Abstract
Despite longstanding interest in parallel evolution, little is known about the genes that control similar traits in different lineages of vertebrates. Pelvic reduction in stickleback fish (family Gasterosteidae) provides a striking example of parallel evolution in a genetically tractable system. Previous studies suggest that cis-acting regulatory changes at the Pitx1 locus control pelvic reduction in a population of threespine sticklebacks (Gasterosteus aculeatus). In this study, progeny from intergeneric crosses between pelvic-reduced threespine and ninespine (Pungitius pungitius) sticklebacks also showed severe pelvic reduction, implicating a similar genetic origin for this trait in both genera. Comparative sequencing studies in complete and pelvic-reduced Pungitius revealed no differences in the Pitx1 coding sequences, but Pitx1 expression was absent from the prospective pelvic region of larvae from pelvic-reduced parents. A much more phylogenetically distant example of pelvic reduction, loss of hindlimbs in manatees, shows a similar left–right size bias that is a morphological signature of Pitx1-mediated pelvic reduction in both sticklebacks and mice. These multiple lines of evidence suggest that changes in Pitx1 may represent a key mechanism of morphological evolution in multiple populations, species, and genera of sticklebacks, as well as in distantly related vertebrate lineages.
Keywords: development, limb, parallel evolution, Pitx1, stickleback
Animal evolution abounds with examples of parallelism, the independent evolution of similar traits in separate but related lineages that were not present in their most recent common ancestor (1). Among animals, parallelisms range from traits such as similar wing spot patterns in phylogenetically divergent lineages of butterflies (2) to more substantial changes in body plan, such as the independent loss of limbs in multiple lineages of lizards (3).
The ecological factors that influence the independent evolution of similar traits in different lineages have attracted considerable attention, yet less is known about the genetic basis for parallel evolution (1, 4). A fundamental question in studies of parallel evolution is whether the same gene or genes control similar adaptive phenotypes in different populations and species. Among vertebrates, this issue has been difficult to address because of the paucity of appropriate model organisms; however, recent studies demonstrate that natural populations of organisms, not just laboratory strains, can be used to dissect the genetic and developmental basis of adaptive organismal diversity (5–16).
The stickleback fish family (Gasterosteidae) provides numerous opportunities to study the genetic basis of parallel evolution. Threespine (Gasterosteus aculeatus) and ninespine (Pungitius pungitius) sticklebacks show repeated evolution of similar adaptive traits among different populations within each genus, and these two genera have also evolved similar derived traits in parallel (4, 17, 18). Among the most striking examples of parallel evolution in sticklebacks is the reduction of the pelvic complex, which consists of a large ventral spine (an enlarged fin ray) and the supporting plate-like pelvic girdle. A complete pelvis is present in all marine and most freshwater populations of both genera (Fig. 1a, c, and d); however, heritable reduction or loss of the pelvic girdle occurs in several derived freshwater populations throughout the circumpolar distribution of threespine and ninespine sticklebacks, likely as an adaptive response to reduced piscine predator loads and/or water chemistry (Fig. 1b) (9, 19–25). Pelvic reduction evolved in parallel among freshwater populations within each genus no longer than 10,000–20,000 years ago, at the end of the last glacial period when marine sticklebacks began to colonize new freshwater habitats (26). In contrast, the most recent common ancestor of threespine and ninespine sticklebacks lived at least 10 million years ago, based on fossil data (27).
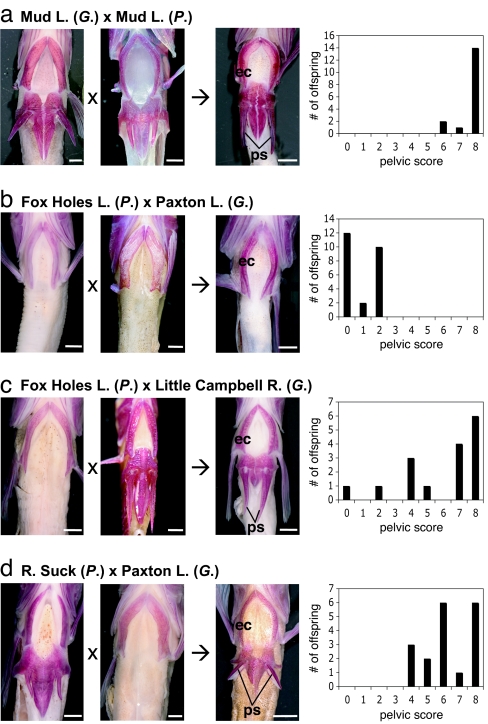
Fig. 1.
