Abstract
Metal ion homeostasis is critical to the survival of all cells. Regulation of nickel concentrations in Escherichia coli is mediated by the NikR repressor via nickel-induced transcriptional repression of the nickel ABC-type transporter, NikABCDE. Here, we report two crystal structures of nickel-activated E. coli NikR, the isolated repressor at 2.1 Å resolution and in a complex with its operator DNA sequence from the nik promoter at 3.1 Å resolution. Along with the previously published structure of apo-NikR, these structures allow us to evaluate functional proposals for how metal ions activate NikR, delineate the drastic conformational changes required for operator recognition, and describe the formation of a second metal-binding site in the presence of DNA. They also provide a rare set of structural views of a ligand-responsive transcription factor in the unbound, ligand-induced, and DNA-bound states, establishing a model system for the study of ligand-mediated effects on transcription factor function.
Keywords: crystallography, DNA complex, nickel, transcription factor, metalloregulator
Metal ions are essential nutrients for all cells, but their intracellular concentrations and distribution must be tightly regulated to avoid toxicity. Nickel ions are particularly important to the physiology of microorganisms such as the model prokaryote Escherichia coli and the human gastric pathogen Helicobacter pylori. In these organisms, incorporation of nickel into enzymes such as [NiFe]-hydrogenase and urease is necessary for metabolic adaptation to changing environmental conditions (1). In E. coli, nickel is acquired via an ABC-type membrane transporter, NikABCDE (2). Transcription of the operon encoding this nickel importer, nikABCDE, is repressed in the presence of nickel by the transcription factor NikR (3, 4). NikR therefore serves as a cytoplasmic nickel sensor, stopping production of the nickel importer when intracellular levels of nickel are sufficient. In H. pylori, which requires the nickel enzyme urease to survive and colonize the acidic gastric mucus of the human stomach, NikR plays a more complex regulatory role. Transcriptome analysis of a deletion mutant and in vitro promoter-binding assays have revealed that H. pylori NikR (HpNikR) regulates transcription of not only the nickel importer and urease enzyme, but also other regulatory networks, some through repression of the gene encoding the ferric uptake regulator (Fur) protein (5–8).
The NikR protein that has been characterized most extensively at the molecular level is the homolog from E. coli. E. coli NikR (EcNikR or NikR) is a homotetramer that binds one nickel(II) ion per subunit with picomolar affinity (9, 10). Stoichiometric nickel ions activate NikR to bind a 28-bp palindromic operator sequence (GTATGA-N16-TCATAC) within the promoter of nikABCDE with low-nanomolar affinity (9, 11). Recently, it was demonstrated that NikR can bind a variety of divalent transition metal ions with high affinity and that stoichiometric amounts of these other ions can activate NikR to bind operator DNA with a potency comparable to that of nickel (10, 11). Excess nickel enhances the affinity of EcNikR for its operator by at least 250-fold, and evidence supports the existence of a second nickel-binding site in the presence of operator DNA with micromolar to nanomolar affinity for nickel (9, 11). The effect is most dramatic for excess Ni2+ compared with Cu2+, Mn2+, Zn2+, Cd+2, and Co2+ (11). Interestingly, the affinity of HpNikR for specific DNA is not significantly affected by the presence of excess nickel ions, and only one nickel-binding site (the high-affinity site) is implicated in the function of that protein (12).
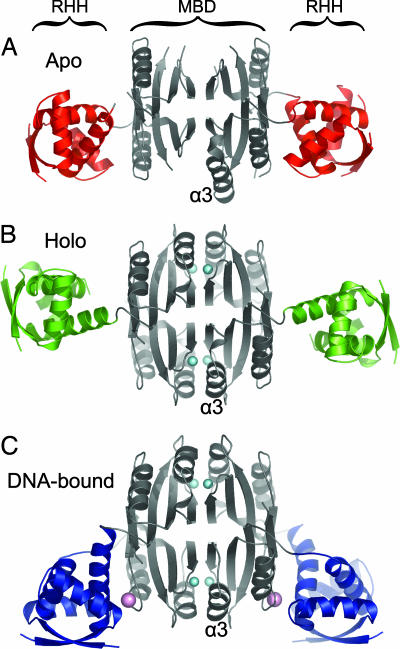
The previously reported crystal structure of apo-EcNikR revealed a modular domain organization, with two dimeric ribbon–helix–helix (RHH) domains flanking a tetrameric metal-binding domain (MBD) (Fig. 1A; ref. 13). This spatial separation of the two DNA-binding domains explained how NikR could recognize two operator subsites distantly separated by two turns of DNA. A crystal structure of the nickel-bound form of the isolated MBD showed that the high-affinity metal-binding site of NikR is located at the subunit interfaces, with the four nickel ions each bridging two subunits of the tetramer. Nickel ions bind with a square-planar coordination geometry, ligated by the protein side chains His-87, His-89, and Cys-95 from one NikR subunit, as well as the side chain of His-76′ from an adjacent subunit. In the absence of bound nickel ions, the α-helix contributing His-76′ (helix α3) to the metal-binding site is structurally flexible and was disordered in the apo-NikR crystal structure. The disordered-to-ordered transition of helix α3 within the MBD upon nickel binding gave preliminary insight into the structural changes that take place upon activation of NikR by metal ions. Recently, several structures of an uncharacterized NikR homolog from Pyrococcus horikoshii (PhNikR) were published that support these features of the EcNikR structure (14).
