Abstract
Zip14 is a member of the SLC39A zinc transporter family, which is involved in zinc uptake by cells. Up-regulation of Zip14 by IL-6 appears to contribute to the hepatic zinc accumulation and hypozincemia of inflammation. At least three members of the SLC39A family transport other trace elements, such as iron and manganese, in addition to zinc. We analyzed the capability of Zip14 to mediate non-transferrin-bound iron (NTBI) uptake by overexpressing mouse Zip14 in HEK 293H cells and Sf9 insect cells. Zip14 was found to localize to the plasma membrane, and its overexpression increased the uptake of both 65Zn and 59Fe. Addition of bathophenanthroline sulfonate, a cell-impermeant ferrous iron chelator, inhibited Zip14-mediated iron uptake from ferric citrate, suggesting that iron is taken up by HEK cells as Fe2+. Iron uptake by HEK and Sf9 cells expressing Zip14 was inhibited by zinc. Suppression of endogenous Zip14 expression by using Zip14 siRNA reduced the uptake of both iron and zinc by AML12 mouse hepatocytes. Zip14 siRNA treatment also decreased metallothionein mRNA levels, suggesting that compensatory mechanisms were not sufficient to restore intracellular zinc. Collectively, these results indicate that Zip14 can mediate the uptake of zinc and NTBI into cells and that it may play a role in zinc and iron metabolism in hepatocytes, where this transporter is abundantly expressed. Because NTBI is commonly found in plasma of patients with hemochromatosis and transfusional iron overload, Zip14-mediated NTBI uptake may contribute to the hepatic iron loading that characterizes these diseases.
Keywords: hemochromatosis, iron transport, liver, zinc transport, inflammation
Infection and inflammation produce changes in zinc and iron metabolism that result in decreased plasma concentrations of both micronutrients (1, 2). Several lines of evidence indicate that the pathophysiologic stimuli responsible for these measurable effects exert their action by altering the expression of iron and zinc transporters. Because the liver plays a central role in iron and zinc metabolism, transporters expressed in this organ would be likely candidates responsible for physiologic changes that have systemic effects. We recently screened transcript abundance of 14 putative zinc transporter genes in murine liver after turpentine-induced inflammation or LPS injection, and we found that Zip14 was the most responsive to these stimuli (2). Transient transfection studies revealed that Zip14 localizes to the plasma membrane and induces cellular zinc accumulation. Furthermore, Zip14 abundance was increased at the sinusoidal plasma membrane of hepatocytes when stimulated with IL-6, suggesting that this transporter contributes to the hypozincemia of inflammation.
Zip proteins have been shown to import zinc into the cytosol from the plasma membrane or intracellular organelles (3, 4). The Zip superfamily name stands for Zrt- and Irt-like proteins. Zrt1 and Zrt2 (zinc-regulated transporters) were identified as zinc transporters of Saccharomyces cerevisiae (5, 6), and Irt1 (iron-regulated transporter) was originally identified in roots of iron-deficient Arabidopsis thaliana (7). Although Irt1 has been shown to restore zinc uptake in zrt1 zrt2 yeast mutants (8), it was originally identified as an iron transporter that seems to be involved in iron uptake from the soil (7). Since the discovery of these Zip transporters, >25 Zip family members have been identified, and, like Irt1, several of them have been shown to transport iron as well as other cations. For example, Arabidopsis Irt2 transports iron and zinc (9), and ZupT from Escherichia coli transports iron, zinc, cobalt, and possibly manganese (10). To date, the predominant substrates of mammalian Zip proteins are largely unknown (3, 4). This background prompted us to investigate a role for Zip14 in iron transport.
Because iron is essential but can be toxic, homeostatic mechanisms tightly control the intestinal absorption, systemic transport, cellular uptake, storage, and cellular efflux of this metal. These regulatory mechanisms respond to iron status as well as to the needs of erythropoiesis (11). Under normal circumstances, iron in plasma is bound to its transport protein transferrin (Tf), which safely sequesters the metal in a noncatalytic form. However, under conditions of iron overload, the iron-binding capacity of plasma Tf can be exceeded, resulting in the appearance of non-Tf-bound iron (NTBI). Plasma NTBI concentrations of humans with hereditary hemochromatosis or β-thalassemia usually range from 0.4 to 20 μM (12, 13). Animal studies demonstrate that NTBI is rapidly cleared from the plasma and taken up mostly by the liver (14). Although it is generally believed that NTBI is a major contributor to tissue iron loading in iron-overload states, the precise molecular mechanisms involved remain poorly defined. In the present work, we demonstrate that Zip14 can mediate the uptake of iron into cells and that suppression of its endogenous expression reduces the uptake of NTBI by hepatocytes.
Results
Overexpression of Mouse Zip14 (mZip14) Increases Cellular Accumulation of Iron and Zinc.
