Abstract
Mammalian CLC proteins function as Cl− channels or as electrogenic Cl−/H+ exchangers and are present in the plasma membrane and intracellular vesicles. We now show that the ClC-6 protein is almost exclusively expressed in neurons of the central and peripheral nervous systems, with a particularly high expression in dorsal root ganglia. ClC-6 colocalized with markers for late endosomes in neuronal cell bodies. The disruption of ClC-6 in mice reduced their pain sensitivity and caused moderate behavioral abnormalities. Neuronal tissues showed autofluorescence at initial axon segments. At these sites, electron microscopy revealed electron-dense storage material that caused a pathological enlargement of proximal axons. These deposits were positive for several lysosomal proteins and other marker proteins typical for neuronal ceroid lipofuscinosis (NCL), a lysosomal storage disease. However, the lysosomal pH of Clcn6−/− neurons appeared normal. CLCN6 is a candidate gene for mild forms of human NCL. Analysis of 75 NCL patients identified ClC-6 amino acid exchanges in two patients but failed to prove a causative role of CLCN6 in that disease.
Keywords: acidification, anion transport, Batten disease, channelopathy, Kufs' disease
The mammalian genome encodes nine CLC proteins. The first branch of this gene family encodes plasma membrane Cl− channels. Their importance is evident from human diseases like myotonia congenita with mutations in ClC-1 and certain forms of renal salt loss with mutations in ClC-Kb or its β-subunit barttin (1).
Proteins encoded by the two other branches (ClC-3, -4, and -5 and the separate ClC-6/-7 branch) are found on intracellular vesicles. It is thought that the conductance of vesicular CLC proteins supports the acidification of endosomal-lysosomal compartments by compensating currents of vesicular H+ ATPases (1, 2), a mechanism that would work with either Cl− channels or electrogenic Cl−/H+ antiport. In fact, ClC-4 and -5 function not as Cl− channels but as electrogenic Cl−/H+ exchangers (3, 4). An impaired acidification of endosomes or synaptic vesicles was found in mice lacking ClC-3 (5, 6) or -5 (7, 8). Probably as a consequence of defective endosomal acidification, the loss of ClC-5 leads to a stark reduction in proximal tubular endocytosis (7, 9) and to human Dent's disease (10).
It is unclear whether the two members of the third branch, ClC-6 and -7 (11), function as Cl− channels or Cl−/H+ antiporters. ClC-7 resides on late endosomes and lysosomes (12, 13) and can be coinserted with the H+-ATPase into the ruffled border of osteoclasts (12). ClC-7 mutations cause osteopetrosis, because osteoclasts become unable to acidify the resorption lacuna (12). The lack of ClC-7 also entails a severe lysosomal storage disease (13). Because ClC-7 requires Ostm1 as a β-subunit for proper function, the loss of Ostm1 causes osteopetrosis and lysosomal storage disease as well (14).
ClC-6 is the least-understood mammalian CLC protein. Its mRNA is found in many tissues, including brain and kidney (11). Upon heterologous expression, the ClC-6 protein was reported to colocalize with markers either of the endoplasmic reticulum (15) or of endosomes (16). We now show that native ClC-6 resides in late endosomes and have explored its physiological role by generating and analyzing ClC-6 knockout (KO) mice. These mice display a progressive neuropathy of the central and peripheral nervous systems with features of neuronal ceroid lipofuscinosis (NCL), a subtype of human lysosomal storage disease. Their neurological phenotype is surprisingly mild and comprises an impairment of pain sensation.
Results
Disruption of the Clcn6 Gene in Mice.
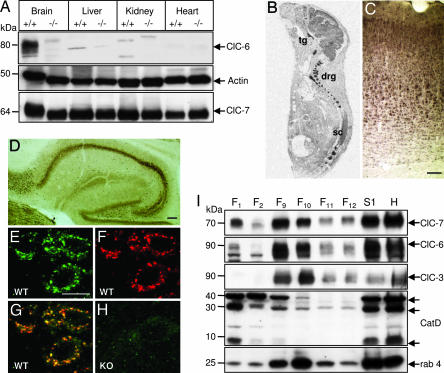
We disrupted the Clcn6 gene by inserting the lacZ gene in-frame into exon 7 (see Fig. 6, which is published as supporting information on the PNAS web site). This predicts a protein in which ClC-6 is fused to β-galactosidase after intramembrane helix D and that lacks the functionally important helices E–R. Western blot analysis showed that ClC-6 was absent from all KO tissues examined (Fig. 1A) and confirmed the specificity of the antiserum. Previous analysis indicated a broad transcription of ClC-6 (11, 17), but in the present study, the ClC-6 protein was nearly exclusively detected in the nervous system. The disruption of ClC-6 did not change the abundance of its closest homolog, ClC-7 (Fig. 1A). Clcn6−/− mice were viable and fertile, had no increased lethality, and were superficially indistinguishable from their WT littermates over their entire life span.
Fig. 1.