Fig. 1.
Conformational flexibility of NikR. (A) Apo-NikR tetramer displayed as a ribbon with the MBD colored gray and the RHH domains colored red. (B) Nickel-activated NikR tetramer displayed as in A, except the RHH domains are colored green and nickel ions are shown as cyan spheres. (C) Operator-bound NikR tetramer displayed as in A and B, except the RHH domains are colored blue and potassium ions are shown as pink spheres. Helix α3, which is stabilized upon high-affinity nickel binding, is labeled in each panel. This figure and other structural figures were made by using PyMOL (DeLano Scientific, San Carlos, CA).
We now present two additional crystal structures of full-length EcNikR activated by stoichiometric nickel ions, both alone and bound to a 30-bp oligonucleotide containing the operator sequence from the nik promoter. These structures allow us to evaluate the possible mechanisms of nickel-induced DNA binding, describe the conformational changes of NikR required for operator recognition, and investigate the formation of additional metal-binding sites in the presence of DNA.
Results
Full-Length Nickel-Activated NikR.
The crystal structure of full-length NikR with stoichiometric nickel reveals domains that are comparable with the two previously determined structures of NikR, with the MBD closely matching the structure of the isolated nickel-bound MBD tetramer (rmsd 0.89–0.90 Å for 304 Cα atoms), and the RHH DNA-binding domains similar to those of the apo-NikR structure (Fig. 1A) (rmsd 0.82–1.21 Å for 84 Cα atoms). The most significant difference in this structure is the relative orientation of the RHH domains with respect to the MBD (Fig. 1). This rigid-body domain movement occurs about flexible linkers connecting the domains, represented by residues 41–52 as described in ref. 13. As shown in Fig. 1B, the RHH domains extend almost completely outward from the MBD, pointing the β-sheets that bind to the DNA major groove in opposite directions. A detergent molecule from an additive crystallization screen (cyclohexyl-propyl-β-d-maltoside) is bound at the interface between the RHH domain and the MBD in this crystal structure (Fig. 6, which is published as supporting information on the PNAS web site). The detergent molecule is likely to stabilize rather than dictate the extended NikR conformation because crystals of Ni-NikR grown in the absence of detergent reveal a structure with a similar orientation and more disorder (E.R.S., unpublished data). Structures of PhNikR also show different relative orientations of the MBD and RHH domains when compared with each other or to the structures of EcNikR, further supporting the idea that NikR is flexible in the absence of DNA (14).
The NikR–Operator Complex.
