Abstract
Biological molecular motors have a number of unique advantages over artificial motors, including efficient conversion of chemical energy into mechanical work and the potential for self-assembly into larger structures, as is seen in muscle sarcomeres and bacterial and eukaryotic flagella. The development of an appropriate interface between such biological materials and synthetic devices should enable us to realize useful hybrid micromachines. Here we describe a microrotary motor composed of a 20-μm-diameter silicon dioxide rotor driven on a silicon track by the gliding bacterium Mycoplasma mobile. This motor is fueled by glucose and inherits some of the properties normally attributed to living systems.
Keywords: glucose, micro actuator, motor protein, nanobiotechnology, Mycoplasma gliding
Nature provides numerous examples of nanometer-scale molecular machines. In particular, motor proteins, which efficiently convert chemical energy into mechanical work, are fascinating examples of functional nanodevices derived from living systems (1). The molecular mechanism underlying the function of these motors has long been a major focus of biophysical research, and the information emerging from those studies should greatly aid in the design and fabrication of novel synthetic micro/nanomotors (2).
At the same time, more application-oriented researchers have initiated projects with the goal of fabricating hybrid micro/nanoactuators driven by biological molecules (3–10). Kinesin, myosin, dynein, and F1-ATPase are well understood motor molecules with the potential to generate mechanical power to drive future microdevices. For instance, earlier in vitro motility systems making use of kinesin or myosin immobilized on cover slips enabled microscopic observation of the random movements of microtubules or actin filaments generated by these motor molecules (11, 12). Those movements did no useful work, however. Thus, a key technology required for the development of hybrid devices using motor proteins is a method to construct ordered systems involving isolated motor proteins in artificial environments. Also, many researchers, including ourselves, have presented preliminary solutions to this problem by integrating motor proteins into lithographic micro- or nanostructures made of inorganic materials, e.g., narrow channels that define the movements of microtubules driven by kinesin (4, 7, 13, 14).
Turning an eye to higher-order biological structures reveals many examples of excellent mechanical devices, including bacterial and eukaryotic flagella and muscle sarcomeres. These motile units are tens of nanometers to several micrometers in size and consist of multiprotein complexes built up with atomic accuracy through the self-assembly and self-organization of protein molecules within cells. In general, these devices work far more efficiently and intelligently than the isolated proteins but, because the principles and mechanisms of self-assembly are only vaguely understood, we are currently unable to assemble higher-order motile units from the isolated component proteins outside the cells. Consequently, research aimed at developing hybrid devices using biological motile units is rare at present.
Mycoplasma mobile, a species of gliding bacteria, is another example of a higher-order unit (cells in this case) with superb motility (15). M. mobile has a pear-shaped cell body ≈1 μm in length and moves continuously over solid surfaces at speeds up to 2–5 μm per second. The mechanism by which it glides remains unknown, although a mechanical walking model that makes use of the rod-like structures protruding from the cell surface has been proposed (16–18). Although three proteins have been identified as essential for gliding, we speculate that this motile system may need a dozen additional proteins, including various cytoskeletal proteins. As a result, it is currently impractical, if not impossible, to reconstitute fully functional motile units from the isolated proteins of M. mobile in vitro. For that reason, we have been attempting to construct micromechanical devices using intact M. mobile cells instead of the isolated proteins. A key benefit of this approach is that hybrid devices into which living cells are integrated enable us to take advantage of preassembled excellent motor units that have the potential for self-repair or self-reproduction when damaged.
We previously reported that M. mobile cells tend to glide along lithographical micropatterns, which enabled us to design patterns that control the direction of their movements (19). Furthermore, by chemically modifying the cell-surface proteins, we were able to bind artificial materials such as polystyrene microbeads to the gliding cells. We next attempted to harness the power generated by the cells to carry out useful work in synthetic environments. Here we describe a microrotary motor powered by M. mobile cells, in which a cogwheel-like silicon dioxide rotor rotates on a silicon track in a predefined direction. To the best of our knowledge, a micromechanical device that integrates inorganic materials with living bacteria has not succeeded until this study.