Tissue distribution and subcellular localization of ClC-6. (A) Western blot analysis of membrane proteins from various tissues. KO tissues lack the ≈85 kDa ClC-6 protein detected by antibody 6N2. Actin and ClC-7 served as controls. (B) In situ hybridization of a P0 mouse for ClC-6 mRNA intensely labeled DRG (drg), trigeminal ganglion (tg), spinal cord (sc), brain, and eye. These signals were absent from Clcn6−/− mice (data not shown). (C and D) Immunostaining of mouse brain for ClC-6 in the diaminobenzidine technique using the 6N2 antibody. Neurons are stained in every neuronal layer of the cortex (C) and in all regions of the hippocampus (D). (E–H) Immunolocalization of ClC-6 in cortical neurons. Most ClC-6-positive structures (E) costained for late endosomal/lysosomal lamp-1 (F), yielding yellow in the overlay (G). 6N2 staining of Clcn6−/− cortex (H) controlled antibody specificity. (I) Subcellular fractionation of mouse brain on a 17% Percoll gradient analyzed by Western blot of selected fractions Fn, postnuclear supernatant S1, and crude homogenate H. Ten microliters of each fraction was loaded. Cathepsin D (CatD) and rab4 are markers for lysosomes and endosomes, respectively. [Scale bars, 100 μm (C and D); 20 μm (E–H).]
Tissue Distribution and Subcellular Localization of ClC-6.
The ClC-6 transcript is broadly expressed during embryogenesis (11). We investigated its expression at postnatal day 0 (P0) by in situ hybridization of whole mouse sections. The nervous system, including brain, trigeminal and dorsal root ganglia (DRG), spinal cord, and eye, was intensely labeled (Fig. 1B). Immunohistochemistry of adult mouse brain revealed a broad expression of the ClC-6 protein, e.g., in the cortex (Fig. 1C) and the hippocampus (Fig. 2D). The staining was almost exclusively observed in neurons.
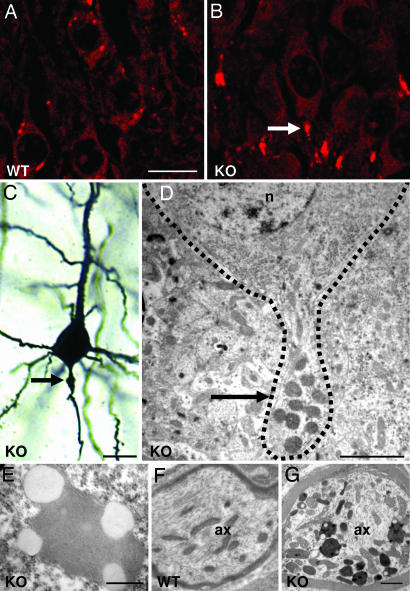
Fig. 2.
Characterization of storage material and axonal swelling in Clcn6−/− mice. (A and B) Autofluorescence in the hippocampus of 3-month-old WT (A) and Clcn6−/− (B) littermates. Note the localization in initial axon segments in the KO (arrow). (C) Golgi staining of a Clcn6−/− cortical neuron reveals proximal axonal swelling (a meganeurite; arrow) that was seen in ≈20% of cortical neurons; age, 6 months. (D) Electron microscopy reveals a similar swelling of a hippocampal axon because of material accumulation (arrow). The neuronal plasma membrane is surrounded by a dashed line; n, nucleus. (E) Avacuolar, lipofuscin-like lipopigment that was nearly always associated with lipid droplets in a KO cortical neuron. (F and G) Electron microscopy of DRG axons (ax) from WT (F) and KO (G) P90 littermates reveals storage material in the KO. [Scale bars, 20 μm (A–C); 2 μm (D); 400 nm (E); and 1 μm (F and G).]
As seen in the immunofluorescence of the cortex (Fig. 1 E–H), DRG, and thalamus (see Fig. 7, which is published as supporting information on the PNAS web site), ClC-6 was present in punctate structures in neuronal somata. These were largely costained for the late endosomal/lysosomal marker lamp-1 (Figs. 1 F and G and 7) and were absent from Clcn6−/− tissue (Fig. 1H). The subcellular distribution of ClC-6 was independently analyzed by fractionating brain membranes using Percoll centrifugation. As indicated by the abundance of cathepsin D (Fig. 1I), lysosomes were enriched in fraction 1. Fractions 9 and 10 contained much less cathepsin D but large amounts of the endosomal marker rab4. ClC-6 was present in endosomal fractions 9 and 10, which also contained ClC-3 and -7. Only ClC-7 was abundant in lysosomal fraction 1, which contained only trace amounts of ClC-6. We conclude that ClC-6 is predominantly expressed in late endosomes.
Axonal Storage in the Central and Peripheral Nervous Systems of Clcn6−/− Mice Is Characteristic of NCL.
Clcn6−/− mice displayed autofluorescence in virtually all brain regions, as shown in Fig. 2B for the hippocampus, which was exclusively found within neurons. Not yet present in 2-week-old mice, it became visible at 4 weeks and was strong at >3 months (for cortex, see Fig. 8 A–D, which is published as supporting information on the PNAS web site). Both the material per cell and the number of affected neurons increased with age. Whereas the weaker autofluorescence of WT brain had the somatic distribution of normal age-dependent accumulation of lipofuscin (Figs. 2A, hippocampus, and 8E, cortex), autofluorescence in Clcn6−/− brain occurred in the proximal axon (Fig. 2B). No autofluorescent material was seen in liver, heart, or kidney. Autofluorescent material (lipofuscin) within neurons is a hallmark of NCL, a subtype of lysosomal storage disease (18).