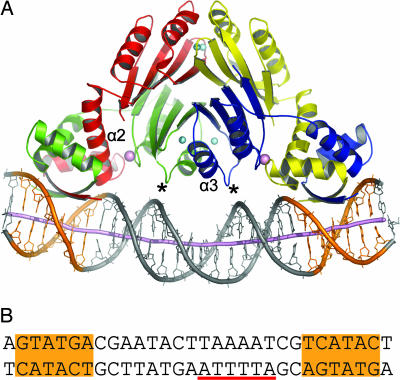
In the crystal structure of the nickel-activated NikR tetramer in complex with its operator DNA (Figs. 1C and 2A), the individual RHH and MBDs are again very similar to the previously determined structures of NikR. The RHH domains align well structurally with both apo-NikR (rmsd 1.16–1.35 Å for 84 Cα atoms) and nickel-bound NikR (rmsd 1.06–1.16 Å for 84 Cα atoms), whereas the MBD closely resembles the nickel-bound forms of both the isolated MBD (rmsd 0.95–1.02 Å for 304 Cα atoms) and full-length NikR (rmsd 0.81–0.84 Å for 304 Cα atoms). However, the orientations of the RHH DNA-binding domains of the repressor relative to the MBD are dramatically different compared with the structures of EcNikR alone (Fig. 1). Specifically, the RHH domains rotate about the flexible interdomain linkers to orient their antiparallel β-strands toward the same face of the repressor, allowing each to occupy the DNA major groove of an operator palindrome half-site (Fig. 2). A similar conformation was observed in one crystal form of nickel-bound PhNikR grown in the absence of DNA (PDB ID code 2BJ9), although in that structure the two RHH domains are oriented toward opposite faces of the MBD (trans conformation as opposed to cis) (14). Superposition of half of the NikR tetramer of these two structures (Fig. 7A, which is published as supporting information on the PNAS web site) gives a rmsd of 1.61 Å for 262 Cα atoms. To allow for the remarkable rotation to the DNA-bound conformation, the linker of one subunit of each RHH domain is extended by the unwinding of the final turn of helix α2 (Fig. 2). This conformation of NikR results in a new protein–protein interface between the RHH and MBD and creates a second metal-binding site at this interface, which is discussed in detail below. The DNA in the operator complex is B form, with no dramatic bends at the interface with the protein as observed in some other protein–DNA complexes (16, 17). The DNA curves smoothly around one face of the NikR tetramer, bending ≈22° over its length and contacting both RHH DNA-binding domains and two of the four MBD subunits (Fig. 2). Each individual RHH–DNA interaction, discussed in more detail below, closely resembles the binding mode of other RHH family members such as Arc (18), MetJ (19), and CopG (20).
Fig. 2.
The NikR–operator DNA complex. (A) NikR–DNA complex with the NikR tetramer colored by subunit, and DNA displayed as sticks with a cartoon tube tracing the phosphorus positions. The dyad-symmetric operator half-sites are colored orange, and the DNA helical axis as calculated in CURVES (15) is shown as a purple tube. Nickel and potassium ions are shown as cyan and pink spheres, respectively. Asterisks indicate the MBD loop that contacts DNA. Helix α2, which contributes the conserved secondary metal site ligands E30 and D34, is labeled on the red NikR molecule. Helix α3, which is structurally stabilized upon high-affinity nickel binding, is labeled on the blue NikR molecule. (B) The dsDNA used for NikR cocrystallization, which includes the wild-type NikR operator sequence. Dyad-symmetric operator half-sites are highlighted in orange, and the -10 region of the nik promoter is underscored with red.
The high-affinity nickel-binding site within the MBD of NikR has the same coordination sphere in the NikR–DNA complex as in the structures of nickel-activated EcNikR and PhNikR alone (Fig. 8, which is published as supporting information on the PNAS web site), with His-76′, His-87, His-89, and Cys-95 bound to the nickel in a square-planar geometry. This result is in contrast to x-ray absorption spectroscopy (XAS) that indicated a coordination change from square planar to a six-coordinate site with loss of thiolate ligation upon specific-DNA binding (21). The discrepancy may result from the more static environment of the crystal relative to the solution phase XAS, although it is not clear from the available EcNikR or PhNikR crystal structures how the high-affinity nickel site could switch from four- to six-coordinate (13, 14). Hydrophobic portions of the molecule block water access to the upper and lower coordination sites of the nickel. In addition, there are no polar side chains nearby for which subtle rearrangement could yield a six-coordinate nickel site, suggesting that a more substantial rearrangement at the tetramer interface than observed in the crystal structures would have to occur.
NikR–Operator Interactions.