Pelvic morphology of intergeneric hybrid progeny. Total bilateral pelvic scores range from 0 (absent pelvis) to 8 (complete pelvis with 4 elements on each side). (a) Parental and representative hybrid fish from Mud Lake control cross showing strong development of pelvic structures. Distribution of scores is skewed toward complete pelvises. (b) In contrast, hybrid progeny from pelvisless parents show very weak or no pelvic development. (c and d) In crosses with one complete-pelvis and one pelvisless parent, distributions of hybrid progeny pelvic scores are skewed toward strong pelvic development. ec, left ectocoracoid; ps, pelvic spine. (Scale bars, 2 mm.)
Previously, we demonstrated that a cis-regulatory change in Pitx1, a homeobox-containing transcription factor that is critical for hindlimb identity and outgrowth (28, 29), was responsible for pelvic reduction in a British Columbian population of threespine sticklebacks (9). Furthermore, complementation tests showed that pelvic reduction has a similar genetic basis in conspecifics from an Icelandic lake (9), and genetic mapping and complementation tests showed that pelvic reduction in several southern Alaskan lakes maps to the linkage group containing Pitx1 (11).
By expanding this complementation approach to different genera, we can test directly whether the same genes control pelvic reduction in Gasterosteus and Pungitius. Although prezygotic behavioral barriers reproductively isolate threespine and ninespine stickleback genera in the limited areas where they cooccur (30, 31), postzygotic barriers are incomplete, and some populations of the two genera can be crossed to produce viable hybrid progeny (30–33), sometimes referred to as “Stichlingsbastarden” (30). This makes a complementation approach feasible.
In this study, we find that pelvic reduction alleles do not complement across genera, thereby suggesting that variation at the Pitx1 locus underlies this trait in both Gasterosteus and Pungitius. We also show that a mammalian example of pelvic modification, hindlimb loss in manatees, shares a morphological signature of Pitx1-mediated pelvic reduction with sticklebacks and genetically modified mice and may, thus, represent an example of convergence in pelvic-reduction mechanisms.
Results
Pelvic Phenotypes in Stickleback Hybrids.
Because threespine and ninespine stickleback lineages diverged millions of years ago, it was conceivable that stickleback hybrids might show morphological defects attributable to developmental instability or other epigenetic factors (34). Furthermore, threespine and ninespine stickleback pelvises have minor structural differences (35). To verify that intergeneric hybrid sticklebacks can have complete and recognizable pelvises, we crossed Gasterosteus and Pungitius with complete pelvises from Mud Lake, AK (Fig. 1a). The resulting hybrid progeny showed expression of a robust, and usually complete, pelvis in all offspring, thereby confirming that hybrid progeny are capable of normal pelvic development.
Hybrid sticklebacks from threespine and ninespine parents show a mix of morphological traits from both genera in this and other studies (30, 32, 33). Nevertheless, previous studies did not show that progeny of intergeneric crosses were, indeed, genetic hybrids. To verify that our hybrid fish inherited alleles from both parents at loci throughout the genome, we genotyped all fish with a series of microsatellite markers described in ref. 5. These assays showed that hybrid progeny in all crosses inherited alleles throughout the genome from both parents, including two markers closely linked to the Pitx1 locus in Gasterosteus.
To test for complementation of pelvic reduction alleles, we crossed Pungitius (no pelvis) from Fox Holes Lakes, Northwest Territories, to Gasterosteus (no pelvis) from the Paxton Lake benthic population, British Columbia (Fig. 1b). Pelvic reduction is not a dominant trait in Paxton benthic Gasterosteus (9) or in several populations of Pungitius (23, 24). If pelvic reduction has a different genetic basis in the parents of the intergeneric hybrid cross, we would expect expression of a complete pelvis in the hybrid progeny. However, in marked contrast to the cross with complete-pelvis parents, all hybrid progeny from pelvisless parents showed severe, bilateral pelvic reduction, suggesting that pelvic-reduction alleles in the two genera failed to complement each other. This finding raised the intriguing possibility that the same genes might underlie pelvic reduction in these two genera of stickleback.
To further explore this hypothesis, we performed additional control crosses to rule out the possibility that pelvic-reduction alleles in one genus were dominant over alleles in the other genus. We performed two small crosses, each with Pungitius (no pelvis) from Fox Holes Lake and Gasterosteus (complete pelvis) from Little Campbell River, British Columbia (Fig. 1c). In contrast to the cross between pelvisless parents of both genera, the crosses with only a pelvisless Pungitius parent produced multiple progeny with strong development of the pelvic complex. Similarly, the reciprocal control cross between a Pungitius (complete pelvis) from the River Suck catchment, Ireland, and a Gasterosteus (no pelvis) from Paxton Lake also yielded progeny with high pelvic scores (Fig. 1d).