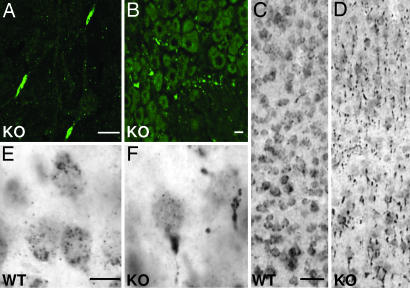
Golgi staining revealed frequent enlargements of the proximal axon of cortical neurons (Fig. 2C). Electron microscopy identified lipopigment deposits as the likely cause of these axonal swellings (Fig. 2D). Similar “meganeurites” were described in various lysosomal storage diseases, including NCL (19, 20). Higher magnification of deposits showed a mixed composition, with lipid droplets associated with amorphous or granular material (granular osmiophilic deposits; GRODs) (Fig. 2E). Unlike WT controls (Fig. 2F), Clcn6−/− DRG axons had abundant storage material (Fig. 2G) that included some curvilinear storage bodies and fingerprints (see Fig. 9 A and B, which is published as supporting information on the PNAS web site). These three types of storage material are also found in NCL patients. A typical feature of NCL is the intraneuronal accumulation of saposin D and/or the subunit c (subc) of the mitochondrial ATP synthase. Axon initial segments of Clcn6−/− neurons were highly immunoreactive for both saposin D (Fig. 3A) and subc (Fig. 3B). In electron microscopy, GRODs of KO proximal axons stained for subc and the lysosomal proteins lamp-1, cathepsin D, and lysosomal acid phosphatase, as well as for the closest homolog of ClC-6, the late endosomal/lysosomal ClC-7 (Fig. 9 C–H).
Fig. 3.
Localization of lysosomal markers in Clcn6−/− neurons. (A and B) Immunostaining for saposin D in the cortex (A) and for subc of ATP synthase (subc) in a DRG of Clcn6−/− mice (Scale bars, 20 μm.) (C–F) Change of cathepsin D immunolocalization in P90 cortical neurons from a cell body staining in WT (C and E) to a strongly localized staining of initial axon segment in the KO (D and F). E and F are higher magnifications. [Scale bars, 50 μm (C and D) and 20 μm (E and F).]
Despite these pathological changes in neuronal morphology, there were obvious signs neither of neuronal death or cell loss nor of degeneration of the retina (data not shown). Peripheral nerves (ischiadicus and femoralis) appeared normal, even in KO mice older than 8 months.
Redistribution of Late-Endosomal/Lysosomal Markers and Lysosomal pH.
Whereas lysosomal cathepsin D was distributed in puncta over cell bodies of WT cortical neurons (Fig. 3 C and E), it was strongly reduced in Clcn6−/− somata, instead being concentrated in proximal axons (Fig. 3 D and F). Similar changes were observed for lamp-1 (data not shown). The lysosomal marker cathepsin D and ClC-7 were similarly mislocalized in Clcn6−/− DRGs (data not shown). Thus, the lack of ClC-6 led to an accumulation of lysosomal proteins, including ClC-7, in initial axon segments.
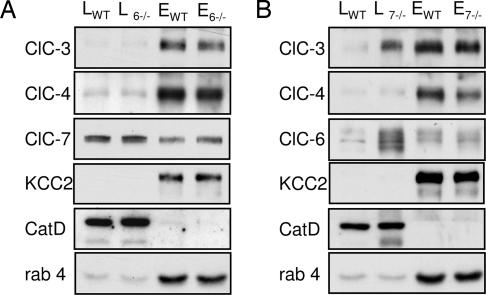
Because Clcn7−/− mice also display a lysosomal storage disease (13), we asked whether the disruption of either ClC-6 or -7 changed the localization of vesicular CLCs expressed in brain along the endosomal/lysosomal pathway. Western analysis of membrane fractions obtained by Percoll centrifugation from WT and Clcn6−/− brain revealed no changed distribution of ClC-7, -3, and -4 or of several controls (Fig. 4A). In contrast, ClC-3 and -6, but not -4, were significantly increased in lysosome-enriched fractions from Clcn7−/− brain (Fig. 4B).
Fig. 4.
Effect of ClC-6 and -7 disruption on subcellular localization of CLC proteins. Western blot analysis of membrane fractions (20 μg of protein per lane) obtained from WT, Clcn6−/− (A), and Clcn7−/− (B) brain separated on Percoll gradients. L, pooled “lysosomal” fractions (F1 + F2), and E, pooled “endosomal” fractions (F9–F12; see Fig. 1I). Subscripts indicate genotypes (WT, Clcn6−/−, and Clcn7−/− littermates). Membranes were probed with antibodies against ClC-3, -4, -6, -7, KCC2, cathepsin D (Cat D), and rab4. Cat D and rab4 identify lysosomes and endosomes, respectively, but endosomal fractions may also contain plasma membranes (as revealed by KCC2). Similarly changed localization of ClC-3 and -6 in Clcn7−/− mice was observed in more than five independent experiments.