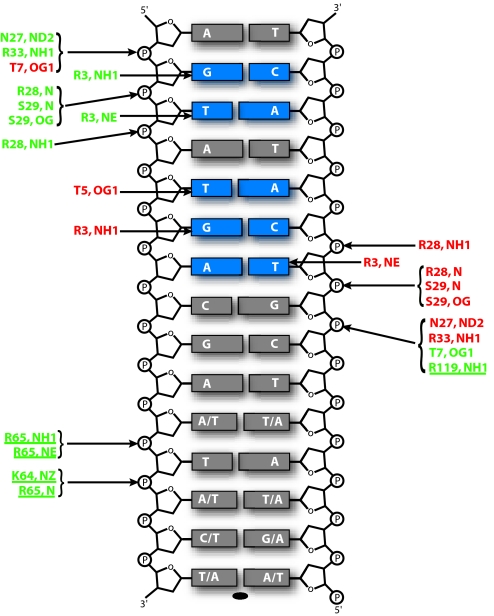
An extensive set of polar interactions is made between NikR and its operator (Figs. 3 and 4). These interactions can be described in three sets for clarity: specific interactions from the RHH domain, nonspecific interactions from the RHH domain, and nonspecific interactions from the MBD. First, specific hydrogen bonding interactions are made by the side chains of Arg-3 and Thr-5 from the β-sheets of the RHH domains to nucleotide bases in the operator major groove, contacting the 5 bp of each operator half-site that abrogate NikR binding when mutated (ref. 4; Figs. 3 and 4). This observation confirms that the crystal structure represents a specific NikR–operator complex. Second, there are ≈14 nonspecific polar interactions between each RHH domain and the phosphate backbone of DNA (Fig. 3). These interactions include hydrogen bonds between two consecutive backbone amide nitrogens at the N terminus of helix α2 and DNA phosphates on either side of the major groove. Analogous positive helix dipole–negative phosphate backbone interactions have been described previously for other RHH family members and serve to anchor the domain to DNA and properly orient the specificity-determinant β-sheets within the major groove (18–20). Other nonspecific interactions with the DNA phosphate backbone are made by the conserved side chains of Thr-7, Asn-27, Arg-28, Ser-29, and Arg-33 from the RHH domain. The final set of interactions, also nonspecific, is from the MBDs of two NikR subunits. The side chain of Arg-119, in a loop connecting the last α-helix and β-strand of NikR, extends between the phosphates of the DNA minor groove. Interactions also are made between a loop leading into helix α3 of the MBD and the center of the NikR operator, including the backbone amide nitrogen of Arg-65 and the side chains of Lys-64 and Arg-65, which make polar interactions with DNA backbone phosphates (asterisk in Figs. 2A and 4). This portion of NikR is disordered in the absence of bound nickel (Fig. 1A), implying that positioning of these residues upon metal binding is of functional importance.
Fig. 3.
NikR–operator DNA interactions. Schematic representation of polar interactions between NikR and operator DNA. Only half of the operator DNA is shown because the interactions made with the other half are symmetric and equivalent. Base pairs colored blue were shown to abrogate NikR binding when mutated (4). The protein atoms involved in these interactions are colored by protein subunit and correspond to the color scheme in Fig. 2A. Interactions contributed by the MBD are underlined.
Fig. 4.
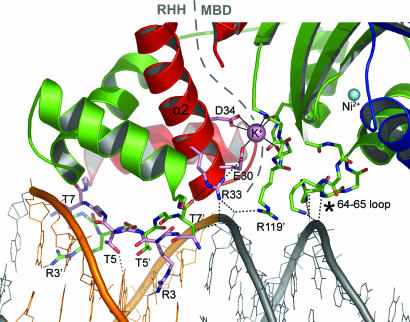
The second metal-binding site. Protein, DNA, and metal ions are displayed as in Fig. 2. A dashed gray line delineates the RHH domain and MBD. Bonds to the potassium ion at the domain interface are shown as solid black lines, and hydrogen bonds as dashed black lines. Portions of NikR contacting DNA or the secondary metal site are shown as sticks, and important sidechains are labeled. An asterisk indicates the MBD loop, containing Lys-64 and Arg-65, which contacts DNA.
A Second Metal-Binding Site.
In the presence of DNA, a second metal-binding site is observed at the interface generated between the RHH domain and the MBD of EcNikR (Figs. 1C, 2A, and 4). The metal in this second metal-binding site is coordinated by two strictly conserved amino acid side chains, Glu-30 and Asp-34, from the RHH DNA-binding domain, and by the three backbone carbonyl oxygens of Ile-116, Gln-118, and Val-121, from a loop of the MBD (Fig. 4). Coordination of the metal ion in this site by both the RHH domain and MBD should serve to lock these domains into the DNA-bound conformation. This metal-binding site is linked through conserved residues Glu-30 and Arg-33 to residues responsible for both nonspecific and specific DNA binding (Fig. 4). Glu-30, Arg-33, and Asp-34 originate from the second helix of the RHH domain, explaining the high degree of conservation along this helix among NikR sequences that is not seen for other RHH repressors.