Together, these crosses demonstrate that hybrid progeny can develop a pelvis if at least one parent from either genus has a pelvis. Hence, pelvic-reduction alleles from one genus do not show simple dominance over complete pelvis alleles from the other genus. Consequently, the severe pelvic reduction observed in the Fox Holes Lakes Pungitius (no pelvis) by Paxton Lake Gasterosteus (no pelvis) cross is not due to dominant alleles in either genus but, rather, to a failure to complement pelvic-reduction alleles at a similar locus or loci.
Conservation of Pitx1 Coding Sequences.
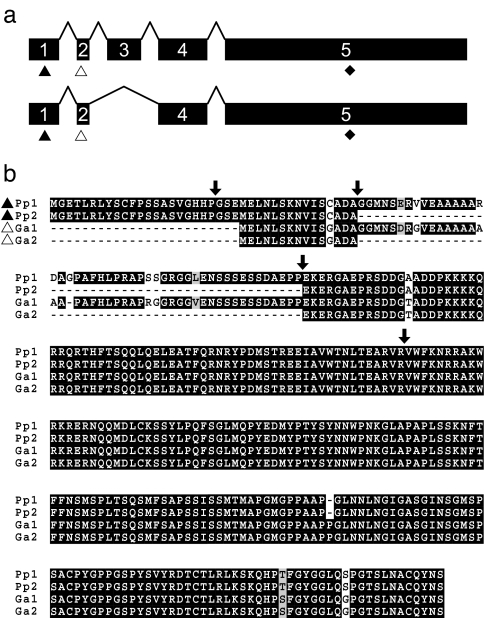
To test for coding changes between Pitx1 alleles from pelvisless and complete-pelvis Pungitius populations that might be responsible for pelvic reduction, we isolated and sequenced mRNA transcripts from Fox Holes Lakes and control populations. The Pitx1 transcript in Pungitius comprises 5 exons and shows high sequence conservation with its ortholog in Gasterosteus (Fig. 2) and other vertebrates (9). Two splice variants of the Pitx1 transcript were identified in Pungitius, and neither variant contained any amino acid coding differences in the Fox Holes Lakes population relative to the control population. Therefore, we could rule out coding changes in Pitx1 as a potential molecular basis for pelvic reduction in Fox Holes Pungitius. The five-exon genomic structure and alternate splice forms of Pitx1 are unique among described vertebrate orthologs for this gene, with other organisms having only three exons (36, 37).
Fig. 2.
Genomic structure and amino acid sequence of Pitx1 in Pungitius and Gasterosteus. (a) The coding sequence comprises five exons; both genera have splice variants missing exon 3. Putative translation start sites vary between Pungitius (black triangles) and Gasterosteus (white triangles), and stop sites are conserved (black diamonds). Exon 1 is noncoding in Gasterosteus. (b) Predicted amino acid sequences of Pitx1 splice variants in Pungitius (Pp1, Pp2) and Gasterosteus (Ga1, Ga2). Arrows mark exon boundaries.
Previously, we identified only the three 3′-most exons of Pitx1 in Gasterosteus (9). However, further sequencing studies confirm that the Gasterosteus Pitx1 transcript also has 5 exons and splice variants similar to those in Pungitius (Fig. 2). Our findings suggest greater diversity in Pitx1 genomic and transcript structure in sticklebacks, and this diversity may extend to other fish. For example, a nonannotated EST clone from medaka (Oryzias latipes; GenBank accession no. BJ732832) suggests a splicing scheme similar to that seen in sticklebacks. The amino acid sequence of all five exons in Gasterosteus is conserved between the Paxton Lake benthic and complete pelvis (control) populations, also ruling out coding changes as a potential molecular basis for pelvic reduction in the former population.
Expression Changes in Pitx1 in Pelvic-Reduced Sticklebacks.
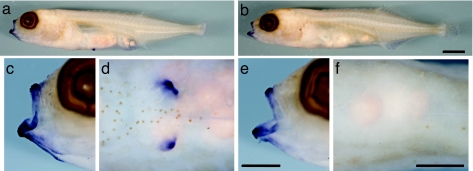
In Gasterosteus, Pitx1 expression is missing from the prospective pelvic region of larvae from populations with severe pelvic reduction (9, 38). To test for similar changes in Pungitius, we examined larval expression patterns of Pitx1 in Fox Holes Lakes and a control population. Pitx1 transcripts were detected in the mouth and lower jaw of larvae of both populations (Fig. 3), confirming that the gene can be expressed in both pelvic-reduced and control fish. In control larvae, Pitx1 is also expressed robustly in the pelvic buds (Fig. 3d). However, in Fox Holes Pungitius larvae, as in pelvic-reduced Gasterosteus, Pitx1 expression is absent from the prospective pelvic region (Fig. 3f). These expression differences, coupled with the coding-sequence conservation between Fox Holes and control fish, are consistent with site-specific regulatory changes in Pitx1 that affect expression in the pelvis, but not other regions, of Fox Holes larvae.
