual GSAM x-ray structures. Besides the strictly conserved active-site Lys-273 (forming the Schiff base linkage with vitamin B6), His-153 (gating loop), together with Glu-125, Arg-132, and Arg-153 (interface helix), was the subject of site-directed mutagenesis (see Table 2, which is published as supporting information on the PNAS web site). Only the K273A mutant led to a complete loss of enzymatic activity, which can be explained by the essential role of the lysine side chain during catalysis (see also Scheme 1). The central role of the asymmetric crossover interaction between Glu-125–His-153–Arg-132 in relaying from one subunit to the other the information about active-site occupancy and gating-loop conformation suggests that signaling can be interrupted by single-point mutations. His-153, which can assume two side chain conformations, is located at the switch point of the communication triad (Fig. 3 B and C). The finding that a H153D mutant showed only 2% enzymatic activity can be explained by the limited spatial flexibility of the interface helix. Modeling of the aspartate side chain into GSAM suggested the formation of a salt bridge with Arg-132 impairing the piston-like helix movement. Therefore, cross-talk in GSAM might be driven by a change in the protonation state of this imidazole, which after protonation is repulsed by Arg-132. These results support our proposed model, in which the Glu-125–His-153–Arg-132 triad at the subunit interface couples the rigid-body displacement of the interface helix.
The truncated α-β motif creating the diminished binding cup of the 5′-phosphate moiety of the cofactors in GSAM is composed of the N-terminal anchoring part of the interface helix (residues 122–125) and a short adjacent β-strand (301*–306*) (Fig. 3A). Remarkably, a comparison of the cofactor in different GSAM structures reveals that the phosphate group of the cofactor is rotated significantly during catalysis (Figs. 2B and 3 B and C). The most extraordinary feature is, however, the unusual intersubunit fixation by the phosphate group of the cofactor (Fig. 3A) (32, 33). The negative charge of the phosphate moiety is thereby largely balanced by means of first and second layer interactions making use of the positive pole of both interface helical dipoles and a complex intersubunit ionic network. A possible contribution of energy for the allosteric communication may result from repulsive electrostatic interactions between both negatively charged phosphate groups lying across the positive field of both interface helix dipole moments. This finding is supported by cofactor fluorescence quenching of GSAM after titration experiments with 10–50 mM K+ or Na+, indicative for a charge-shielding effect (data not shown). In addition, singular value decomposition revealed that reduction of the cofactor aldimine with NaBH3CN in one subunit lowers the pK of the aldimine in the other subunit (4).
Conclusion and Perspective
The data in our study provide a molecular basis for the reaction trajectory and intersubunit communication between the allosteric pair in GSAM. It remains elusive, however, why the enzyme performs this extensive, energy-consuming motion. The most attractive explanation for this is the formation of a ternary complex with the V-shaped GluTR, to perform substrate channeling between two subsequent enzymes in the tetrapyrrol biosynthesis. To answer this question, cocrystallization of the GluTR–GSAM complex would be an ideal approach.
Because no counterpart of GSAM exists in animals, and the pathway of ALA synthesis is ubiquitous, the enzyme is a promising target for highly selective herbicides. The crystal structures presented here could assist in the structure-based design of such compounds.
Materials and Methods
Protein Preparation and Crystallization.
Synechococcus GSAM was expressed in Escherichia coli and purified as described in ref. 34. All preparation steps and crystallization setups were performed at 4°C. For all modifications of GSAM, solution spectra were recorded on a Uvikon 860 double-beam spectrophotometer. Before spectral measurements, the noninteracting cofactor was removed from the enzyme by buffer exchange with ultrafiltration cells.
Preparation 1.
The apo form of GSAM was obtained by long-term dialysis of the enzyme against phosphate buffer (pH 6.8) over 2 weeks. The lapse of cofactor release was monitored spectroscopically until only a peak at 280 nm was retained.
Preparation 2.
Double PMP form was obtained by mixing the apoenzyme (200–250 μM) with the amine form of the cofactor in molar excess. To avoid unwanted shift to the PLP form, the double PMP form of the enzyme was incubated with a molar excess of DAVA (1–5 mM) (2).
Preparation 3.
The double PLP form was obtained by mixing the apoenzyme (170–200 μM) with the aldimine form of the cofactor in equimolar amounts. Provided that the enzyme remained in the double PLP form, titration experiments with different concentrations of DAVA solutions (10–100 μM) were performed. This preparation yielded catalytic intermediates EA-5 (Scheme 1, step iii) and DAVA-IA (Scheme 1, step iv).
Preparation 4.
The PMP/PLP form of GSAM was obtained by reconstitution of the apoenzyme with both forms of the cofactor. To obtain an equimolar distribution of both peaks for PMP (330 nm) and PLP (420 nm), the enzyme was titrated with DAVA. This preparation yielded catalytic intermediates KE-4 (Scheme 1, step vi) and DAVA-IA (Scheme 1, step iv), respectively.