Because other vesicular CLC proteins facilitate vesicular acidification (5, 6, 8), we determined the lysosomal pH of cultured hippocampal neurons by ratiometric fluorescence imaging (see Fig. 10, which is published as supporting information on the PNAS web site). There was no significant difference between WT and Clcn6−/− neurons [pH 4.67 ± 0.06 and 4.58 ± 0.03, respectively (SEM, n > 20)].
Gene Expression Profiling in Hippocampus.
Changes in hippocampal gene expression were evaluated by microarray analysis using RNA from P14 mice that lack overt morphological changes. Several up-regulated genes could be classified as being involved in inflammatory processes or microglia activation or as genes of the extracellular matrix (see Table 1, which is published as supporting information on the PNAS web site). Although neither microglial activation nor astrogliosis was detected by immunocytochemistry in older KO mice, these results suggest a minor inflammatory response with microglial activation in KO brains even before morphological changes set in.
Impaired Nociception and Mild Behavioral Abnormalities in Clcn6−/− Mice.
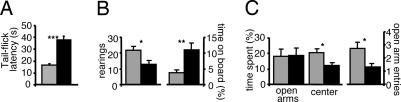
Because DRGs prominently express ClC-6 and in the KO accumulate impressive amounts of storage material, 3- to 5-month-old mice were tested for pain sensation in the tail-flick assay. Tail-flick latencies of Clcn6−/− mice were doubled compared with WT, indicating a defect in nociception (Fig. 5A). The modified hole-board test revealed reduced vertical exploratory activity, as indicated by the number of rearings (Fig. 5B). The time spent in the center of the test arena was strongly increased, possibly suggesting a decrease in anxiety. However, specific analysis of anxiety-related behavior in the elevated plus-maze test, as reflected in the time spent in the open arms, failed to reveal differences between the genotypes (Fig. 5C). The time spent in the center, thought to be related to decision making as well as the number of open arm entries, was reduced in KO mice. The number of closed-arm entries, regarded as reflecting locomotor activity, was almost identical in both genotypes (data not shown). Motor skills as tested by rotarod experiments were unaffected (not shown). Clcn6−/− mice also showed an increased acoustic startle response (Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 5.
Behavioral analysis of Clcn6−/− mice and WT littermates. (A) Latencies from heat application to tail flicking of 3- to 5-month-old female mice (n = 14 per genotype). (B) Modified hole-board test. Rearing activity and percentage of time spent on the board of 5- to 10-month-old male and female mice (n = 14 or 15 per genotype). (C) Elevated plus-maze test. Percentage of time spent in open arms, center, and number of open-arm entries of 3- to 16-month-old male and female mice (n = 24 per genotype). Gray, WT; black, Clcn6−/− mice. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
Mutational Analysis of Patients with Late-Onset NCL.
The NCL-like neuropathy of Clcn6−/− mice suggested that CLCN6 might be mutated in a subtype of human NCL. Because the neurological deficits of ClC-6 KO mice are rather mild and do not include blindness, we focused on late-onset forms of NCL and Kufs' disease, an adult-onset variant. In a sample of 75 patients, we identified two heterozygous missense mutations. In patient A (from Poland), a mutation in exon 17 changed Val-580 to a Met in helix R. In patient B (described in ref. 21), a mutation in exon 18 changed Thr-628 to Arg in the first cytoplasmic cystathionine β-synthase (CBS) domain (Fig. 12, which is published as supporting information on the PNAS web site). Neither mutation was found in 200 control chromosomes. However, no second mutation was found in either patient, and none of the respective parents were known to have had NCL.
Discussion
We have shown here that ClC-6, the least-understood mammalian CLC Cl− transport protein, is predominantly expressed in the nervous system, where it resides in late endosomes. Its disruption led to a lysosomal storage disease that differed markedly from that observed upon disruption of ClC-7, its closest homolog. Although the life span of ClC-6 KO mice was not compromised, these mice showed reduced pain sensitivity and mild behavioral abnormalities.
Localization and Cellular Function of ClC-6.
Previous studies showed a broad expression of the ClC-6 mRNA (11, 17), but the ClC-6 protein is predominantly expressed in the nervous system. The expression of ClC-6 agrees well with the exclusively neuronal phenotype observed upon its disruption. Whereas mice lacking the broadly expressed ClC-7 accumulated storage material in renal proximal tubules in addition to neurons (13), this was not observed in Clcn6−/− mice (data not shown).
Our work revealed that ClC-6 resides in endosomes, agreeing with a recent report (16) but contrasting with a previous one (15). Thus, ClC-3, -4, -5, -6, and -7 are all present in the endosomal/lysosomal system. ClC-5 is expressed in early (and likely also recycling) endosomes and may reach the plasma membrane (2, 7). The same may be true for ClC-4 (16). Unlike ClC-4 and -5, ClC-7 is prominently expressed in lysosomes and late endosomes (12, 13). ClC-3 is thought to reside in late endosomes (5). In immunofluorescence, ClC-6 colocalized largely but not completely with lamp-1 and was found in endosomal, but not lysosomal, subcellular fractions. This result suggests that ClC-6 resides in late endosomes as well. The presence of ClC-3 and -6 in late endosomes is further supported by their changed localization in Clcn7−/− brain (Fig. 4B), where a significant proportion of both these CLCs, but not of ClC-4, had “leaked” into lysosomes. The lack of ClC-7 might impair the lysosomal degradation of ClC-3 and -6 that, unlike the “earlier” ClC-4, more easily reach lysosomes from late endosomes.