Results and Discussion
Our strategy for constructing microrotary motors driven by the gliding bacterium M. mobile had two basic components (Fig. 1). One was to induce a majority of the M. mobile cells within the circular tracks to circle in one direction, and the other was to dock a rotor that just fit into the track and bind the rotor to the circling M. mobile cells. M. mobile cells tend to move along the bottom of lithographic walls, and we were previously able to take advantage of this property to introduce the cells into circular tracks in an asymmetric manner, resulting in unidirectional circling of the majority of the cells within the tracks (19). Using the same principle, we designed a pattern for microrotary motors so that cells moving randomly within large square depressions in the lithographic patterns were introduced asymmetrically into circular tracks, to which rotors will be docked. Our initial attempts using this pattern did not work well, because, unlike our previous pattern, our new pattern has a large elevated portion around the circular tracks, and cells gliding on the elevated portion kept falling into the tracks and moving in unwanted directions (data not shown). It thus became apparent that we needed to modify the lithographic surfaces such that the cells would not attach to and glide on the elevated portions of the patterns; the movements of the cells would be restricted to the depressions within the patterns.
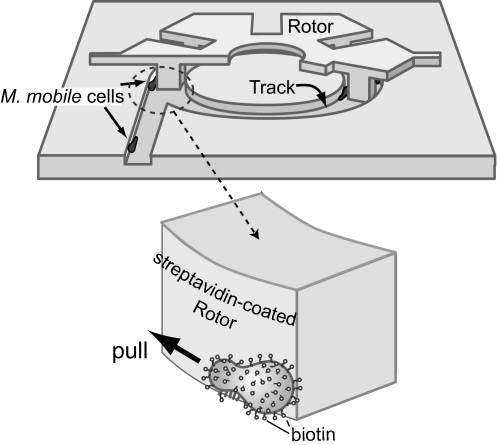
Fig. 1.
Schematic illustrations of a microrotary motor driven by the gliding bacterium M. mobile. The microrotary motor consists of three parts, an Si circular track, an SiO2 rotor whose protrusions just fit into the groove of the track, and living cells of M. mobile that circle unidirectionally within the track. Unidirectional circling of the M. mobile cells is achieved by asymmetric introduction of the cells into the circular track along the track walls (19). The rotor is docked onto the track and binds to the circling M. mobile cells by biotin–streptavidin interactions. As a result, the rotor is pulled by the cells and rotates in one direction.
It has been reported that Mycoplasma pneumoniae, a relative of M. mobile, requires a sialic acid-containing glycoprotein (sialic protein) such as fetuin, laminin, or human CG to adhere to and glide over solid surfaces. This finding suggested that, for these cells to glide, sialic proteins first need to be adsorbed onto a surface, after which the cells adhere to the immobilized proteins by recognizing the sialic acid residue (20). More recently, Nagai and Miyata (21) found that M. mobile also depends on sialic protein for attachment and suggested that, in our previous experiments, sialic proteins present in the culture medium were adsorbed onto the surface, to which M. mobile cells attached. Thus, we decided to take advantage of this property to restrict the movements of M. mobile by selectively depositing sialic protein on the surface of the depressions within the patterns (Fig. 5, which is published as supporting information on the PNAS web site). This deposition of sialic protein was achieved by covalently crosslinking fetuin molecules to the self-assembled monolayer of mercaptoundecanic acid on the gold surface deposited on the bottom of the micropatterned vertical grooves that were prepared as summarized in Fig. 2A. With these improvements (Fig. 3A–D), we were able to induce a majority of the cells in the circular tracks to circle in a clockwise direction (65.7%, n = 134).
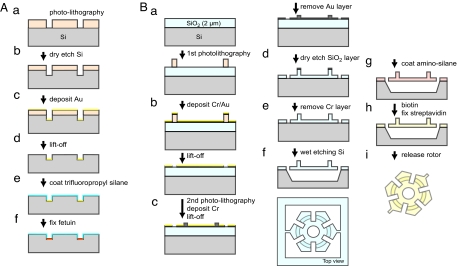
Fig. 2.