Fig. 3.
Pelvic expression of Pitx1 differs between control and Fox Holes Lakes Pungitius larvae. (a, c, and d) Whole-mount in situ hybridization of larvae from a complete-pelvis (control) population shows Pitx1 expression in the mouth and lower jaw (enlarged lateral view in c) and the prospective pelvic region (enlarged ventral view in d). (b, e, and f) Pitx1 expression is also detected in the mouth and lower jaw of Fox Holes Lakes larvae, but expression is absent from the prospective pelvic region. [Scale bars, 1 mm (a and b) and 0.5 mm (c–f).]
Directional Asymmetry in Manatees.
A signature of Pitx1-mediated pelvic reduction is the tendency for any remaining pelvic rudiments to be larger on the left than on the right side. This directional asymmetry likely arises from early expression of the Pitx2 gene, a closely related family member that is expressed preferentially on the left-hand side of developing embryos (29, 39). Mice that are homozygous for a knockout allele of Pitx1 show greater hindlimb reduction on the right than on the left side. A similar asymmetry is seen in many pelvic-reduced stickleback populations (9, 20, 21, 38), and this asymmetry has been mapped genetically to the Pitx1 locus in a cross with pelvic-reduced Paxton benthic fish (9). Directional asymmetry could not be assessed in the Fox Holes Lakes Pungitius population because of the complete absence of pelvic structures (19). However, a nearby pelvic-reduced Pungitius population has been described from Pine Lake, located only 55 kilometers from Fox Holes. Of the fish from this population that had a pelvic spine on only one side, significantly more were left-sided than right-sided (19), as expected if pelvic reduction in this population is also mediated by loss of Pitx1.
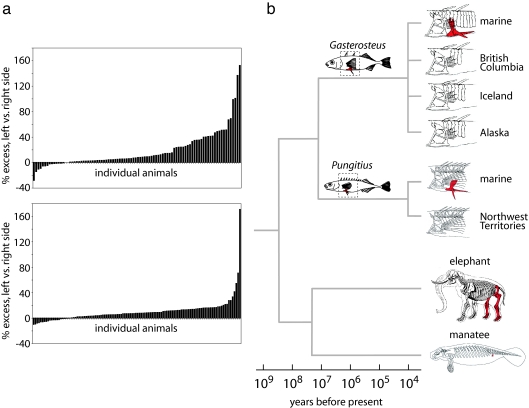
Gasterosteus and Pungitius last shared a common ancestor at least 10 million years ago, suggesting that mechanisms of pelvic reduction may have evolved in parallel over large evolutionary distances. To test whether pelvic reduction in mammals may also share common features, we examined the directional asymmetry of pelvic reduction in natural populations of Florida manatees (Trichechus manatus latirostris), which retain only small pelvic rudiments and no external hindlimbs. Eighty-one and a half percent of individuals showed larger pelvic vestiges on the left than on the right side (n = 114, Fig. 4a). This percentage is remarkably similar to the percentage of sticklebacks from an F2 laboratory cross (9) that inherited two Paxton benthic alleles at the Pitx1 locus and that show larger pelvic rudiments on the left than on the right side (83.5%, n = 91, Fig. 4a). The mean ratio of left to right pelvic size was significantly different from one in both manatees and in pelvic-reduced sticklebacks from the Paxton benthic cross. In both cases, the sign of asymmetry was the same as that observed in mice missing a functional Pitx1 gene (mean L/R ratio: manatees, 1.10 ± 0.018, P < 0.0001, 95% confidence interval, 1.07 to 1.14; sticklebacks, 1.13 ± 0.025, P < 0.0001, 95% confidence interval, 1.08 to 1.18).
Fig. 4.
Asymmetry is a morphological signature of pelvic reduction in multiple, distantly related vertebrates. (a) In both an F2 threespine stickleback cross (Upper) and a natural population of Florida manatees (Lower), pelvic remnants tend to be larger on the left side of the body than on the right. Each bar on the histograms represents a different individual; negative values indicate a larger right remnant. (b) Complementation and mapping crosses suggest that Pitx1 is involved repeatedly in the evolution of pelvic reduction in different populations of threespine (Gasterosteus) and ninespine (Pungitius) sticklebacks. Freshwater populations diverged from marine populations 10,000–20,000 years ago, whereas the two genera shared a complete-pelvis common ancestor at least 10 million years ago. Similar genetic mechanisms may underlie pelvic reduction in manatees, which diverged hundreds of millions of years ago from the lineage that includes sticklebacks and tens of millions of years ago from complete-pelvis relatives, such as elephants. Animal drawings in (b) are modified after refs. 19, 35, 52, 64.