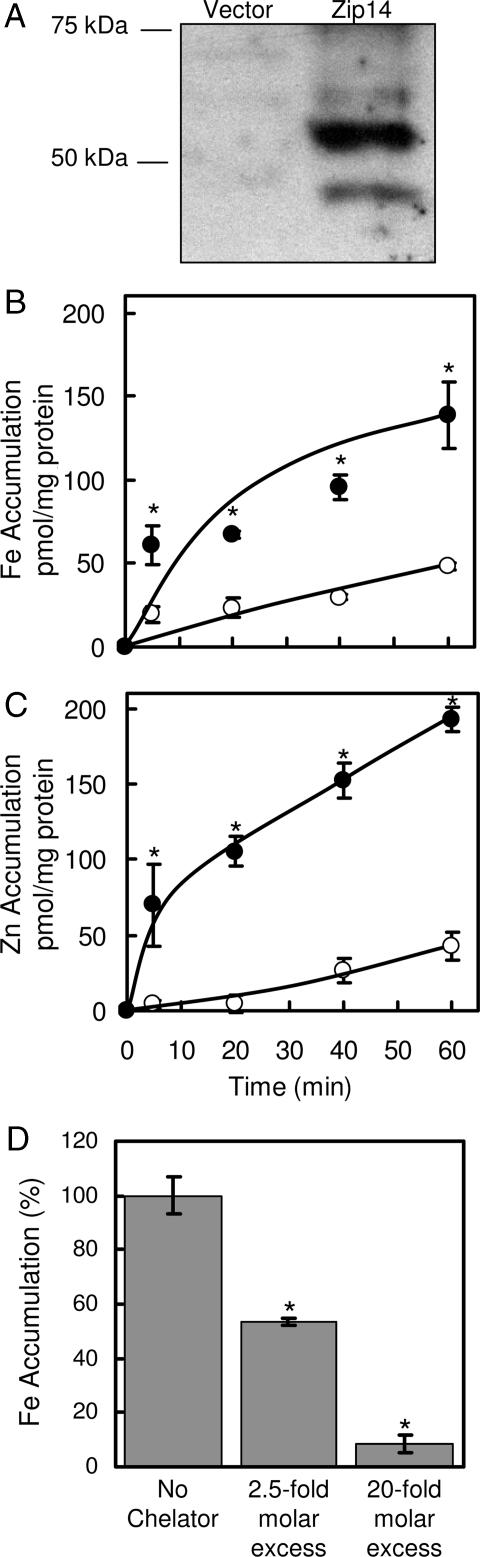
HEK 293H cells were transiently transfected with either empty vector or vector containing mZip14 cDNA. Western blot analysis (Fig. 1A) revealed the presence of two immunoreactive bands of ≈50 kDa, which are similar to the predicted molecular mass of Zip14 (54 kDa). Bands were also detected at ≈100 and 150 kDa (data not shown), likely representing mZip14 multimers. Overexpression of mZip14 increased the uptake of iron and zinc by 2- and 4-fold, respectively, after 60 min (Fig. 1 B and C). Uptake of iron from ferric citrate by HEK 293H cells overexpressing mZip14 was inhibited in a dose-responsive manner by adding bathophenanthroline sulfonate (BPS) (Fig. 1D), a membrane-impermeant ferrous-iron chelator. This finding suggests that mZip14 mediates iron uptake in the ferrous form.
Fig. 1.
Overexpression of mZip14 increases iron and zinc accumulation by HEK 293H cells. (A) HEK 293H cells were transfected with either empty vector (pCMV-Sport6) or the same plasmid containing mZip14 and grown for 48 h. Western blots of total cell lysates (15 μg of protein per lane) were prepared by using SDS/PAGE and immunoblotting with anti-mZip14 antibody. (B) Time course of iron accumulation measured with 59Fe in medium containing 2 μM ferric citrate (pH 7.4). (C) Time course of zinc accumulation measured with 65Zn in medium containing 2 μM ZnCl2 (pH 7.4), transfected with mZip14 (●) or vector alone (○). Nonspecific 59Fe and 65Zn binding to the cell surface was estimated by measuring uptake at 4°C. (D) Effect of BPS at 2.5- and 20-fold molar excess on mZip14-dependent iron accumulation after 60 min. Values are mean ± SD (n = 3; ∗, P < 0.05 compared with control) and are representative of multiple experiments.