Fabrication processes of the microtrack (A) and the rotor (B). See Materials and Methods for details.
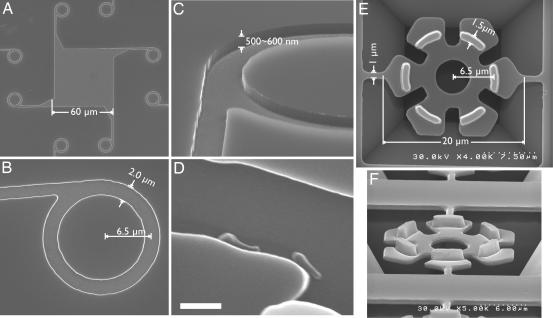
Fig. 3.
Scanning electron micrographs of the Si track (A–D) and the SiO2 rotor (E and F). (A) Overview of the track. When the M. mobile cells were settled onto the substrate, most attached within the square-shaped central depression (60 × 60 μm) created by Si etching and coated with fetuin molecules. A cell moving around in the square would eventually reach a side wall and begin to move along it until it was introduced into a circular track. (B) Enlarged image of the circular track. A straight track extended from the square area is asymmetrically connected to the circular track. A cell moving from the straight track enters the circular track and circles in a clockwise direction, jumping the gap formed by the straight track, guided by the sharp tip. (C) Tilted view of the circular track. The track was steeply carved by anisotropic etching with reactive ion. The depth of the etching was ≈500–600 nm. (D) Two cells are gliding along the side wall of the track. (Scale bar, 1 μm.) (E) Hexagonal microrotors during the process of fabrication (top view). Rotors are tethered to the Si base by two thin bridges designed to break upon sonication, releasing the rotors. (F) Rotors have protrusions that fit the circular grooves shown in A–C (tilted view). The heights of the protrusions are ≈1.4–1.5 μm.
The rotors were fabricated by using a general surface manufacturing process for Si and SiO2 (Fig. 2B). The rotor bodies (20 μm in diameter) were made by reactive ion etching of a 2-μm thermal SiO2 layer on an Si substrate with a Cr mask (Fig. 3E). Then the Si under the rotors was anisotropically etched by wet etching, so that the rotors were tethered to the Si base by two thin (1-μm-wide) bridges designed to break upon sonication. The rotors also have protrusions (1.4 μm tall) designed to fit into the circular groove of the tracks (Fig. 3F). To bind the protrusions to the cells moving in the groove, the entire surface of the rotors was coated with streptavidin. Upon sonication, 20–80% of the rotors were successfully released. Because there were 20,000 rotors on a 5 × 5-mm2 silicon chip, we were routinely able to harvest at least 4,000 rotors from one chip, which was sufficient for the following processes.
Although wild-type M. mobile cells glide normally on glass surfaces to which fetuin is adsorbed, the gliding cells often detached from gold surfaces to which fetuin was covalently crosslinked (data not shown). This observation implies that either the fetuin density on the gold surfaces was lower than that achieved by adsorption, or the fetuin was immobilized such that the majority of sialic acid residues were inaccessible to the cells. Therefore, to minimize detachments of the cells from the fetuin-coated gold substrates, we used a mutant strain of M. mobile that harbors a missense mutation in its gli521 gene (22). This strain moves slower than the wild-type strain (≈2 vs. ≈5 μm per second), but it binds to the substrate more stably. To connect M. mobile cells to the streptavidin-coated rotor, the cell-surface proteins were chemically biotinylated by using biotin-polyethylene glycol-succinimide. This biotin reagent has a long flexible linker composed of polyethylene glycol (stretched length, 20 nm), which increased the collision frequency to the poorly diffusive rotors so that the cells efficiently attached to the rotor when the two came into close proximity.