Discussion
Generation of interspecific hybrids can be a useful tool for testing the genetic mechanisms underlying evolutionary change in natural populations (40, 41). Previous studies suggest that viable hybrids can be generated between different mammalian species that have diverged within the last 2–3 million years and between fish, frog, and bird species separated by >20 million years (42–44).
Gasterosteus and Pungitius diverged ≥10 million years ago. Although hybrids have never been reported in the wild (31), these genera can be crossed by using artificial fertilization in the laboratory (30, 32, 33). Here, we used intergeneric crosses to compare the genetic basis of pelvic reduction in these two distantly related stickleback genera. Intergeneric hybrids can clearly form a robust pelvis, as long as one of the parents of the cross comes from a population with complete pelvises. Some phenotypic heterogeneity is present in control crosses with one pelvic-reduced parent, which could arise from either heterozygosity still present in wild populations or from variable penetrance and expressivity of genes controlling pelvic reduction in the hybrid state. In contrast, all of the hybrid progeny are severely pelvic-reduced in the Paxton benthic Gasterosteus-by-Fox Holes Pungitius cross. Previous genome-wide linkage mapping and expression studies suggest that Pitx1 is the major locus controlling pelvic reduction in Paxton benthic fish (9), and absence of complementation between Paxton benthic Gasterosteus and Fox Holes Pungitius strongly suggests that a similar genetic mechanism involving Pitx1 underlies pelvic reduction in these two separate genera.
We cannot exclude the possibility that failure of complementation is due to nonallelic noncomplementation, perhaps because of heterozygosity for different components of single or parallel pathways controlling pelvic development. However, pelvic reduction in Northwest Territories Pungitius populations shows several other characteristic features that match those of Pitx1-mediated pelvic reduction in threespine sticklebacks: (i) loss of Pitx1 expression at the site where the pelvic fin would normally develop, (ii) retention of normal coding sequence and normal expression at other sites, and (iii) directional asymmetry, with greater morphological reduction on the right than on the left side (19).
Pelvic reduction has evolved repeatedly in many other groups, including fish, amphibians, reptiles, and mammals (3, 35, 45). Previous marker gene-expression studies show significant alteration of Hoxb9, Pitx1, and Tbx4 expression in pufferfish; Shh, Hox, and apical ridge marker expression in snake; and Hand2, Shh, and Fgf8 expression in whale embryos (46–48). However, crosses between forms with and without a pelvis are difficult or impossible in each of these groups. As a result, outside of sticklebacks, it is not clear whether the phenotypic trait of pelvic reduction maps genetically to the Pitx1 locus or to any other candidate loci that have been examined by expression analysis.
Pelvic reduction mediated by changes in Pitx1 function has a characteristic morphological signature that is relatively easy to examine in any group for which large population samples are present. Sticklebacks and Pitx1 knockout mice both show greater reduction of pelvic structures on the right than on the left side of the body, a directional asymmetry thought to arise because of preferential expression of the related gene Pitx2 on the left but not the right side of developing embryos (29). Left-biased expression of Pitx2 has been observed in organisms as distantly related as fish and mammals, suggesting an ancient role in patterning of the left–right body axis, including multiple tissues outside the hindlimb (39, 49, 50).
Several groups of marine mammals evolved from four-legged ancestors during the Tertiary. Fossil sirenians dating to the Eocene and Miocene document several stages in the overall transition from terrestrial to fully aquatic lifestyle, including complete loss of the external hindlimb as part of a series of adaptations for body streamlining (51). Modern manatees have a vestigial pelvic apparatus consisting of small, free-floating, paired pelvic bones located in the body-wall musculature and lacking the femur, tibia, fibula, tarsals, and digits (51, 52). The left–right pelvic pairs studied here clearly show significant directional asymmetry. Both the direction of asymmetry and the overall proportion of animals that show greater pelvic size on the left than on the right side closely resemble the morphological features of Pitx1-mediated pelvic reduction seen in mice and sticklebacks.
Further study of the molecular basis of pelvic reduction in sticklebacks, manatees, and other animals will require the identification of the cis-acting regulatory sequences that control Pitx1 expression in the developing hindlimb. In this and other examples in which morphological evolution has been traced to regulatory rather than coding-region changes, it has been difficult to locate the precise regulatory modules that underlie functional changes in the corresponding gene (53–55). However, enhancer studies of the Drosophila yellow gene have recently identified particular sequences that have been gained and lost in fruit fly lineages with different color variants (56, 57). Similar studies should now be possible in mice and fish to identify elements controlling hindlimb-specific expression of the Pitx1 gene.