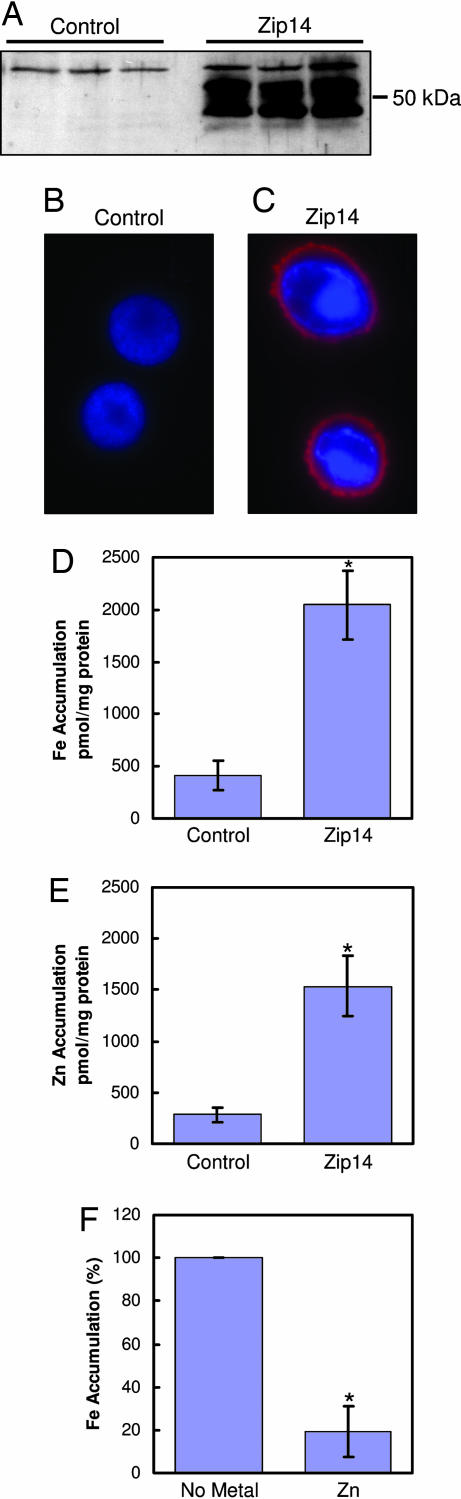
To establish further that mZip14 functions as an iron and zinc transporter, this protein was heterologously expressed in Sf9 insect cells. Western analysis demonstrates robust mZip14 expression in Sf9 cells infected with baculovirus containing mZip14 cDNA (Fig. 2A). The bands detected were comparable in mass with those detected by using HEK cell expression (Fig. 1A). Immunofluorescence revealed abundant mZip14 expression at the plasma membrane (Fig. 2C) that was absent in the control-infected cells (Fig. 2B). mZip14 expression (Fig. 2C) was associated with a 3-fold increase in iron and zinc uptake after 15 min (Fig. 2 D and E), respectively. The addition of a 100-fold molar excess of zinc to the uptake medium of the Sf9 cells reduced mZip14-mediated iron uptake by 80% (Fig. 2F). Zinc excess produced a lower reduction (60%) of mZip14-dependent iron uptake in HEK 293H cells (data not shown).
Fig. 2.
mZip14 localizes to the plasma membrane of Sf9 cells and increases iron and zinc accumulation. (A) Sf9 cells were infected with wild-type baculovirus (control) or baculovirus containing the cDNA for mZip14. After 64 h, total cell lysates (15 μg of protein per lane) were analyzed by Western blotting as above. Lanes represent three independent experiments. (B and C) The same cells were analyzed by immunofluorescence microscopy. Cells were fixed and incubated with an anti-mZip14 antibody followed by goat anti-rabbit IgG-Alexa Fluor 594. Cells were counterstained with DAPI to visualize nuclei. (D) Iron accumulation measured with 59Fe in medium containing 2 μM FeCl2. (E) Zinc accumulation measured with 65Zn in medium containing 2 μM ZnCl2. (F) mZip14-dependent iron accumulation measured with 59Fe in medium containing 2 μM FeCl2 and 200 μM ZnCl2. Cells were incubated at 27°C for 15 min in uptake medium. Values are the mean ± SD of three separate experiments, each with n = 3. ∗, P < 0.05 compared with control.
Suppression of Zip14 Expression Reduces the Uptake of Iron and Zinc by Mouse Hepatocytes.
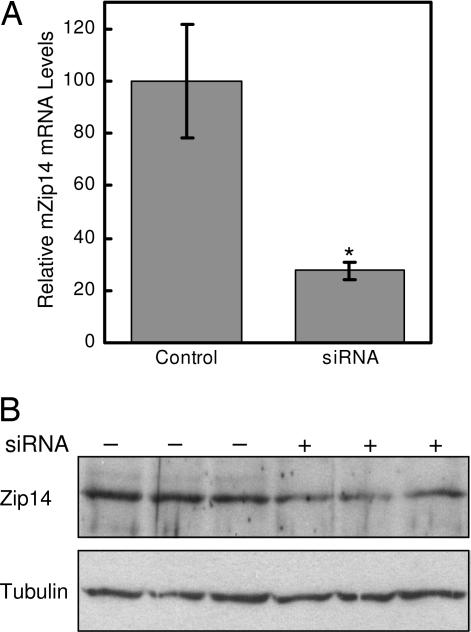
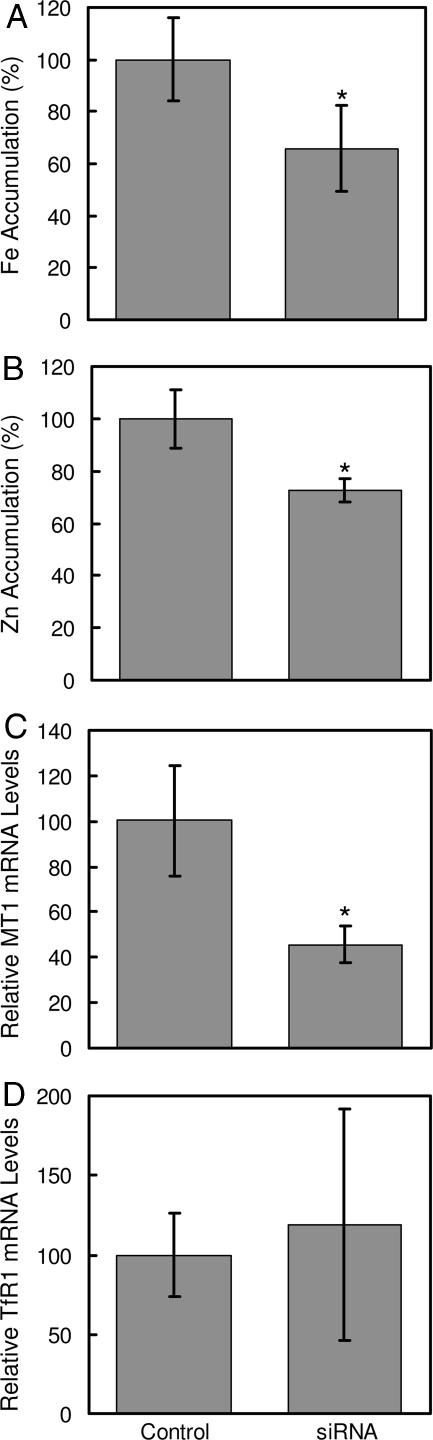