This same secondary metal-binding site was observed in the structure of nickel-activated PhNikR determined from crystals soaked in 5 mM Ni2+ and 1 mM phosphate in the absence of DNA (Fig. 7 B and C) (14). Although under these conditions PhNikR has nickel in this metal-binding site, two pieces of evidence indicate that this second site is not occupied by nickel in our EcNikR-DNA structure: metal analysis and anomalous dispersion. Specifically, transition metal content assayed by using the indicator 4-(2-pyridylazo) resorcinol revealed only four Ni2+ ions per tetramer. Also, an anomalous difference Fourier map that was calculated by using data collected at the nickel peak wavelength (1.4845 Å) demonstrates that these four nickel ions are located in the high-affinity sites exclusively (Fig. 9 A and B, which is published as supporting information on the PNAS web site). Instead, the metal in this second site is best modeled as a potassium ion from the 200 mM KCl included in the crystallization buffer based on the size of the difference electron density peak, the refined bond lengths, and the refined B factors. Unfortunately potassium is not amenable to an anomalous dispersion experiment prohibiting confirmation via x-ray data.
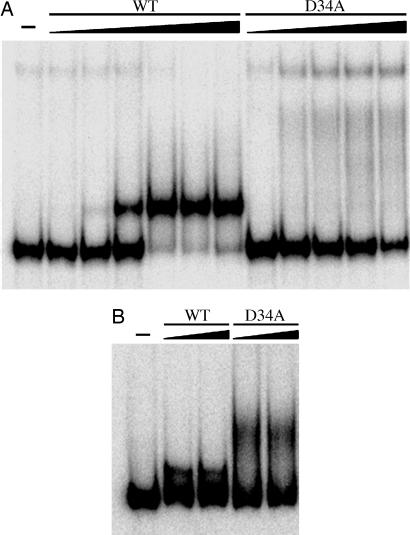
To examine whether this second metal-binding site is functionally relevant to the formation of the NikR–DNA complex, a D34A mutation was introduced into NikR and mobility-shift assays were performed. As previously reported (4, 11), DNA binding is observed in this assay only if there is excess metal present, so experiments were performed with NikR loaded with stoichiometric amounts of nickel and in the presence of excess Ni2+ or K+. For WT-NikR, both 35 μM Ni2+ and K+ result in a shift of the DNA (Fig. 5), which is caused by the binding of the protein tetramer to the DNA (11). Although the shift is cleaner for Ni2+, the results with K+ are impressive given that the concentration used is 1,000-fold less than what is found in the crystal or in an E. coli cell (22). These results indicate that potassium ions are sufficient to produce detectable DNA binding by holo-NikR. In contrast, neither metal affords tight DNA binding by D34A-NikR, and no shift indicative of a 4:1 NikR–DNA complex is observed (Fig. 5), demonstrating that Asp-34 is critical for the DNA-binding response of NikR. A lower mobility band is observed in mobility-shift assay experiments with D34A NikR (Fig. 5) and may be due to multiple NikR tetramers binding to the DNA with lower affinity. The difference in affinity of D34A NikR for the nik operator does not appear to be due to changes in secondary structure content (as reported by circular dicroism) or high-affinity nickel-binding properties for the mutant protein (S.C.W. and D.B.Z., data not shown). Mutation of the other ligand Glu-30 to Ala also disrupts DNA binding (ref. 14; S.C.W. and D.B.Z., preliminary data not shown).
Fig. 5.
DNA binding in the presence of excess metal. (A) WT NikR (1, 10, 100 pM, 1, 10, and 100 nM) or the D34A mutant (10 pM, 1, 10, 100 nM, and 1 μM) were preincubated with stoichiometric nickel and incubated subsequently with 100-bp nik DNA in the presence of 35 μM NiSO4. The reactions were analyzed on a 7% native gel with 35 μM NiSO4 in the gel and running buffer. (B) WT NikR (0.5 and 1 μM) and D34A NikR (0.5 and 1 μM) were preincubated with stoichiometric nickel and incubated subsequently with 100-bp nik DNA in the presence of 35 μM KCl. The reactions were analyzed on a 7% native gel with 35 μM KCl in the gel and running buffer. In all mobility-shift assay experiments, KCl (100 mM) was present in the binding buffer.