Crystallization experiments were performed by using the vapor diffusion technique, mixing 4 μl of protein solution (180–200 μM protein concentration) with 21.5% (wt/vol) PEG 20000 and 150 mM magnesium acetate in 100 mM Na-cacodylate buffer (pH 7.2). Microcrystals appear after 2–3 h and were used in a stepwise protocol as macroseeds to obtain full-size crystals within 12 h. The crystals belong to space group P212121 and contain one dimer per asymmetric unit.
Polarized Single-Crystal Absorption Microspectrophotometry.
GSAM crystals were stored at 25% (wt/vol) PEG 20000 and 100 mM magnesium acetate in 100 mM Na-cacodylate buffer (pH 7.2). Single-crystal absorption spectra were collected in the range of 250–500 nm with a single-beam Zeiss microspectrophotometer. Anisotropic absorbances with the electric vector oriented along the three orthogonal crystal axes (Aa, Ab, and Ac) were measured in a crystal with plane-polarized light by using different crystal faces. The isotropic crystal absorption spectra were approximated by using the equation Aiso = 0.3 × (Aa + Ab + Ac) (35). Identical GSAM crystals used for x-ray diffraction experiments were analyzed spectroscopically in advance. To trap certain intermediate steps, crystals were soaked with DAVA, analyzed spectroscopically, and immediately flash-frozen in liquid nitrogen without the need of additional cryoprotectant solutions. Double PLP-GSAM crystals were also soaked with 10 mM DAVA in the stabilizing solution to analyze, spectroscopically and structurally, gating-loop shifts in the crystals.
Structure Determination and Refinement.
The data sets were collected at beamlines BW7B (Deutsches Elektronen-Synchrotron, Hamburg, Germany) and PX-I (Swiss Light Source, Villigen, Switzerland). The diffraction images were processed by using the MOSFLM program suite (36), and the structure factors were scaled and reduced by using SCALA from the CCP4 package (37). Statistics of the merged data are given in Table 1. Analysis of the self-rotation and native Patterson function suggested significant deviations from local twofold symmetry (data not shown). A 180° self-rotation map, calculated in different resolution ranges with the strongest 35% of the reflections, shows a resolution-independent disturbance of 222 symmetry. Additionally, the difference in relative peak height grows even with increasing resolution, thus allowing exclusion of low-resolution phenomena.
The 2.2-Å-resolution crystal structure of the apoenzyme of GSAM was determined with Patterson search methods by using the AMORE program of the CCP4 package (37). A model of previously published GSAM (PDB entry 2GSA) subdivided in the N-terminal segment (residues 7–69), the cofactor-binding domain (residues 70–326), and the C-terminal region (residues 327–433) was used as a search model (4). The gating-loop regions (residues 150–183), together with the cofactors of both active sites, were excluded from the search model. All subsequent structures (double PMP and double PLP form as well as the PMP/PLP form of GSAM) were determined by using apo-GSAM as a search model to avoid model bias within the active sites and both gating-loop regions in subunits A and B, respectively. Individual structures were refined with CNS (38) and alternated with manual electron density interpretation by using MAIN (39). For additional details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Site-Directed Mutagenesis and Activity Assay.
The plasmid pSAT 1.4, which contains an EcoRI fragment of Synechococcus (PCC 6301), was used as a template to introduce mutations based on oligonucleotide primers by PCR. The inserts were cloned into the expression vector, and the sequences were confirmed. Synechococcus GSAM was expressed in E. coli and purified as described in ref. 34. All of the GSAM-mutant proteins were obtained in yields comparable with that of the wild-type protein (34 mg/liter culture medium), indicating that the mutations did not interfere with proper folding. Protein fractions from each purification step were analyzed for purity and proper folding by SDS/PAGE, analytical ultracentrifugation, CD spectroscopy, and limited tryptic digestion (40). Wild-type enzyme and all mutant forms were assayed for GSAM activity by measuring the rate of ALA synthesis (41).
Supplementary Material
Acknowledgments
We thank Roberto Contestabile, Rob John, and Johan Jansonius for helpful discussions and support during the project; Ariel Lustig for performing analytical ultracentrifugation of several GSAM modifications and mutants; Sascha Popow (Deutsches Elektronen-Synchrotron) and Clemens Schulze-Briese (Swiss Light Source) for assistance during data collection; and George Orriss, Victoria Robinson, and Suat Oezbek for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation.
Abbreviations
- DAVA
4,5-diaminovalerate
- GluTR
glutamyl-tRNA reductase
- GSA
glutamate-1-semialdehyde
- GSAM
glutamate-1-semialdehyde aminomutase
- PLP
pyridoxal 5′-phosphate
- PMP
pyridoxamine 5′-phosphate.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2HOY (apo-GSAM), 2HOZ (double PMP form), 2HP1 (double PLP form), and 2HP2 (PMP/PLP-GSAM)].