The contribution of endogenous Zip14 to zinc and iron uptake by hepatocytes was examined by using Zip14-specific siRNA. As shown in Fig. 3, treatment of AML12 mouse hepatocytes with Zip14 siRNA for 96 h decreased Zip14 mRNA levels by 75% and protein levels by ≈50%. Zip14 protein in AML12 cells was detected as a band of ≈70 kDa, perhaps reflecting glycosylation (15). Suppression of Zip14 expression decreased the uptake of iron and zinc by 30% and 25%, respectively (Fig. 4A and B). Diminished Zip14 expression also resulted in a decrease in mRNA levels of Mt1, a zinc-responsive gene (Fig. 4C). However, transferrin receptor 1 (TfR1) mRNA levels were unaffected by Zip14 siRNA treatment, indicating that the cellular iron status did not change (Fig. 4D).
Fig. 3.
mZip14 siRNA decreases Zip14 expression in AML12 mouse hepatocytes. (A) QRT-PCR analysis of Zip14 mRNA levels in AML12 cells treated for 96 h with a nontargeting (control) siRNA or siRNA targeted to mZip14. Values are the mean ± SD of three separate experiments, each with n = 3. ∗, P < 0.05 compared with control. (B) Cells in parallel were analyzed for Zip14 protein levels by Western blotting of total cell lysates (50 μg of protein per lane). To confirm equivalent protein loading, the Western blot was stripped and reprobed with anti-tubulin antibody. A representative experiment is shown.
Fig. 4.
Zip14 knockdown in mouse hepatocyte AML12 cells reduces iron and zinc accumulation and MT1 (metallothionein 1) expression without affecting TfR1 mRNA levels. AML12 cells were treated with nontargeting (control) siRNA or Zip14 siRNA for 96 h. (A) Iron accumulation measured with 59Fe in medium containing 2 μM ferric citrate. (B) Zinc accumulation measured with 65Zn in medium containing 2 μM ZnCl2. (C and D) QRT-PCR analysis of MT1 (C) and TfR1 (D) mRNA levels. For 59Fe and 65Zn accumulation, cells were incubated with uptake medium for 15 min. Values are mean ± SD of three separate experiments, each with n = 3. ∗, P < 0.05 compared with control.
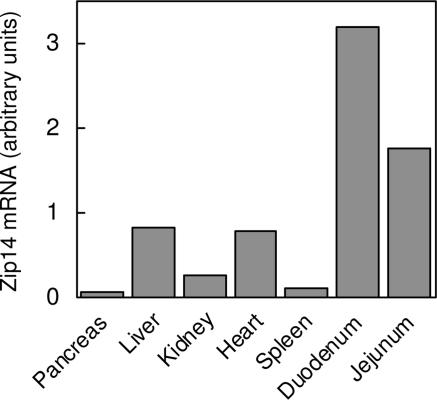
Zip14 mRNA Expression in Murine Tissues.
Quantitative real-time PCR (QRT-PCR) analysis of RNA isolated from various mouse tissues important in zinc and iron metabolism indicates higher Zip14 expression in liver and heart, relative to the spleen, kidney, and pancreas (Fig. 5). In addition, the abundant Zip14 expression in duodenum and jejunum is of interest from a nutritional perspective.
Fig. 5.
Tissue-expression profile of mZip14. Individual tissues were excised from two CD-1 mice and pooled before isolating total RNA. Zip14 transcript abundance was determined by QRT-PCR starting with an equal amount of total RNA from each tissue.