These partial shifts of ClC-3 and -6 to lysosomal fractions might be interpreted as a compensatory mechanism. However, preliminary data show that ClC-6 and -7 double KOs resemble Clcn7−/− mice. The transcription of other CLC genes was not changed in Clcn3−/− (5), Clcn7−/− (13), or Clcn6−/− mice (this work). Because the disruption of either ClC-3 (5) or -6 led to neuropathy, these CLCs cannot functionally replace each other. This might be related to their additional expression in other compartments [ClC-3 is also found on synaptic vesicles (5)] or to so-far-unknown functional differences. Vesicular CLC proteins may facilitate luminal acidification by shunting proton pump currents (1), as experimentally confirmed for ClC-5 (8) and -3 (5, 6). Whereas the acidification of the osteoclast resorption lacuna was impaired in the ClC-7 KO (12), the lysosomal pH of Clcn7−/− neurons was normal despite severe lysosomal storage disease (13). Likewise, lysosomal pH was unchanged in mice lacking the ClC-7 β-subunit Ostm1 (14). It was similarly unchanged in Clcn6−/− neurons. We cannot rule out, however, a changed pH of late endosomes, the main site of ClC-6 expression.
Whereas ClC-0, -1, -2, and -K are Cl− channels, the bacterial ClC-ec1 (22) and mammalian ClC-4 and -5 proteins (3, 4) operate as electrogenic Cl−/H+ exchangers. It is unknown whether ClC-6 functions as an exchanger, because its absence from the cell surface precluded biophysical studies. If so, the direct coupling of Cl− to H+ transport might change vesicular Cl− concentrations in the KO even in the presence of normal luminal pH. Our knowledge of possible roles for vesicular Cl− is sparse. For instance, the lysosomal enzyme cathepsin C is strongly activated by Cl− (23). We conclude that the lysosomal pathology observed in Clcn6−/− mice most likely occurs because of a change in the H+ or Cl− concentration, or both, in a late endosomal/prelysosomal compartment.
Neuronal Phenotype of Clcn6−/− Mice Resembles Mild Forms of Human NCL.
The neurological deficits of Clcn6−/− mice were mild and did not reduce their life span under laboratory conditions. These deficits consisted primarily of an impaired pain perception and moderate behavioral abnormalities that may reflect an unspecific cognitive disorder. The apparent absence of neuronal cell loss suggests these abnormalities were caused by a functional impairment of neurons that had accumulated storage material. As with other neurodegenerative disorders, it is unclear how intracellular deposits might compromise cellular functions. The conspicuous “storage” in initial axon segments, which was associated with axonal “ballooning” and meganeurites, is compatible with a hindrance of trafficking into or from the axon (24).
The deposits had a mixed composition and contained various lysosomal proteins. Surprisingly, these proteins had almost disappeared from neuronal cell bodies, accumulating instead in initial axon segments. Of note, the treatment of brain slices with inhibitors of lysosomal cathepsins entailed a similar redistribution of lysosomes and axonal enlargements (24). The intraneuronal deposits were autofluorescent and contained saposin D and subc of the mitochondrial ATP synthase. Both findings, as well as the morphology of storage material, are typical for human lysosomal storage diseases named NCL or sometimes Batten disease (25–27). NCL is a genetically and phenotypically heterogeneous group of progressive neurodegenerative disorders with at least eight subtypes (28). Clinical features variably include progressive mental and visual decline, motor disturbances, epilepsy, and premature death (18). As in Clcn6−/− mice, genes involved in inflammatory response and microglial activation were up-regulated in NCL (29). Enlargements of initial axon segments like those found here were described in human NCL (19, 30). Hence, the phenotype of ClC-6 KO mice can be confidently classified as NCL.
CLCN6 as Candidate Gene for Human NCL.
The present KO phenotype suggested CLCN6 as a candidate gene for human NCL, in particular because genes underlying several forms of this disease remain unknown. There is considerable clinical variability in NCL, with late-onset forms being generally less severe. NCL is mostly associated with visual impairment or blindness, but vision appears normal in the adult-form Kufs' disease. As with Clcn6−/− mice, patients with Kufs' disease accumulate storage material in enlarged initial axon segments (19, 30). For these reasons, and because gene(s) underlying this disorder remain to be identified, we focused our mutational analysis on Kufs' disease and other late-onset forms of NCL.
CLCN6 mutations were found in only 2 of 75 patients with late-onset NCL. The V580M mutation changed a residue in helix R, which lines the Cl− permeation path to the cytoplasmic opening (31). The Val side chain is likely to protrude into the ion pathway. Val is not totally conserved at this position, however, because it is sometimes replaced by isoleucine (Fig. 12B). The other mutation (T628R) introduced a positive charge. This Thr is located in the first cytoplasmic CBS domain and is conserved in all mammalian CLCs (Fig. 12C). CBS domains may bind nucleotides (32) and modulate the gating of some CLC Cl− channels (33). Disease-causing mutations in CBS domains are not unprecedented. Several missense mutations in CBS1 and CBS2 of ClC-7 were found in osteopetrosis, mostly associated with the dominant form of the disease (12, 34).