After biotinylated cells were spread on the patterned surface and began to move along the circular tracks, we placed a rotor onto the track and docked it using a micromanipulator (Fig. 4A). Typically, rotors began to rotate within a few minutes and, as expected, the majority (84%, n = 51) rotated clockwise. Continuous rotations lasting >1 min were observed (Fig. 4B; see Movie 1, which is published as supporting information on the PNAS web site), although the rotations were mostly intermittent. The rotation rate was 1.5–2.6 rpm (Fig. 4C). On rare occasions, a continuously rotating rotor would abruptly reverse direction (see Movie 2e, which is published as supporting information on the PNAS web site), suggesting that the motion was driven by only a few cells. Torque generated by an individual cell circling in this pattern was 1.8 × 10−16 N·m, assuming a stall force of 27 pN (23), so that the torque of each motor is probably in the range of 2–5 × 10−16 N·m. This torque is ≈4 orders of magnitude smaller than that of the electrostatic microactuators of Micro Electro Mechanical Systems (MEMS) (24). A more precise estimate of the torque requires visualization and counting of the cells that are driving the rotation, which is not possible under the current setup. We next estimated the viscous drag in a narrow gap between the rotor and the track in a simplified model, in which a disk with a circular rim rotates above a surface having a circular depressed track onto which the rim fits. We calculated the friction between the disk and the surface and that between the rim and the track separately, assuming the equation:
where r is the distance from the center of rotation, μ is the viscosity of the solution (0.84 × 10−3 Pa·s), ω is the rotational rate, and h is the width of the gap between the object and the surface (500 nm for the disk). Although it is difficult to estimate the gap between the rim and the track surface, when the gap is assumed to be 50 nm, the sum of the two frictional forces was 1.5 × 10−17 N·m (see Supporting Text, which is published as supporting information on the PNAS web site, for details). This is at least 1 order of magnitude smaller than the torque generated by a single M. mobile cell, so the rotational force generated by one or a few M. mobile cell(s) is sufficient to turn the rotor against the friction between the rotor and the surface. Consistent with this, the rotation rate of 1.5–2.6 rpm relates to a linear velocity of 1.2–2.1 μm per second along the grooves where the cells move, which is comparable to the unloaded gliding velocity of the cells on flat surfaces. But if the gap is assumed to be 2.5 nm, the sum of the frictional force is 1.8 × 10−16 N·m, which is comparable to the torque generated by a single cell. So we speculate that the continuous rotating rotor would float at least 2.5 nm above the surface.
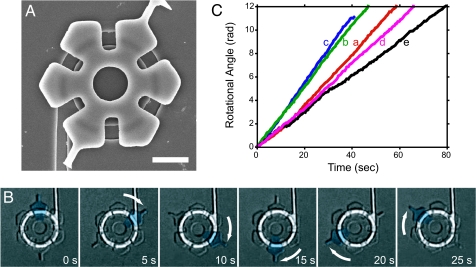
Fig. 4.
Image of a rotor docked on the track and examples of the rotation of a rotor driven by the bacteria. (A) Scanning electron micrograph of a rotor docked onto a circular groove after placement using a micromanipulator. (Scale bar, 5 μm.) (B) Time-lapse photomicrographs of a rotating rotor taken at 5-s intervals. A portion of the rotor was pseudocolored in cyan to enable tracking. This rotor continuously rotated ≈60 degrees in 5 s (2.0 rpm). (C) Rotational speed of individual rotors. The rotational angles of continuously rotating rotors were measured from images captured at 0.5-s intervals and plotted against time. The traces marked a, b, c, and d correspond to the rotors shown in Movie 1 in sections a, b, and c of Movie 2, respectively.