References
- 1.Jordan PM, Shemin D. J Biol Chem. 1973;248:1019–1024. [PubMed] [Google Scholar]
- 2.Pugh CE, Harwood JL, John RA. J Biol Chem. 1992;267:1584–1588. [PubMed] [Google Scholar]
- 3.Smith MA, Kannangara CG, Grimm B. Biochemistry. 1992;31:11249–11254. doi: 10.1021/bi00160a041. [DOI] [PubMed] [Google Scholar]
- 4.Hennig M, Grimm B, Contestabile R, John RA, Jansonius JN. Proc Natl Acad Sci USA. 1997;94:4866–4871. doi: 10.1073/pnas.94.10.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraut J. Science. 1988;242:533–540. doi: 10.1126/science.3051385. [DOI] [PubMed] [Google Scholar]
- 6.Pauling L. Nature. 1948;161:707–709. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 7.Knowles JR. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- 8.Berendsen HJ, Hayward S. Curr Opin Struct Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 9.Jardetzky O. Prog Biophys Mol Biol. 1996;65:171–219. doi: 10.1016/s0079-6107(96)00010-7. [DOI] [PubMed] [Google Scholar]
- 10.Monod J, Wyman J, Changeux JP. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 11.Frauenfelder H, Parak F, Young RD. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 12.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 13.Volkman BF, Lipson D, Wemmer DE, Kern D. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 14.Stevens SY, Sanker S, Kent C, Zuiderweg ER. Nat Struct Biol. 2001;8:947–952. doi: 10.1038/nsb1101-947. [DOI] [PubMed] [Google Scholar]
- 15.Alexander FW, Sandmeier E, Mehta PK, Christen P. Eur J Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- 16.Sanner MF, Olson AJ, Spehner JC. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Bruice TC. Acc Chem Res. 2002;35:139–148. doi: 10.1021/ar0001665. [DOI] [PubMed] [Google Scholar]
- 18.Hol WG. Prog Biophys Mol Biol. 1985;45:149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 19.Lockless SW, Ranganathan R. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 20.Suel GM, Lockless SW, Wall MA, Ranganathan R. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 21.First EA, Fersht AR. Biochemistry. 1993;32:13658–13663. doi: 10.1021/bi00212a034. [DOI] [PubMed] [Google Scholar]
- 22.Joseph D, Petsko GA, Karplus M. Science. 1990;249:1425–1428. doi: 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- 23.Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Nat Struct Biol. 2001;8:963–967. doi: 10.1038/nsb1101-963. [DOI] [PubMed] [Google Scholar]
- 24.Fetrow JS. FASEB J. 1995;9:708–717. [PubMed] [Google Scholar]
- 25.Li J, Wynn RM, Machius M, Chuang JL, Karthikeyan S, Tomchick DR, Chuang DT. J Biol Chem. 2004;279:32968–32978. doi: 10.1074/jbc.M403611200. [DOI] [PubMed] [Google Scholar]
- 26.Moser J, Schubert WD, Beier V, Bringemeier I, Jahn D, Heinz DW. EMBO J. 2001;20:6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop K, Gough K, Mahoney S, Smith A, Rogers L. FEBS Lett. 1999;450:57–60. doi: 10.1016/s0014-5793(99)00465-2. [DOI] [PubMed] [Google Scholar]
- 28.Koshland DE., Jr Science. 1963;142:1533–1541. doi: 10.1126/science.142.3599.1533. [DOI] [PubMed] [Google Scholar]
- 29.Contestabile R, Angelaccio S, Maytum R, Bossa F, John RA. J Biol Chem. 2000;275:3879–3886. doi: 10.1074/jbc.275.6.3879. [DOI] [PubMed] [Google Scholar]
- 30.Koshland DE, Jr, Nemethy G, Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 31.Tyacke RJ, Harwood JL, John RA. Biochem J. 1993;293:697–701. doi: 10.1042/bj2930697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denessiouk KA, Denesyuk AI, Lehtonen JV, Korpela T, Johnson MS. Proteins. 1999;35:250–261. doi: 10.1002/(sici)1097-0134(19990501)35:2<250::aid-prot10>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 33.Denesyuk AI, Denessiouk KA, Korpela T, Johnson MS. Biochim Biophys Acta. 2003;1647:234–238. doi: 10.1016/s1570-9639(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 34.Grimm B, Smith AJ, Kannangara CG, Smith M. J Biol Chem. 1991;266:12495–12501. [PubMed] [Google Scholar]
- 35.Hofrichter J, Eaton WA. Annu Rev Biophys Bioeng. 1976;5:511–560. doi: 10.1146/annurev.bb.05.060176.002455. [DOI] [PubMed] [Google Scholar]
- 36.Leslie AGW. MOSFLM Users Guide. Cambridge, UK: Medical Research Council Laboratory of Molecular Biology; 1994. [Google Scholar]
- 37.Collaborative Computing Project Number 4 Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Turk DC. PhD thesis. Munich, Germany: Technische Universität München; 1992. [Google Scholar]
- 40.Fox JW, Mayer U, Nischt R, Aumailley M, Reinhardt D, Wiedemann H, Mann K, Timpl R, Krieg T, Engel J, et al. EMBO J. 1991;10:3137–3146. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauzerall D, Granick S. J Biol Chem. 1956;219:435–446. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.