Discussion
Parenchymal cells are the most abundant cell type in the liver, and they contribute markedly to the regulation of zinc and iron metabolism (16, 17). The molecular mechanisms by which these metals are internalized from blood into hepatocytes are only partially understood, however. Hepatocytes can acquire iron from blood through multiple routes (17). Under normal conditions, circulating iron is internalized into hepatocytes mainly by the TfR pathway (17). The normal range of Tf saturation is 20–50%; however, Tf-binding capacity can be supersaturated during iron-overload conditions such as hereditary hemochromatosis, where plasma NTBI is greatly increased (11, 18). In rats, NTBI is efficiently cleared by the liver; ≈70% can be removed from a single pass (19). That the liver can accumulate NTBI from plasma is supported by the fact that mice with congenital hypotransferrinemia can absorb iron and efficiently accumulate it in the liver (14). These findings suggest NTBI uptake by the liver and other tissues, e.g., pancreas and heart, and they explain the iron accumulation and consequent tissue damage that occur during hereditary hemochromatosis (20).
Until now, divalent metal transporter 1 (DMT1; SLC11A2) was the only transmembrane iron transporter implicated in NTBI uptake. DMT1 transports Fe2+ into cells, and it also mediates the recovery of iron from acidified endosomes during the Tf-TfR recycling process (21, 22). Although DMT1 is able to transport Zn2+ as well, this cation is poorly transported relative to Fe2+ (21). DMT1 expression has been detected at the plasma membrane of hepatocytes (23), and its levels increase in iron-loaded Hfe−/− mice (24). However, the degree to which DMT1-mediated NTBI uptake can occur from the blood plasma remains to be determined, given that DMT1 transport activity proceeds optimally at pH 5.5 (22). Moreover, it was recently shown that ablation of Dmt1 results in neonates that lack any developmental abnormalities but have abnormally high liver stores (25). Administration of iron dextran also caused a marked accumulation of iron in hepatocytes of Dmt1−/− mice. These observations clearly indicate that the liver has alternate mechanisms for iron uptake.
Results presented in this report strongly suggest that mouse Zip14 functions as an iron and zinc importer that is critical for both NTBI and zinc uptake by cultured hepatocytes. The capability of mouse Zip14 to transport both iron and zinc is clearly evidenced by the significant increase in uptake rate for both cations in HEK 293H and Sf9 insect cells expressing this protein. The use of siRNA technology allowed us to examine the contribution of endogenous Zip14 to zinc and NTBI uptake. The AML12 cells used in these experiments are a nontumorigenic cell line that preserves many features of primary hepatocytes, including synthesis of albumin and Tf (26). In this cell line, an siRNA-mediated reduction in Zip14 protein levels by ≈50% resulted in a 40% and 30% reduction in iron and zinc uptake, respectively. It is likely that the residual zinc- and iron-uptake activities were mediated by remaining Zip14 or by other Zip proteins or DMT1. Interestingly, compensatory mechanisms were not sufficient to recover intracellular zinc levels because mRNA levels of Mt1 were reduced. On the other hand, Zip14-mediated iron uptake does not seem to be essential to maintain intracellular iron status because TfR mRNA levels, which respond to cellular iron concentrations, were unchanged.
In the present study, we found that zinc competed for iron uptake in HEK and Sf9 cells expressing mZip14, suggesting that these metals share a common uptake pathway. That endogenous Zip14 mediates the import of both iron and zinc is also supported by the observation that siRNA-targeted suppression of Zip14 expression in AML12 hepatocytes resulted in decreased uptake of both cations. It has long been appreciated that zinc competes for liver NTBI uptake. Using a rat liver-perfusion system, Wright et al. (27) determined that zinc was strongly inhibitory of ferrous iron uptake. Further kinetic studies revealed that zinc increased the apparent Km for iron transport without changing the apparent Vmax, consistent with competitive inhibition. Similarly, in primary hepatocytes isolated from rats and mice, zinc has been shown to inhibit the uptake of iron when presented as ferric citrate, a physiologically relevant form of NTBI (24, 28).
The tissue distribution of Zip14 has been examined in several studies. Analysis of Zip14 mRNA levels by using a human multiple-tissue Northern blot of 16 different tissues revealed most abundant expression in liver, followed by heart and pancreas (29). Zip14 transcript abundance has also been determined by using a human multiple-tissue expression array representing >30 adult tissues and 7 fetal tissues. Similar to the tissue Northern blot, Zip14 was most abundantly expressed in liver, pancreas, and heart (15). Among fetal tissues, highest Zip14 expression was detected in liver and heart. The expression profile of Zip14 in nine different mouse tissues (excluding pancreas) was recently investigated by using QRT-PCR (30). Most abundant Zip14 expression was found in liver, followed by heart, kidney, and white adipose tissue. In the present study, we used QRT-PCR to examine Zip14 transcript levels in seven mouse tissues especially important to zinc and iron homeostasis, and we found strong expression of Zip14 in liver, heart, and kidney but weak expression in pancreas and spleen. Interestingly, we observed most abundant Zip14 expression in duodenum and jejunum. The localization and function of Zip14 in these latter tissues merit further study.