We were often able to initiate rotation of stationary rotors by tapping the microscope or nudging the rotor with a needle attached to the micromanipulator. Apparently, some rotors were unable to move because of a small hitch somewhere in the track. In addition, the density of moving cells appears to be limited by the density of fetuin on the track surfaces. If we can somehow increase the fetuin density or replace fetuin with another protein that has a higher sialic acid content and that binds to the surface more efficiently, larger numbers of cells could drive the rotation of each rotor, resulting in smoother rotation of a larger number of rotors. Moreover, the improved directional uniformity of the cells' movement within the circular tracks should improve the performance of the motor, such as the lifetime and the torque. The directional uniformity of the cells' movement depends on the sharpness of the tip at the entrance of the circular tracks. For instance, in the pattern shown in Fig. 3B, a cell moving to the right along the lower wall of the linear portion would dissociate from the wall at the entrance of the circle, move straight, hit one of the circular walls ahead, and start to circle in the clockwise direction, if the tip at the entrance is sharp. If the tip at the entrance is less sharp, a larger number of cells would make a U turn along the tip and circle in the unwanted counterclockwise direction. In addition, cells circling in the clockwise direction along the outer circular wall would exit the pattern if the tip at the entrance is not sharp. In our previous study, we were able to achieve that 90% of cells moved in one direction within circular tracks, with tips at the entrance whose radius of curvature is 50 nm (19). In this study, we were unable to fabricate such sharp patterns, because the nanolithographic processes are not compatible with the chemical process to fix fetuin onto defined areas, resulting in the compromised directional uniformity (65%). Future improvements in lithographic process or the track geometry should solve this problem.
Recently, several groups reported attempts to integrate living organisms into micromechanical devices in efforts to realize microrobots or microtransport systems. Xi et al. (25) succeeded in developing microrobots whose locomotion was driven by cardiomyocytes attached to specific sites within beam-like microstructures made of SiO2; the periodic contraction of the cells flexed the structures and powered the locomotion. In addition, Darnton et al. (26) suggested that a solid–fluid interface activated by attaching the flagellated bacterium Serratia marcescens to a solid surface has the potential to serve as a pump or mixer in microfluidic systems. More recently, Weibel et al. (27) presented a prototype of a microtransport system in which cells of the phototactic flagellated alga Chlamydomonas reinhardtii served as microtransporters or “microoxen,” moving attached microobjects toward a light source through a channel made of polydimethylsiloxane.
It is noteworthy that current nanobiotechnological methods are far too immature to reconstitute such functions from isolated motor proteins. For instance, the sarcomeric structures of cardiomyocytes necessary for efficient contractions cannot be reconstructed in vitro from isolated components. Likewise, the rotation of bacterial flagella is driven by membrane potential, and the beating of eukaryotic flagella is driven by the interaction of the motor protein dynein with microtubules in 9 + 2 arrangements, neither of which can be reconstituted in vitro from isolated proteins. Moreover, these motile systems have other important functions, including regulatory mechanisms necessary for oscillatory beating of cardiomyocytes or phototaxis of Chlamydomonas. To design and assemble those functions in vitro, which is likely indispensable for realization of intelligent devices, would be even more challenging than merely reconstructing the motor system. By contrast, it is easy to obtain enormous numbers of living motile cells by self-reproduction in a simple nutrient medium within a few days, without any laborious purification processes.
To take further advantage of live cells as microtransporters, the next challenge is to encode useful properties genetically. For instance, it would be useful to genetically modify the surface proteins of M. mobile to enable easy linkage to cargo, such as using the in vivo biotinylation system, or to implement chemotactic regulatory systems, so that the cells migrate directionally by sensing a chemical clue. In that sense, it is fortunate that the mycoplasma genome is one of the simplest (28). On the other hand, the potential biohazard represented by genetically engineered microorganisms should be considered seriously if they are to be used outside the laboratory. In that regard, Uenoyama and Miyata (22) recently succeeded in making gliding M. mobile ghosts by partially dissolving the cell membrane using Triton X-100. These ghosts are not alive, but the motor units are still active, and they glide at the same speed as the intact cells if adenosine triphosphate is supplied exogenously. One might consider using such ghosts as preassembled supramolecular motor units with superb performance to circumvent potential biohazard problems.