Recent genetic and molecular studies have identified several examples of the repeated involvement of the same genes in the evolution of similar traits in independent lineages, including the repeated evolution of similar trichome and pigmentation traits in different species of fruit fly (55–58); pelvic reduction and armor-plate patterning in threespine sticklebacks (9–11, 13); sodium-channel resistance to neurotoxins in snakes and clams (15, 59); Mc1r-mediated changes in pigmentation patterns in birds, mammals, and reptiles (6–8, 12, 60); and albinism in blind Mexican cavefish (16). The current work suggests that common genetic mechanisms may also underlie major structural changes in skeletal patterning and limb formation in very distantly related lineages (Fig. 4).
Why are particular genes involved repeatedly in the evolution of similar phenotypes? Perhaps we should not be surprised that the same genetic pathways are involved in parallel evolution of similar traits (1), because the finite number of genes required to build a structure during development limits the realm of possible evolutionary changes (61). However, the total numbers of genes involved in pigmentation, trichome patterning, and limb outgrowth and patterning are not small, and changes in many different genes are known to produce similar phenotypes in laboratory mutants. It is possible that some genes are preferential hotspots for mutations, perhaps because of target size or genomic features that predispose to insertion, rearrangement, or other sequence changes. One of the most important constraints may be avoidance of negative pleiotropic defects in natural populations. Most known examples of parallel evolution consist of coding-region changes in genes with highly specific expression patterns (Mc1r, Oca2) (6–8, 12, 16, 60), or cis-acting regulatory changes that alter tissue- or region-specific expression of genes that otherwise have complex patterns and multiple functions (Ubx, Ovo/shaven baby, Yellow, Eda, Pitx1) (9, 13, 54–58). For genes with highly restricted expression patterns, either coding or regulatory mutations can generate new phenotypes that are confined to a particular tissue. For genes expressed in multiple tissues, regulatory mutations in highly modular cis-acting control sequences provide a mechanism to avoid pleiotropic effects and confine phenotypic changes to a particular tissue type or body region (9, 57, 61).
More examples are clearly needed to determine whether specificity and modularity are key constraints that lead to reuse of particular genes when similar phenotypes arise in different populations and distantly related species. The large number of examples of parallel phenotypic evolution in sticklebacks provides an excellent system to study the molecular basis of many different traits in multiple populations, species, and genera. As illustrated by studies of pelvic reduction, the genetic mechanisms originally found in studies of local populations of sticklebacks may have broad generality, including the identification of pathways that are used repeatedly to control parallel or convergent phenotypic changes across a wide range of other animals.
Materials and Methods
Fish Collection, Husbandry, and Phenotyping.
Intergeneric hybrid crosses were performed in vitro, and progeny were fixed in ethanol and stained with alizarin red for analysis (5). Pelvic structures in fish with standard lenght (SL) = 22–67 mm were scored for presence of anterior process, posterior process, ascending process, and spine for a maximum pelvic score of 4 on each side (25). All pelvic elements are strongly ossified in complete-pelvis populations of Pungitius by SL = 20 mm (M.D.S., personal observations) and in Gasterosteus by SL = 16.5 mm (62). Larvae for in situ hybridization studies came from crosses from Fox Holes Lakes (Northwest Territories) and an unnamed creek near Anchorage, AK.
Genotyping.
Nine Gasterosteus microsatellite markers (5) that amplified PCR products from Pungitius DNA were used to genotype all crosses to test for inheritance of alleles from both parents in hybrids: Stn329 (linkage group (LG) 1 in Gasterosteus), Stn79 and Stn81 (LG7), Stn85 (LG8), Stn102 and Stn108 (LG9), Stn294 and Stn315 (LG16), and Stn194 (LG19). Five markers amplified PCR products from parents and progeny in most, but not all, crosses: Stn242 (LG1), Stn336 (LG7), Stn144 and Stn287 (LG12), and Stn186 (LG19). Genotyping reactions were performed as described (5) and analyzed by using an ABI 3730xl automated sequencer (Applied Biosystems, Foster City, CA).
Pitx1 mRNA Transcript Analysis.
We isolated total RNA from larval stickleback progeny of complete-pelvis adult fish from an unnamed creek (Alaska), generated RACE-ready cDNA (SMART kit; Clontech, Mountain View, CA) and amplified the complete 5′ UTR, coding region, and part of the 3′ UTR (5′-to-3′ primer sequences: CTGCTCGGGGCTCTCGGTAAGTGAA for 5′ RACE; CACACGCATGAAGTGGATTTACTG and CTCCCGTCAGCTGTTGTACTG for the coding region; TTCAACTCCATGAGCCCGCTCACCT for 3′ RACE; universal primers from the kit also used for RACE). Amplification products were cloned (pCR-TOPO2.1; Invitrogen, Carlsbad, CA) and sequenced. The same protocol and primers were also used for marine (complete pelvis) and Paxton Lake benthic (no pelvis) Gasterosteus.
In Situ Hybridization.