Although neither mutation was found in 200 control chromosomes, we cannot be certain that they are disease-causing. The lack of ClC-6 plasma membrane expression precluded functional tests. Moreover, in the absence of known NCL in one of the parents, we expected a recessive mode of inheritance, but no CLCN6 mutations were found on the other allele of either patient. We may have missed mutations in the promoter region, or the mutants may exert dominant negative effects with incomplete penetrance. Dominant forms of adult-onset NCL are known (35). CLCN6 remains a candidate gene for subtypes of human NCL, but our analysis failed to prove a causative role of CLCN6 in NCL and suggests that mutations in this gene would be rare.
Phenotypic Comparison to Mice Lacking ClC-3 or -7.
The disruption of two other vesicular CLCs, ClC-3 and -7, led to neuropathies that differ markedly from the present pathology. Loss of ClC-3 led to retinal degeneration and massive hippocampal cell loss (5). Although Yoshikawa et al. (36) reported NCL-like features in their Clcn3−/− mice, we found them to be negligible compared with Clcn7−/− (13) and Clcn6−/− mice (this study). ClC-7 KO mice display a severe NCL-like neurodegeneration in addition to osteopetrosis (13). Although ClC-6 and -7 are close homologs and are both present in late endosomes of brain, the neuropathologies ensuing from their disruption differ. Autofluorescence in Clcn7−/− mice was stronger and was mainly associated with microglia. In electron microscopy, Clcn7−/− osmiophilic material was stained more homogeneously and was not associated with lipid droplets. Somata of Clcn7−/− neurons were filled with storage material that did not penetrate neuronal processes, again in striking contrast to Clcn6−/− mice. Clcn7−/− and Clcn3−/−, but not Clcn6−/−, mice displayed a drastic neuronal cell loss. Further, in vitro activities of lysosomal enzymes that are typically increased in NCL (37) were strongly increased in Clcn7−/− but were almost unchanged in Clcn6−/− animals (see Fig. 13, which is published as supporting information on the PNAS web site). Whereas Clcn7−/− mice lived no more than 10 weeks, even when their osteopetrosis was rescued by transgenic ClC-7 expression (13), Clcn6−/− mice had a normal life span. These differences might be related to the pronounced lysosomal expression of ClC-7 or to distinct transport properties. It should also be mentioned that, unlike Clcn7−/− mice, Clcn6−/− mice lacked any signs of osteopetrosis.
This work has addressed the physiological role of ClC-6, leading to the discovery that ClC-6 is important for lysosomal function. Its disruption led to a lysosomal storage phenotype that is most likely related to changes in prelysosomal H+ or Cl− concentrations. Both ions influence endosomal and lysosomal function through various mechanisms (38). Because the present phenotype is strikingly different from the neuropathology observed with the lack of ClC-3 or -7, our results suggest that late endosomal/lysosomal CLC proteins have more complex roles in endosomal/lysosomal compartments than previously recognized.
Experimental Procedures
Generation of Clcn6−/− Mice.
The Clcn6 gene was disrupted by homologous recombination in ES cells and blastocyst injection as detailed in Supporting Text, which is published as supporting information on the PNAS web site.
In Situ Hybridization.
In situ hybridization with a 250-nt cRNA probe of the 3′ region of Clcn6 was performed as described in ref. 5.
Antibodies.
Polyclonal rabbit antibodies (6N2) were raised against a peptide (INDPYLEVLETMDNKC) from the ClC-6 N terminus and affinity-purified. Other antibodies were rabbit anti-ClC-7 (12), rabbit anti-ClC-3 (5), rabbit anti-KCC2 (39), rabbit anti-cathepsin D (Oncogene, Carpinteria, CA), rat anti-lamp-1 (BD PharMingen, San Diego, CA), rabbit anti-rab4 (Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-saposin D (gift of K. Sandhoff, Universität Bonn, Bonn, Germany), and rabbit anti-subc (gift of E. Kominami, Juntendo University, Tokyo, Japan). Secondary antibodies coupled to Alexa Fluor 488 or 546 (Molecular Probes, Carlsbad, CA) were used for immunofluorescence and coupled to HRP for Western blots. For detailed procedures, see Supporting Text.
Subcellular Fractionation on Percoll Gradients.
Brains were homogenized in 250 mM sucrose/3 mM imidazole (pH 7.4). Homogenates were centrifuged 10 min at 1,000 × g, and supernatants were subjected to centrifugation on 17% Percoll as described (14).
Neuronal Cultures.
Cultures of hippocampal neurons were prepared from P0 embryos, as described in detail in Supporting Text.
Morphology.
Autofluorescence was excited at 546 nm. Golgi staining was done with the FD Rapid GolgiStain Kit (FD NeuroTechnologies, Ellicott City, MD). For details of histology, immunocytochemistry, and electron microscopy, see Supporting Text.
Lysosomal pH Measurements.
Lysosomal pH was measured by using the dextran-coupled pH-sensitive ratiometric dye Oregon green 488 (Molecular Probes; refs. 13 and 14). Procedures for loading lysosomes and ratiometric imaging are given in Supporting Text.