One day, combinations of MEMS or lithographic materials with nanocomponents, including synthetic molecular machines and protein devices, may be used to construct hybrid microsystems with diverse functions. However, we still do not have efficient methods for assembling complex molecular machines on lithographic materials. Using living cells or organelles as preassembled functional units with the potential for self-reproduction and self-repair circumvents some of these problems, and our hybrid motor represents a step in that direction.
Materials and Methods
Strain and Culture.
A mutant strain of M. mobile that harbors a missense mutation in its gli521 gene (22) was used in this study. The cells were cultured in Aluotto medium (2.1% heart infusion broth/0.56% yeast extract/10% horse serum/0.025% thallium acetate/0.005% ampicillin) at 25°C (29).
Biotinylation of M. mobile Cells.
After collecting M. mobile cells by centrifugation and resuspending them in PBS, their surface proteins were functionalized by chemical modification induced by exposing the cells to 25–100 μM biotin-polyethylene glycol-succinimide (Nektar, Huntsville, AL) for 15 min at room temperature. The cells were then washed in fresh PBS containing 50 mM glucose. The extent of the biotinylation reaction was qualitatively checked by staining the cells with fluorescent Alexa-594-conjugated streptavidin (Molecular Probes, Eugene, OR). The biotinylation did not affect their gliding speed (19).
Fabrication of the Track.
The fabrication process is shown schematically in Fig. 2A. In Fig. 2Aa, after applying a coating of hexamethyldisilazane (HMDS) to promote adhesion, a 1.2-μm-thick layer of photoresist AZ5214E (Clariant, Tokyo, Japan) was spin-coated onto a silicon (Si) substrate. The sample was then baked at 90°C for 2 min and exposed to UV light through a photo mask (Fig. 6A, which is published as supporting information on the PNAS web site). A postexposure bake at 120°C for 1 min was followed by a flood exposure to UV light, after which the sample was developed for 1 min with 2.38% tetramethylammonium hydroxide. In Fig. 2Ab, the resist patterns were transferred onto the silicon substrate by using reactive-ion etching with SF6/CF4 gas. The flow rates of the SF6 and CF4 were 2.5 and 5 sccm, respectively, and the pressure was 1 Pa. The etching depth was 500–600 nm. Thereafter (Fig. 2Ac), a 15-nm-thick layer of chromium (Cr) and a 30-nm-thick layer of gold (Au) were deposited on the substrate by vacuum evaporation. The Au/Cr layer was then lifted off in acetone (Fig. 2Ad), leaving a gold surface (Au/Cr layer) at the bottom of the troughs. After removing the HMDS layer using an oxygen plasma asher (Fig. 2Ae), the silicon surface was silanized by soaking the substrate overnight in 3% trifluoropropyl ethoxyl silane dissolved in toluene, after which it was baked at 120°C for 5 min. To define areas where M. mobile cells were allowed to glide (Fig. 2Af), we selectively immobilized the sialic protein fetuin, which M. mobile cells require to glide, on the gold surface as follows. First, the sample was soaked overnight in ethanol containing 10 mM 11-mercaptoundecanoic acid and then rinsed with ethanol. The carboxyl residues of the self-assembled monolayer of mercaptoundecanoic acid on the Au surface were activated by soaking the substrate in 2% 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide in phosphate buffer (pH 4.0) for 30 min, after which fetuin was bound by soaking the substrate for 2 h in a solution of 0.5 mg/ml fetuin in borate buffer (pH 8.0) with 0.001% Pluronic F127 (BASF, Mount Olive, NJ). Pluronic F127 is a surfactant and effectively reduced nonspecific adsorption of fetuin onto silanized surfaces. The reaction was stopped by addition of 5 mM 2-aminoethanol. The sample was stable for at least 1 week in PBS at 4°C.
Fabrication of the Rotor.