Pungitius Pitx1 coding fragments were cloned into pCR-TOPO4 (Invitrogen), transcribed (DIG mix; Roche, Indianapolis, IN), and hydrolyzed. Stage-30 larvae (63) from Fox Holes and unnamed creek (Alaska) populations were fixed in 4% PFA, and whole-mount in situ hybridization was performed as described (9). Both splice variants gave comparable results; Fig. 3 shows the variant without exon 3.
Manatee Pelvic Measurements.
Left and right pelvic vestiges were prepared in a large necropsy study of Florida manatees (52). Weights for each side were determined to the nearest 0.01 gram on an electronic balance. Left–right ratio, and percent excess of left over right side was calculated as (left mass/right mass) and [(left–right)/right × 100], respectively. Asymmetry in the Paxton benthic cross was determined from the anterior–posterior length of the pelvic girdle as described (9). Mean ratios of left to right pelvic size were tested for significant difference from unity by two-tailed t test.
Acknowledgments
We thank Dolph Schluter (University of British Columbia, Vancouver, BC, Canada) for Paxton fish; members of the Kingsley lab, Katie Peichel, Frank von Hippel, and Caitlyn Ellis, and the Shannon Regional Fisheries Board for collecting assistance and discussions; and Sentiel Rommel and the Florida Fish and Wildlife Research Institute for providing manatee pelvis samples. This work was supported, in part, by a Helen Hay Whitney Foundation fellowship and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences (to M.D.S.), a National Science Foundation grant (to M.A.B.), and National Institutes of Health Grant 1P50HG02568 (to D.M.K). D.M.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
References
- 1.Futuyma DJ. Evolutionary Biology. Sunderland, MA: Sinauer; 1986. [Google Scholar]
- 2.Rensch B. Evolution Above the Species Level. New York: Columbia Univ Press; 1959. [Google Scholar]
- 3.Greer AE. J Herpetol. 1991;25:166–173. [Google Scholar]
- 4.Schluter D, Clifford EA, Nemethy M, McKinnon JS. Am Nat. 2004;163:809–822. doi: 10.1086/383621. [DOI] [PubMed] [Google Scholar]
- 5.Peichel CL, Nereng K, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- 6.Theron E, Hawkins K, Bermingham E, Ricklefs RE, Mundy NI. Curr Biol. 2001;11:550–557. doi: 10.1016/s0960-9822(01)00158-0. [DOI] [PubMed] [Google Scholar]
- 7.Nachman MW, Hoekstra HE, D'Agostino SL. Proc Natl Acad Sci USA. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eizirik E, Yuhki N, Johnson WE, Menotti-Raymond M, Hannah SS, O'Brien SJ. Curr Biol. 2003;13:448–453. doi: 10.1016/s0960-9822(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro MD, Marks ME, Peichel CL, Nereng K, Blackman BK, Jonsson B, Schluter D, Kingsley DM. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 10.Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. PLoS Biol. 2004;2:635–641. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, Bell MA, Kimmel CB, Postlethwait JH. Proc Natl Acad Sci USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mundy NI, Badcock NS, Hart T, Scribner K, Janssen K, Nadeau NJ. Science. 2004;303:1870–1873. doi: 10.1126/science.1093834. [DOI] [PubMed] [Google Scholar]
- 13.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 14.Kimmel CB, Ullmann B, Walker C, Wilson C, Currey M, Phillips PC, Bell MA, Postlethwait JH, Cresko WA. Proc Natl Acad Sci USA. 2005;102:5791–5796. doi: 10.1073/pnas.0408533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geffeney SL, Fujimoto E, Brodie ED, III, Brodie ED, Jr, Ruben PC. Nature. 2005;434:759–763. doi: 10.1038/nature03444. [DOI] [PubMed] [Google Scholar]
- 16.Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 17.Wootton RJ. The Biology of the Sticklebacks. London: Academic; 1976. [Google Scholar]
- 18.Bell MA, Foster SA. The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford Univ Press; 1994. p. 571. [Google Scholar]
- 19.Nelson JS. Copeia. 1971;1971:707–717. [Google Scholar]
- 20.Reimchen TE. Can J Zool. 1980;58:1232–1244. [Google Scholar]
- 21.Bell MA, Francis RC, Havens AC. Copeia. 1985;1985:437–444. [Google Scholar]
- 22.Takata K, Goto A, Hamada K. Jpn J Ichthyol. 1985;32:100–103. [Google Scholar]
- 23.Zyuganov VV, Rosanov AS. Doklady Akademii Nauk SSR. 1987;293:155–159. [Google Scholar]