Discussion
The DNA-binding activity of many transcription factors is regulated by the binding of small molecule effectors. In very few cases, however, is the mechanism of activation understood at the molecular level. Our structures of apo-NikR, nickel-activated NikR and the NikR–DNA complex allow us to evaluate mechanistic hypotheses regarding the metal ion-activated repressor function of EcNikR. In the absence of nickel, NikR has no measurable affinity for an oligonucleotide containing its operator sequence (9, 11). Addition of stoichiometric nickel ions leads to a half-maximal protection of the nik operator from DNase at a concentration of 5–30 nM NikR (9, 11).
Numerous structures of ligand-activated transcription factors support the model that ligand binding directly alters the conformation of the DNA-binding domain, leading to an increase or decrease in affinity for DNA (23–26). For many prokaryotic transcription factors that are dimeric, this allosteric transition results in a change in the spacing of two DNA-binding domains, leaving them better suited to interact with multiple operator subsites along the DNA molecule (23–26). Our crystal structures of NikR in the unliganded (apo), nickel-bound (activated), and operator-bound forms provide insight into allosteric transitions upon ligand binding (Fig. 1) and do not support such a mechanism. The apo-NikR structure revealed a conformation that is not suitable for interaction with operator DNA (13). The nickel-activated NikR structure demonstrates that stoichiometric nickel binding alone does not preorganize a conformation capable of operator binding (Fig. 1B). Structures of apo and nickel-activated PhNikR are in agreement with our findings (14). Indeed, the RHH domains of NikR are connected to the MBD by flexible linkers (Figs. 1 and 2) that vary significantly in length and amino acid sequence as a function of species, making it unlikely that an allosteric signal could be propagated via this route.
Although binding of nickel to the high-affinity site of NikR does not directly alter the conformation of the RHH domains for DNA binding, it does have a very important short-range effect on the conformation of NikR. As described previously, residues 62–80 of NikR are disordered in the absence of nickel but form a loop and a stable α-helix (helix α3) when nickel is bound because of the direct involvement of His-76 in nickel coordination (13). These structural rearrangements are consistent with limited proteolysis experiments, in which nickel induced protection from digestion at Lys-64 and Arg-65 in solution (27). The NikR–DNA complex structure shows the importance of this conformational change in creating 6–8 polar interactions between the loop containing residues Lys-64 and Arg-65 and the phosphate backbone of operator DNA (Fig. 4). Because this loop and the following helix are disordered in the absence of nickel, binding of the nickel corepressor creates a surface of the MBD suitable for interacting with DNA by stabilization of these secondary structure elements. Complementary interaction of the nickel-stabilized MBD with DNA would localize NikR to the DNA helix and allow cooperative interaction of both RHH domains. Stable binding to the operator then occurs when the specific operator base sequence is recognized and additional specific base contacts can be made.
The secondary metal-binding site, formed at the interface between the RHH and MBD domains in the DNA-bound conformation of NikR and occupied by potassium in our structure, adds an additional level of complexity to the function of this transcription factor. Because the concentration of K+ in the NikR–DNA complex crystallization solution is similar to that within an E. coli cell (22), we expect that under physiological conditions this site would be occupied by K+ if no metal with a higher affinity for this site is available. Previous experiments to measure the affinity of nickel for the “low-affinity” nickel-binding site demonstrated that nanomolar concentrations of excess nickel are sufficient to increase the affinity of NikR for its operator, even in the presence of 100 mM potassium (11). Although it is possible that excess nickel ions displace potassium at the second metal-binding site, thereby increasing operator affinity, our Ni2+-soaking experiments do not support this idea. In our hands, soaking the NikR–DNA complex crystals with 5 mM Ni2+ does not result in nickel displacing potassium at this binding site. Instead, nickel is observed at several nonconserved sites on the surface of the protein (Fig. 9 C and D). It is conceivable that the conditions under which we soaked the NikR–DNA complex crystals somehow prevented exchange of nickel for potassium. Unfortunately, we were unable to crystallize the NikR–DNA complex in the presence of excess nickel ions. As discussed above, we know from crystallographic studies of PhNikR that Ni2+ can bind to this second metal site under certain soaking conditions (14). Regardless of whether the site is occupied by Ni2+ or K+, the ligands are the same (all oxygen atoms) (Figs. 4 and 7 B and C) and the bond distances are long (2.7–3.2 Å; ref. 14). Whereas this type of coordination is common for potassium binding sites in proteins (28), it is not for nickel (29).