Behavioral Analysis.
Clcn6−/− mice and WT littermate controls were 3–10 months old. For the tail-flick test, mice were loosely wrapped in an adsorbent towel, and their tails were placed on the tail-flick analgesia apparatus. Rotarod performance was assessed with an accelerating procedure. These assays, the modified hole-board test, and the elevated plus-maze experiments are described in detail in Supporting Text.
Expression Profiling.
RNA was extracted from hippocampi of P14 Clcn6−/− mice and control littermates, converted into double-stranded cDNA, labeled with biotin, and hybridized to Affymetrix (Santa Clara, CA) murine genome U74v2 microarrays, as detailed in Supporting Text.
NCL Patients and Mutational Analysis.
CLCN6 exons were amplified from human genomic DNA and analyzed by single-stranded conformation analysis and sequencing. Descriptions of this analysis and of the patients are found in Supporting Text.
Supplementary Material
Acknowledgments
We thank H. Maier (Universitätsklinik Eppendorf, Hamburg, Germany) for measuring auditory brainstem responses; N. Krönke, E. Orthey, and C. Kitzmüller for technical assistance; K. Sandhoff (University of Bonn, Bonn, Germany) for the anti-saposin D antibody; and E. Kominami (Juntendo University, Tokyo, Japan) for the anti-subc antibody; and many colleagues who provided samples from NCL patients, in particular A. Gal (Universitätsklinik Eppendorf), K. E. Wisniewski (New York State Institute for Basic Research in Developmental Disabilities, New York, NY), H. H. Goebel (Universität Mainz, Mainz, Germany), A. Janecke (Medizinische Universität Innsbruck, Innsbruck, Austria), B. Dermaut (University of Antwerpen, Antwerpen, Belgium), M. Burmeister (University of Michigan, Ann Arbor, MI), M. A. Farrell (Beaumont Hospital, Dublin, Ireland), and J. Zaremba (Institute of Psychiatry and Neurology, Warsaw, Poland), for sending DNA and cell lines from patient A. This work was supported in part by the Ernst-Jung-Preis für Medizin, the Nationale Genomforschungsnetz program of the Bundesministerium für Bildung und Forschung, the Eumorphia (European Union), the Wellcome Trust, and the Batten Disease Support and Research Association. M.P. and R.P.-C. were supported by Marie Curie Fellowships of the European Union.
Abbreviations
- KO
knockout
- NCL
neuronal ceroid lipofuscinosis
- Pn
postnatal day n
- DRG
dorsal root ganglia
- subc
subunit c
- CBS
cystathionine β-synthase.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The expression profiling results presented in this paper have been deposited in the ArrayExpress database (accession no. E-MEXP-731).
References
- 1.Jentsch TJ, Poët M, Fuhrmann JC, Zdebik AA. Annu Rev Physiol. 2005;67:779–807. doi: 10.1146/annurev.physiol.67.032003.153245. [DOI] [PubMed] [Google Scholar]
- 2.Günther W, Lüchow A, Cluzeaud F, Vandewalle A, Jentsch TJ. Proc Natl Acad Sci USA. 1998;95:8075–8080. doi: 10.1073/pnas.95.14.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheel O, Zdebik A, Lourdel S, Jentsch TJ. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 4.Picollo A, Pusch M. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- 5.Stobrawa SM, Breiderhoff T, Takamori S, Engel D, Schweizer M, Zdebik AA, Bösl MR, Ruether K, Jahn H, Draguhn A, et al. Neuron. 2001;29:185–196. doi: 10.1016/s0896-6273(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 6.Hara-Chikuma M, Yang B, Sonawane ND, Sasaki S, Uchida S, Verkman AS. J Biol Chem. 2005;280:1241–1247. doi: 10.1074/jbc.M407030200. [DOI] [PubMed] [Google Scholar]
- 7.Piwon N, Günther W, Schwake M, Bösl MR, Jentsch TJ. Nature. 2000;408:369–373. doi: 10.1038/35042597. [DOI] [PubMed] [Google Scholar]
- 8.Günther W, Piwon N, Jentsch TJ. Pflügers Arch. 2003;445:456–462. doi: 10.1007/s00424-002-0950-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang SS, Devuyst O, Courtoy PJ, Wang XT, Wang H, Wang Y, Thakker RV, Guggino S, Guggino WB. Hum Mol Genet. 2000;9:2937–2945. doi: 10.1093/hmg/9.20.2937. [DOI] [PubMed] [Google Scholar]
- 10.Lloyd SE, Pearce SH, Fisher SE, Steinmeyer K, Schwappach B, Scheinman SJ, Harding B, Bolino A, Devoto M, Goodyer P, et al. Nature. 1996;379:445–449. doi: 10.1038/379445a0. [DOI] [PubMed] [Google Scholar]
- 11.Brandt S, Jentsch TJ. FEBS Lett. 1995;377:15–20. doi: 10.1016/0014-5793(95)01298-2. [DOI] [PubMed] [Google Scholar]