The rotor was fabricated in a 2.0-μm thermal SiO2 layer on an Si substrate <001>. The fabrication process is shown schematically in Fig. 2B. The rotor body patterns (Figs. 2B a and b and 6B) were transferred to the Cr/Au layer (20 nm of Cr, 25 nm of Au) on the silicon substrate by using the liftoff process. The Cr layer served as a mask for the dry etching of the SiO2 layer mentioned below, whereas the Au layer enabled us to easily align the pattern during the second photolithographic process. To make the protrusions on the rotor (Fig. 2Bc), which fit into the grooves of the tracks, photolithographic patterns were made on the substrate, which already had the rotor body patterns, using the photomask illustrated in Fig. 6C. The Au layer was etched by soaking the sample for 10 s in Au etchant (5.2% potassium iodide/2.8% iodine) after removing the resist residue with an oxygen plasma asher. Then an additional Cr pattern (60 nm thick) was defined on the sample by using the liftoff process, followed by removal of the remaining Au layer. The Cr patterns (Fig. 2Bd) were transferred to the SiO2 layer by reactive ion etching for 24 min (SF6 gas flow rate, 25 sccm; pressure, 5 Pa; plasma power, 40 W), which completely etched the unmasked regions. The heights of the fabricated protrusions were 1.4–1.5 μm. The residual Cr layer (Fig. 2Be) was removed with a Cr etchant [7.5% HClO4/12.1% (NH4)2Ce(NO3)6/80.4% H2O]. The Si layer under the rotor (Fig. 2Bf) was then etched with an Si etchant (320 mg of pyrocatechol/6 mg of pyrazine/320 μl of H2O/1 ml of ethylendiamine) for 5 min at 110°C. Because this wet etching process is not isotropic, the holes designed in the rotor are necessary to efficiently etch the Si portion under the rotor.
Streptavidin Coating and Harvesting of the Rotor.
To connect the rotor and the biotinylated M. mobile cells, the rotor surface was coated with streptavidin by using the process shown schematically in Fig. 2B g–i. The rotors, tethered to the silicon substrate (Fig. 2Bg), were silanized by soaking them for 2 h in a solution of aminopropyl triethoxyl silane (3% aminopropyl triethoxyl silane/2% acetic acid/5% water/90% ethanol), then rinsed with ethanol, dried with nitrogen gas, and baked at 90°C for 5 min. The amino-coated rotors (Fig. 2Bh) were then reacted with 1 mM succinimidyl-6′-(biotinamido)-6-hexanamido hexanoate (EZ-Link NHS-LC-LC-biotin; Pierce, Rockford, IL) dissolved in 40 mM phosphate buffer (pH 8.0) for 1 h at 37°C. After rinsing with PBS, the substrate was soaked in a solution of 0.02 mg/ml Alexa-488-conjugated streptavidin for 30 min. Finally (Fig. 2Bi), a 5 × 5-mm2 Si chip having rotors was dipped into 300–500 μl of solution in a 1.5-ml microtube and sonicated by inserting the probe of a 70-W hand-held ultrasonic processor at 40% of the maximum power for 10–30 s. The released rotors were collected by centrifugation. The streptavidin coating was checked under a fluorescence microscope (Fig. 7, which is published as supporting information on the PNAS web site).
Docking the Rotor onto the Track and Rotation.
Biotinylated cells suspended in medium containing 0.001% Pluronic F127 were perfused into a flow chamber constructed by using a track substrate (5 × 5 mm2) with spacers that was inverted on a glass slide and were incubated for 15 min to allow attachment onto the track. The chamber was then rinsed with PBS containing 50 mM glucose to remove unattached cells. Next, the substrate was detached from the flow chamber and set on a holder constructed on a glass slide such that the patterned surface faced up. PBS containing the released rotors was then dripped onto the track substrate, and the rotors were allowed to settle onto the track surface. Using a micromanipulator and a microscope equipped with a LUMPlanFL/IR ×60/0.90-W water immersion objective (Olympus, Tokyo, Japan), a rotor was moved to a point above a circular track and then docked by fitting the protrusions of the rotor into the groove of the track (Fig. 7C). Initially, this operation was often hindered by an unknown adhesive material in the solution, but the problem was alleviated by adding 1 μg/ml DNase (DNase I) and 10 mM magnesium chloride to the buffer solution.