- 24.Blouw DM, Boyd GJ. Heredity. 1992;68:33–42. [Google Scholar]
- 25.Bell MA, Orti G, Walker JA, Koenings JP. Evolution (Lawrence, Kans) 1993;47:906–914. doi: 10.1111/j.1558-5646.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 26.McPhail JD, Lindsey CC. Bull Fish Res Board Can. 1970;173:1–381. [Google Scholar]
- 27.Bell MA, Baumgartner JV, Olson EC. Paleobiology. 1985;11:258–271. [Google Scholar]
- 28.Logan M, Tabin CJ. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- 29.Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Development (Cambridge, UK) 2003;130:45–55. doi: 10.1242/dev.00192. [DOI] [PubMed] [Google Scholar]
- 30.Leiner M. Z Tierpsychol. 1940;4:167–169. [Google Scholar]
- 31.Wilz KJ. Copeia. 1970;1970:587–590. [Google Scholar]
- 32.van Oordt GJ. Proc Sect Sci Koninklijke Akad Wetenschappen; 1925. pp. 470–474. [Google Scholar]
- 33.Kobayasi H. J Hokkaido Gakugei Univ. 1959;10:385–405. [Google Scholar]
- 34.Levin DA. Evolution (Lawrence, Kans) 1970;24:613–624. doi: 10.1111/j.1558-5646.1970.tb01795.x. [DOI] [PubMed] [Google Scholar]
- 35.Nelson JS. J Fish Res Board Can. 1971;28:427–442. [Google Scholar]
- 36.Crawford MJ, Lanctot C, Tremblay JJ, Jenkins N, Gilbert D, Copeland N, Beatty B, Drouin J. Mamm Genome. 1997;8:841–845. doi: 10.1007/s003359900589. [DOI] [PubMed] [Google Scholar]
- 37.Tremblay JJ, Goodyer CG, Drouin J. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- 38.Cole NJ, Tanaka M, Prescott A, Tickle CA. Curr Biol. 2003;13:R951–R952. doi: 10.1016/j.cub.2003.11.039. [DOI] [PubMed] [Google Scholar]
- 39.Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. Development (Cambridge, UK) 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- 40.Sucena E, Stern DL. Proc Natl Acad Sci USA. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parichy DM, Johnson SL. Dev Genes Evol. 2001;211:319–328. doi: 10.1007/s004270100155. [DOI] [PubMed] [Google Scholar]
- 42.Prager EM, Wilson AC. Proc Natl Acad Sci USA. 1975;72:200–204. doi: 10.1073/pnas.72.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 44.Bolnick DI, Near TJ. Evolution (Lawrence, Kans) 2005;59:1754–1767. [PubMed] [Google Scholar]
- 45.Bejder L, Hall BK. Evol Dev. 2002;4:445–458. doi: 10.1046/j.1525-142x.2002.02033.x. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka M, Hale LA, Amores A, Yan YL, Cresko WA, Suzuki T, Postlethwait JH. Dev Biol. 2005;281:227–239. doi: 10.1016/j.ydbio.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Cohn MJ, Tickle C. Nature. 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- 48.Thewissen JG, Cohn MJ, Stevens LS, Bajpai S, Heyning J, Horton WE., Jr Proc Natl Acad Sci USA. 2006;103:8414–8418. doi: 10.1073/pnas.0602920103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CR, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 50.Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 51.Abel O. Abh K K Geol Reichsanstalt. 1904;19:1–223. [Google Scholar]
- 52.Fagone DM, Rommel SA, Bolen ME. Fla Sci. 2000;63:177–181. [Google Scholar]
- 53.Goodyer CG, Tremblay JJ, Paradis FW, Marcil A, Lanctot C, Gauthier Y, Drouin J. Neuroendocrinology. 2003;78:129–137. doi: 10.1159/000072794. [DOI] [PubMed] [Google Scholar]
- 54.Stern DL. Nature. 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sucena E, Delon I, Jones I, Payre F, Stern DL. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 56.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 57.Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 58.Gompel N, Carroll SB. Nature. 2003;424:931–935. doi: 10.1038/nature01787. [DOI] [PubMed] [Google Scholar]
- 59.Bricelj VM, Connell L, Konoki K, Macquarrie SP, Scheuer T, Catterall WA, Trainer VL. Nature. 2005;434:763–767. doi: 10.1038/nature03415. [DOI] [PubMed] [Google Scholar]
- 60.Rosenblum EB, Hoekstra HE, Nachman MW. Evolution (Lawrence, Kans) 2004;58:1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 61.Stern DL. Evolution (Lawrence, Kans) 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 62.Bell MA, Harris EI. Copeia. 1985;1985:789–792. [Google Scholar]
- 63.Swarup H. J Embryol Exp Morphol. 1958;6:373–383. [PubMed] [Google Scholar]
- 64.McGowan C. Dinosaurs, Spitfires, and Sea Dragons. Cambridge, MA: Harvard Univ Press; 1991. [Google Scholar]