Zip14 clusters within the LZT (LIV-1 zinc transporter) subfamily, whose members all contain a unique metalloprotease motif, EXPHEXGD, where X is any amino acid. Most mammalian Zip proteins are grouped into this subfamily, including Zip4, the acrodermatitis enteropathica-linked gene (31). LZT proteins are phylogenetically distant from Irt1 and Irt2, expressed in A. thaliana, and ZupT, from E. coli, which have been found to be capable of transporting iron (8–10). The substrate specificity of several mammalian members of the Zip family, including human/mouse Zip1 (h/mZip1), h/mZip2, mZip3, mZip4, and mZip5, have been tested by using metal-competition studies (32–36). In this regard, only h/mZip1 and hZip2 exhibited zinc-uptake activity, which appears to be partially inhibited by the addition of excess iron. Nevertheless, overexpression of hZip1 and hZip2 in cells does not seem to affect iron uptake (32, 33). These observations suggest that only few mammalian Zip proteins could have iron-transport activity.
In this work, we demonstrate the capability of Zip14 to facilitate cellular iron uptake. Furthermore, we demonstrate that this transporter is involved in zinc and iron uptake by hepatocytes. Further studies on the properties and regulation of Zip14 are needed to establish its role in normal iron homeostasis and disorders of iron metabolism.
Materials and Methods
Cell Culture.
HEK 293H cells (human embryonic kidney cells; Invitrogen, Carlsbad, CA) were cultured in DMEM supplemented with 10% FBS (Mediatech, Herndon, VA). AML12 mouse hepatocytes (American Type Culture Collection, Manassas, VA) were grown in DMEM/F-12 containing 10% (vol/vol) FBS, 40 ng/ml dexamethasone, and ITS (insulin, Tf, selenium) supplement (BD Biosciences, San Jose, CA). Medium also contained penicillin, streptomycin, and amphotericin B (Sigma, St. Louis, MO). Mammalian cell lines were grown at 37°C in 5% CO2. Sf9 insect cells (American Type Culture Collection) were cultured at room temperature in HyQ TNM-FH medium (HyClone, Logan, UT) supplemented with 10% (vol/vol) FBS, penicillin, and streptomycin.
Expression of mZip14 in HEK 293H Cells.
Effectene reagent (Qiagen, Valencia, CA) was used for transient transfection of HEK 293H cells with either empty pCMV-Sport6 or pCMV-Sport6 containing mouse Zip14 cDNA (GenBank accession no. BC021530). mZip14 overexpression was assessed by Western blotting 48 h after transfection. These methods were described in detail in ref. 2.
Expression of mZip14 in Insect Cells.
mZip14 was expressed in Sf9 insect cells by using the baculovirus expression system with Gateway technology (Invitrogen). Briefly, the same mouse Zip14 cDNA was PCR-amplified and cloned by TOPO-TA cloning (Invitrogen) into pENTR/D-TOPO vector for subsequent recombination into pDEST8 vector. pDEST8-Zip14 was then transformed into DH10Bac cells for transposition of Zip14 into the bacmid. Recombinant bacmid DNA was isolated, its sequence was verified, and it was transfected into Sf9 cells for production of infectious recombinant baculovirus particles, which were amplified and used to infect Sf9 cell monolayers. Expression of mZip14 in Sf9 monolayers was confirmed by Western blotting 64 h after infection. Wild-type baculovirus was used as a control.
Measurement of Iron and Zinc Uptake.
HEK 293H cells were washed with Hanks' balanced salt solution (HBSS) and incubated at 37°C in serum-free DMEM with 2% (wt/vol) BSA for 60 min to deplete Tf from the cells (37). 59FeCl3 (PerkinElmer, Wellesley, MA) was diluted with ferric citrate (molar ratio 1:10) to a final concentration of 2 μM Fe. After incubation with 59Fe-labeled ferric citrate for the times indicated, the cells were washed three times with 0.9% NaCl/1 mM BPS/1 mM diethylenetriaminepentaacetate (pH 7.4). To measure iron uptake in Sf9 cells, cells were washed with DMEM and incubated for 15 min at 25°C in DMEM containing 59FeCl3, 2 μM FeCl3, and 1 mM ascorbate (pH 7.0). Thereafter, cells were washed three times with 0.9% NaCl/1 mM BPS (MP Biomedicals, Solon, OH). To measure zinc uptake, HEK 293H cells were washed with HBSS and incubated at 37°C with DMEM containing 65ZnCl2 (Oak Ridge National Laboratory, Oak Ridge, TN) and 2 μM ZnCl2. After incubation with 65Zn, the cells were washed three times with 0.9% NaCl/10 mM EDTA. A similar procedure was carried out to assay zinc uptake by Sf9 cells at 25°C. All cells were harvested and then digested in a solution containing 0.2% SDS and 0.2 M NaOH before measuring cpm in a γ-ray spectrometer (Packard, Meriden, CT). Uptake data were expressed as mass by using respective specific activities (2).
siRNA-Mediated Suppression of mZip14 Expression.