- 12.Kornak U, Kasper D, Bösl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. Cell. 2001;104:205–215. doi: 10.1016/s0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 13.Kasper D, Planells-Cases R, Fuhrmann JC, Scheel O, Zeitz O, Ruether K, Schmitt A, Poët M, Steinfeld R, Schweizer M, et al. EMBO J. 2005;24:1079–1091. doi: 10.1038/sj.emboj.7600576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange PF, Wartosch L, Jentsch TJ, Fuhrmann JC. Nature. 2006;440:220–223. doi: 10.1038/nature04535. [DOI] [PubMed] [Google Scholar]
- 15.Buyse G, Trouet D, Voets T, Missiaen L, Droogmans G, Nilius B, Eggermont J. Biochem J. 1998;330:1015–1021. doi: 10.1042/bj3301015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T, Rai T, Hayama A, Sohara E, Suda S, Itoh T, Sasaki S, Uchida S. J Cell Physiol. 2006;206:792–798. doi: 10.1002/jcp.20516. [DOI] [PubMed] [Google Scholar]
- 17.Kida Y, Uchida S, Miyazaki H, Sasaki S, Marumo F. Histochem Cell Biol. 2001;115:189–194. doi: 10.1007/s004180000245. [DOI] [PubMed] [Google Scholar]
- 18.Goebel HH, Wisniewski KE. Brain Pathol. 2004;14:61–69. doi: 10.1111/j.1750-3639.2004.tb00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkovic SF, Carpenter S, Andermann F, Andermann E, Wolfe LS. Brain. 1988;111:27–62. doi: 10.1093/brain/111.1.27. [DOI] [PubMed] [Google Scholar]
- 20.Braak H, Braak E. Clin Neuropathol. 1987;6:116–119. [PubMed] [Google Scholar]
- 21.Goebel HH, Schochet SS, Jaynes M, Gutmann L. Acta Anat (Basel) 1998;162:127–132. doi: 10.1159/000046477. [DOI] [PubMed] [Google Scholar]
- 22.Accardi A, Miller C. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 23.Cigic B, Pain RH. Eur J Biochem. 1999;264:944–951. doi: 10.1046/j.1432-1327.1999.00697.x. [DOI] [PubMed] [Google Scholar]
- 24.Bednarski E, Ribak CE, Lynch G. J Neurosci. 1997;17:4006–4021. doi: 10.1523/JNEUROSCI.17-11-04006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer DN, Fearnley IM, Walker JE, Hall NA, Lake BD, Wolfe LS, Haltia M, Martinus RD, Jolly RD. Am J Med Genet. 1992;42:561–567. doi: 10.1002/ajmg.1320420428. [DOI] [PubMed] [Google Scholar]
- 26.Ezaki J, Kominami E. Brain Pathol. 2004;14:77–85. doi: 10.1111/j.1750-3639.2004.tb00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyynelä J, Palmer DN, Baumann M, Haltia M. FEBS Lett. 1993;330:8–12. doi: 10.1016/0014-5793(93)80908-d. [DOI] [PubMed] [Google Scholar]
- 28.Mole SE. Brain Pathol. 2004;14:70–76. doi: 10.1111/j.1750-3639.2004.tb00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jalanko A, Vesa J, Manninen T, von Schantz C, Minye H, Fabritius AL, Salonen T, Rapola J, Gentile M, Kopra O, Peltonen L. Neurobiol Dis. 2005;18:226–241. doi: 10.1016/j.nbd.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Goebel HH, Braak H, Seidel D, Doshi R, Marsden CD, Gullotta F. Clin Neuropathol. 1982;1:151–162. [PubMed] [Google Scholar]
- 31.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 32.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Estévez R, Pusch M, Ferrer-Costa C, Orozco M, Jentsch TJ. J Physiol (London) 2004;557:363–378. doi: 10.1113/jphysiol.2003.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frattini A, Pangrazio A, Susani L, Sobacchi C, Mirolo M, Abinun M, Andolina M, Flanagan A, Horwitz EM, Mihci E, et al. J Bone Miner Res. 2003;18:1740–1747. doi: 10.1359/jbmr.2003.18.10.1740. [DOI] [PubMed] [Google Scholar]
- 35.Nijssen PC, Ceuterick C, van Diggelen OP, Elleder M, Martin JJ, Teepen JL, Tyynelä J, Roos RA. Brain Pathol. 2003;13:574–581. doi: 10.1111/j.1750-3639.2003.tb00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshikawa M, Uchida S, Ezaki J, Rai T, Hayama A, Kobayashi K, Kida Y, Noda M, Koike M, Uchiyama Y, et al. Genes Cells. 2002;7:597–605. doi: 10.1046/j.1365-2443.2002.00539.x. [DOI] [PubMed] [Google Scholar]
- 37.Tyynelä J, Sohar I, Sleat DE, Gin RM, Donnelly RJ, Baumann M, Haltia M, Lobel P. EMBO J. 2000;19:2786–2792. doi: 10.1093/emboj/19.12.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faundez V, Hartzell HC. Sci STKE 2004. 2004;re8 doi: 10.1126/stke.2332004re8. [DOI] [PubMed] [Google Scholar]
- 39.Hübner CA, Stein V, Hermanns-Borgmeyer I, Meyer T, Ballanyi K, Jentsch TJ. Neuron. 2001;30:515–524. doi: 10.1016/s0896-6273(01)00297-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.