Taken together, results from crystallography and mobility-shift assays suggest that this conserved second metal site is physiologically relevant, critical for the function of NikR, and well suited to bind a metal such as K+. Although the existence of this second site can explain the increase in affinity of NikR for DNA in the presence of K+ (Fig. 5B), it does not seem intuitive that replacing K+ with Ni2+, a metal less suited to this type of site, would further increase the affinity of NikR for DNA. Thus, we hypothesize that K+ normally occupies this site and that the effect of excess Ni2+ is electrostatic, increasing the affinity of the protein for DNA by increasing its positive charge via binding to nonconserved sites on the protein surface (Fig. 9 C and D). If this is the case, it could be expected that an equivalent response to excess nickel ions in other NikR homologs would not be observed. Indeed, recent characterization of HpNikR revealed that the presence of excess nickel ions does not substantially increase the affinity of that protein for the ureA promoter (12). Binding of nickel ions to the high-affinity sites of NikR and the subsequent increase in affinity for specific DNA therefore seems to be the primary conserved mechanism of function of the NikR family. Additional lower-affinity binding sites may serve to tune the response of NikR to excess metal ions in an organism-specific manner.
Metal-responsive transcription factors play key roles in intracellular physiology, and NikR is important for microorganisms that rely on nickel ions for specialized metabolic processes. The crystal structures described here suggest that NikR employs a different mechanism of ligand induction than has been hypothesized for most transcription factors. Instead of having a direct effect on the orientation of the RHH DNA-binding domains, the biggest role of high-affinity nickel binding seems to be localization of the repressor to the DNA helix via a short-range allosteric effect that creates a DNA-complementary surface on the MBD. The conserved secondary metal-binding site created between the RHH and MBD domains can be occupied by the intracellularly abundant metal ion K+ and can lock the otherwise flexible NikR in a conformation suitable for DNA binding. We hope future studies will reveal the organism-specific nuances of the response of NikR to metal ions.
Materials and Methods
Crystallography.
Nickel-bound selenomethionine-labeled NikR was produced and purified as described in ref. 13. Deoxyoligonucleotides containing the EcNikR operator sequence within the nik promoter were obtained from Integrated DNA Technologies (Coralville, IA) and purified by reverse-phase HPLC. The oligo used for cocrystallization with NikR is depicted in Fig. 2B. Formation of the NikR–DNA complex and crystallization of both the complex and Ni-NikR were carried out as described (Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Diffraction data were reduced and scaled in DENZO and SCALEPACK (30), respectively. The Ni-NikR structure was solved by molecular replacement by using the program EPMR (31) with a model of a dimer of the MBD (PDB ID code 1Q5Y) and a dimer of the RHH domain from the apo-NikR structure (ID code 1Q5V). The NikR–DNA complex was solved by a combination of molecular replacement in the program Phaser (32) with the models described above and selenium single-wavelength anomalous dispersion (Se-SAD) methods by using the program SOLVE (33). Density improvement for the NikR–DNA complex structure was carried out in DM (34) from the CCP4 suite (35). Iterative cycles of model building/rebuilding and refinement were carried out in Xfit (36) and CNS (37), respectively. The final Ni-NikR model has Rwork/Rfree of 23.9%/27.5%, whereas the NikR–DNA complex model has Rwork/Rfree of 26.1%/30.3%. Data collection and refinement statistics are given in Table 1, which is published as supporting information on the PNAS web site, and additional details regarding the structure determination and refinement are provided in the Supporting Materials and Methods.
NikR Mutants and Mobility-Shift Assays.
Mutants were constructed from the pNIK103 parent vector (4) by using the QuikChange PCR mutagenesis method (Stratagene, La Jolla, CA). The D34A mutation was prepared by using the primers 5′-GTTCCGAAGCTATCCGCGcCATTCTGCGTAGCGC-3′ and 5′-GCGCTACGCAGAATGgCGCGGATAGCTTCGGAAC-3′. The fidelity of the mutagenesis was confirmed by DNA sequencing (ACGT, Toronto, ON, Canada). Wild-type NikR and the D34A mutant were purified on a Ni(II)-NTA column (Qiagen, Valencia, CA), followed by anion exchange chromatography, as described in refs. 4 and 10. The molecular mass of the D34A mutant was confirmed by electrospray mass spectrometry (Mr-calc = 15,049.8 Da, Mr-obs = 15,049.2 Da). An assay with Ellman's reagent was used to show that both wild-type NikR and the D34A mutant were fully reduced, and an HPLC assay was used to confirm that both proteins were apo (38). Nickel-binding experiments were performed as described in ref. 11. Electrophoretic mobility-shift assays were performed with a 100-bp DNA fragment (≤0.2 nM) containing the nik promoter as described in ref. 11, the indicated concentrations of protein loaded with stoichiometric nickel, and a binding buffer containing 20 mM Tris (pH 7.5), 100 mM KCl, 3 mM MgCl2, 0.1% IGEPAL, 5% glycerol, and 0.1 mg/ml sonicated herring sperm DNA (Promega, Madison, WI). Excess nickel or potassium was added to the binding buffer, running buffer, and the gel before polymerization to a final concentration of 35 μM.