AML12 cells were seeded 1 × 105 cells per well and immediately transfected with 15 nM (final concentration) nontargeting siRNA 1 siCONTROL (Dharmacon, Lafayette, CO) or mouse Zip14 SMARTpool siRNA. HiPerFect transfection reagent (Qiagen) was used for transfection of siRNA, and Zip14 expression was assessed 96 h later. Uptake of iron and zinc by AML12 cells was determined as described above for HEK 293H cells, except that serum-free DMEM/F-12 50:50 medium was used.
Animals.
Adult male (weighing ≈25 g) CD-1 mice were obtained from Charles River Laboratories (Wilmington, MA) and acclimated for several days on standard rodent diet (Harlan Teklad, Indianapolis, IN) and tap water. Mice were anesthetized by using isoflurane, and tissues were quickly excised and immersed in RNAlater (Ambion, Austin, TX) for subsequent RNA isolation. Protocols were approved by the University of Florida Institutional Animal Care and Use Committee.
mRNA Quantitation.
Total RNA was isolated by using RNA-Bee reagent (TelTest, Austin, TX) and treated with DNase (Ambion). QRT-PCR for mZip14 and metallothionein 1 was performed by using a one-step reverse transcriptase reaction (Applied Biosystems, Foster City, CA) as described in ref. 2. TfR1 mRNA levels were determined by using two-step QRT-PCR and SYBR Green (Applied Biosystems), with primers 5′-TCATGAGGGAAATCAATGATCGTA-3′ (forward) and 5′-GCCCCAGAAGATATGTCGGAA-3′ (reverse). Transcript abundances were normalized to levels of 18S rRNA.
Western Blot Analysis.
Total cell lysates were prepared as described in ref. 38. Fifty micrograms of protein was mixed with Laemmli buffer and incubated for 15 min at 37°C. Proteins were separated electrophoretically on an SDS/7.5% polyacrylamide gel, transferred to nitrocellulose, and incubated for 1 h in blocking solution [5% nonfat dry milk in Tris-buffered saline-Tween 20 (TBS-T)]. Blots were incubated overnight at 4°C in blocking buffer containing 2.5 μg/ml affinity-purified rabbit anti-mZip14 antibody (2). After washing in TBS-T, blots were incubated for 40 min with a 1:2,000 dilution of HRP-conjugated donkey anti-rabbit IgG (Amersham Biosciences, Piscataway, NJ). To confirm equivalent loading, blots were stripped for 5 min in 0.5 M glycine, pH 2.8/0.5 M NaCl, blocked for 1 h in blocking buffer, and reprobed with mouse monoclonal anti-tubulin clone B-5-1-2 (Sigma) followed by HRP-conjugated goat anti-mouse IgG (Zymed, San Francisco, CA). Cross-reactivity was visualized by using enhanced chemiluminescence (Pierce, Rockford, IL) and x-ray film.
Immunofluorescent Localization of mZip14.
Sf9 cells were fixed with 4% paraformaldehyde and incubated for 1 h in PBS containing 10% normal goat serum and 10 μg/ml affinity-purified anti-mZip14 antibody. Thereafter, cells were washed three times with PBS and incubated for 45 min in 3% BSA (wt/vol) with anti-Alexa Fluor 594 (Invitrogen) conjugate to anti-rabbit IgG. Zip14 localization was visualized with an Axiovert microscope (Zeiss).
Statistical Analysis.
Data are presented as the means ± SD and were analyzed by Student's t test. The significance level was set at P < 0.05.
Acknowledgments
We thank Louis Lichten for helpful discussions. This work was supported in part by National Institutes of Health (NIH) Grant DK 31127 and Boston Family Endowment Funds (to R.J.C.) and NIH Grant DK 065064 (to M.D.K.).
Abbreviations
- BPS
bathophenanthroline sulfonate
- DMT1
divalent metal transporter 1
- hZip
human Zip
- mZip
mouse Zip
- NTBI
non-transferrin-bound iron
- QRT-PCR
quantitative real-time PCR
- Tf
transferrin
- TfR
Tf receptor
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Weiss G. Best Pract Res Clin Haematol. 2005;18:183–201. doi: 10.1016/j.beha.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, Ganz T, Cousins RJ. Proc Natl Acad Sci USA. 2005;102:6843–6848. doi: 10.1073/pnas.0502257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eide DJ. Pflügers Arch. 2004;447:796–800. doi: 10.1007/s00424-003-1074-3. [DOI] [PubMed] [Google Scholar]
- 4.Liuzzi JP, Cousins RJ. Annu Rev Nutr. 2004;24:151–172. doi: 10.1146/annurev.nutr.24.012003.132402. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Eide D. Proc Natl Acad Sci USA. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Eide D. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 7.Eide D, Broderius M, Fett J, Guerinot ML. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. Plant Mol Biol. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- 9.Vert G, Briat JF, Curie C. Plant J. 2001;26:181–189. doi: 10.1046/j.1365-313x.2001.01018.x. [DOI] [PubMed] [Google Scholar]
- 10.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. J Bacteriol. 2005;187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentze MW, Muckenthaler MU, Andrews NC. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 12.Cabantchik ZI, Breuer W, Zanninelli G, Cianciulli P. Best Pract Res Clin Haematol. 2005;18:277–287. doi: 10.1016/j.beha.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs EM, Hendriks JC, van Tits BL, Evans PJ, Breuer W, Liu DY, Jansen EH, Jauhiainen K, Sturm B, Porter JB, et al. Anal Biochem. 2005;341:241–250. doi: 10.1016/j.ab.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Craven CM, Alexander J, Eldridge M, Kushner JP, Bernstein S, Kaplan J. Proc Natl Acad Sci USA. 1987;84:3457–3461. doi: 10.1073/pnas.84.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor KM, Morgan HE, Johnson A, Nicholson RI. FEBS Lett. 2005;579:427–432. doi: 10.1016/j.febslet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 16.King J, Cousins RJ. In: Modern Nutrition in Health and Disease. Shils ME, Shike M, Ross AC, Caballero B, Cousins RJ, editors. Baltimore: Lippincott Williams & Wilkins; 2005. pp. 271–285. [Google Scholar]
- 17.Anderson GJ, Frazer DM. Semin Liver Dis. 2005;25:420–432. doi: 10.1055/s-2005-923314. [DOI] [PubMed] [Google Scholar]
- 18.Grootveld M, Bell JD, Halliwell B, Aruoma OI, Bomford A, Sadler PJ. J Biol Chem. 1989;264:4417–4422. [PubMed] [Google Scholar]
- 19.Brissot P, Wright TL, Ma WL, Weisiger RA. J Clin Invest. 1985;76:1463–1470. doi: 10.1172/JCI112125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olynyk JK. Liver. 1999;19:73–80. doi: 10.1111/j.1478-3231.1999.tb00014.x. [DOI] [PubMed] [Google Scholar]
- 21.Mackenzie B, Ujwal ML, Chang MH, Romero MF, Hediger MA. Pflügers Arch. 2006;451:544–558. doi: 10.1007/s00424-005-1494-3. [DOI] [PubMed] [Google Scholar]
- 22.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 23.Trinder D, Oates PS, Thomas C, Sadleir J, Morgan EH. Gut. 2000;46:270–276. doi: 10.1136/gut.46.2.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chua AC, Olynyk JK, Leedman PJ, Trinder D. Blood. 2004;104:1519–1525. doi: 10.1182/blood-2003-11-3872. [DOI] [PubMed] [Google Scholar]
- 25.Gunshin H, Fujiwara Y, Custodio AO, Direnzo C, Robine S, Andrews NC. J Clin Invest. 2005;115:1258–1266. doi: 10.1172/JCI24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JC, Merlino G, Fausto N. Proc Natl Acad Sci USA. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright TL, Brissot P, Ma WL, Weisiger RA. J Biol Chem. 1986;261:10909–10914. [PubMed] [Google Scholar]
- 28.Baker E, Baker SM, Morgan EH. Biochim Biophys Acta. 1998;1380:21–30. doi: 10.1016/s0304-4165(97)00120-7. [DOI] [PubMed] [Google Scholar]
- 29.Nomura N, Nagase T, Miyajima N, Sazuka T, Tanaka A, Sato S, Seki N, Kawarabayasi Y, Ishikawa K, Tabata S. DNA Res. 1994;1:223–229. doi: 10.1093/dnares/1.5.223. [DOI] [PubMed] [Google Scholar]
- 30.Tominaga K, Kagata T, Johmura Y, Hishida T, Nishizuka M, Imagawa M. FEBS J. 2005;272:1590–1599. doi: 10.1111/j.1742-4658.2005.04580.x. [DOI] [PubMed] [Google Scholar]
- 31.Taylor KM, Nicholson RI. Biochim Biophys Acta. 2003;1611:16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 32.Gaither LA, Eide DJ. J Biol Chem. 2001;276:22258–22264. doi: 10.1074/jbc.M101772200. [DOI] [PubMed] [Google Scholar]
- 33.Gaither LA, Eide DJ. J Biol Chem. 2000;275:5560–5564. doi: 10.1074/jbc.275.8.5560. [DOI] [PubMed] [Google Scholar]
- 34.Dufner-Beattie J, Langmade SJ, Wang F, Eide D, Andrews GK. J Biol Chem. 2003;278:50142–50150. doi: 10.1074/jbc.M304163200. [DOI] [PubMed] [Google Scholar]
- 35.Dufner-Beattie J, Wang F, Kuo YM, Gitschier J, Eide D, Andrews GK. J Biol Chem. 2003;278:33474–33481. doi: 10.1074/jbc.M305000200. [DOI] [PubMed] [Google Scholar]
- 36.Wang F, Kim BE, Petris MJ, Eide DJ. J Biol Chem. 2004;279:51433–51441. doi: 10.1074/jbc.M408361200. [DOI] [PubMed] [Google Scholar]
- 37.Barisani D, Cairo G, Ginelli E, Marozzi A, Conte D. Hepatology. 1999;29:464–470. doi: 10.1002/hep.510290205. [DOI] [PubMed] [Google Scholar]
- 38.Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Blood. 2003;102:4191–4197. doi: 10.1182/blood-2003-04-1250. [DOI] [PubMed] [Google Scholar]