Supporting Information.
See additional details in Fig. 10 and Tables 2 and 3, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
This research is supported in part by National Institutes of Health Grant GM69857 (to C.L.D.), a Lester Wolfe Predoctoral Fellowship, and the Natural Sciences and Engineering Research Council (Canada). Advanced Light Source beamline 5.0.2 and National Synchrotron Light Source beamline X29A are funded by the U.S. Department of Energy.
Abbreviations
- RHH
ribbon–helix–helix
- MBD
metal-binding domain.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors for crystal structures of Ni-NikR and its complex with operator DNA have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2HZA and 2HZV, respectively).
References
- 1.Mulrooney SB, Hausinger RP. FEMS Microbiol Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 2.Navarro C, Wu LF, Mandrand-Berthelot MA. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 3.De Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. J Bacteriol. 1999;181:670–674. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chivers PT, Sauer RT. J Biol Chem. 2000;275:19735–19741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 5.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Mol Microbiol. 2003;49:947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 6.van Vliet AHM, Ernst FD, Kusters JG. Trends Microbiol. 2004;12:489–494. doi: 10.1016/j.tim.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, Kusters JG, van Vliet AH. Infect Immun. 2005;73:7252–7258. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V. J Bacteriol. 2005;187:7703–7715. doi: 10.1128/JB.187.22.7703-7715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chivers PT, Sauer RT. Chem Biol. 2002;9:1141–1148. doi: 10.1016/s1074-5521(02)00241-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang SC, Dias AV, Bloom SL, Zamble DB. Biochemistry. 2004;43:10018–10028. doi: 10.1021/bi049405c. [DOI] [PubMed] [Google Scholar]
- 11.Bloom SL, Zamble DB. Biochemistry. 2004;43:10029–10038. doi: 10.1021/bi049404k. [DOI] [PubMed] [Google Scholar]
- 12.Abraham LO, Li Y, Zamble DB. J Inorg Biochem. 2006;100:1005–1014. doi: 10.1016/j.jinorgbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, Drennan CL. Nat Struct Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 14.Chivers PT, Tahirov TH. J Mol Biol. 2005;348:597–607. doi: 10.1016/j.jmb.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 16.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 17.Schultz SC, Shields GC, Steitz TA. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 18.Raumann BE, Rould MA, Pabo CO, Sauer RT. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 19.Somers WS, Phillips SE. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 20.Gomis-Ruth FX, Sola M, Acebo P, Parraga A, Guasch A, Eritja R, Gonzalez A, Espinosa M, del Solar G, Coll M. EMBO J. 1998;17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrington PE, Chivers PT, Al-Mjeni F, Sauer RT, Maroney MJ. Nat Struct Biol. 2003;10:126–130. doi: 10.1038/nsb890. [DOI] [PubMed] [Google Scholar]
- 22.Epstein W, Schultz SG. J Gen Physiol. 1966;49:469–481. doi: 10.1085/jgp.49.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Aalten DM, DiRusso CC, Knudsen J. EMBO J. 2001;20:2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 25.Zhang RG, Joachimiak A, Lawson CL, Schevitz RW, Otwinowski Z, Sigler PB. Nature. 1987;327:591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 27.Dias AV, Zamble DB. J Biol Inorg Chem. 2005;10:605–612. doi: 10.1007/s00775-005-0008-2. [DOI] [PubMed] [Google Scholar]
- 28.Harding MM. Acta Crystallogr D. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 29.Rulisek L, Vondrasek J. J Inorg Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Kissinger CR, Gehlhaar DK, Fogel DB. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 32.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger TC, Berendzen J. Acta Crystallogr D. 1999;55:1174–1178. doi: 10.1107/S0907444999003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowtan KD, Main P. Acta Crystallogr D. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project No 4 Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 36.McRee DE. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Atanassova A, Lam R, Zamble DB. Anal Biochem. 2004;335:103–111. doi: 10.1016/j.ab.2004.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.