Supplementary Material
Acknowledgments
We thank Mr. Wei-Heong Tan for advice on the estimation of friction and Dr. Shoji Takeuchi for helpful discussion. This work was supported in part by the Talent in Nanobiotechnology Course, Promotion Budget for Science and Technology (Y.H. and T.Q.P.U.) and by grants-in-aid for Scientific Research of Priority Areas (to M.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vale RD, Milligan RA. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 2.Balzani V, Credi A, Raymo FM, Stoddart JF. Angew Chem Int Ed. 2000;39:3349–3391. doi: 10.1002/1521-3773(20001002)39:19<3348::aid-anie3348>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 3.Hess H, Vogel V. J Biotechnol. 2001;82:67–85. doi: 10.1016/s1389-0352(01)00029-0. [DOI] [PubMed] [Google Scholar]
- 4.Hiratsuka Y, Tada T, Oiwa K, Kanayama T, Uyeda TQP. Biophys J. 2001;81:1555–1561. doi: 10.1016/S0006-3495(01)75809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limberis L, Magda JJ, Stewart RJ. Nano Lett. 2001;1:277–280. [Google Scholar]
- 6.Böhm KJ, Stracke R, Mühlig P, Unger E. Nanotechnology. 2001;12:238–244. [Google Scholar]
- 7.Jia LL, Moorjani SG, Jackson TN, Hancock WO. Biomed Microdevices. 2004;6:67–74. doi: 10.1023/b:bmmd.0000013368.89455.8d. [DOI] [PubMed] [Google Scholar]
- 8.Soong RK, Bachand GD, Neves HP, Olkhovets AG, Craighead HG, Montemagno CD. Science. 2000;290:1555–1558. doi: 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- 9.Liu H JJ S, Bachand GD, Rizk SS, Looge LL, Hellinga HW, Montemagno CD. Nat Mater. 2002;1:173–177. doi: 10.1038/nmat761. [DOI] [PubMed] [Google Scholar]
- 10.Limberis L, Stewart RJ. Nanotechnology. 2000;11:47–51. [Google Scholar]
- 11.Vale RD, Reese TS, Sheetz MP. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kron SJ, Spudich JA. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess H, Clemmens J, Qin D, Howard J, Vogel V. Nano Lett. 2001;1:235–239. [Google Scholar]
- 14.van den Heuvel MG, Butcher CT, Smeets RM, Diez S, Dekker C. Nano Lett. 2005;5:1117–1122. doi: 10.1021/nl0506554. [DOI] [PubMed] [Google Scholar]
- 15.Rosengarten R, Kirchhoff H. J Bacteriol. 1987;169:1891–1898. doi: 10.1128/jb.169.5.1891-1898.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uenoyama A, Kusumoto A, Miyata M. J Bacteriol. 2004;186:1537–1545. doi: 10.1128/JB.186.5.1537-1545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seto S, Uenoyama A, Miyata M. J Bacteriol. 2005;187:3502–3510. doi: 10.1128/JB.187.10.3502-3510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyata M. In: Mycoplasmas: Molecular Biology, Pathogenicity, and Strategies for Control. Blanchard A, Browning G, editors. Norwich, UK: Horizon Bioscience; 2005. pp. 137–163. [Google Scholar]
- 19.Hiratsuka Y, Miyata M, Uyeda TQP. Biochem Biophys Res Commun. 2005;331:318–324. doi: 10.1016/j.bbrc.2005.03.168. [DOI] [PubMed] [Google Scholar]
- 20.Roberts DD, Olson LD, Barile MF, Ginsburg V, Krivan HC. J Biol Chem. 1989;264:9289–9293. [PubMed] [Google Scholar]
- 21.Nagai R, Miyata M. J Bacteriol. 2006;188:6469–6475. doi: 10.1128/JB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uenoyama A, Miyata M. Proc Natl Acad Sci USA. 2005;102:12754–12758. doi: 10.1073/pnas.0506114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyata M, Ryu WS, Berg HC. J Bacteriol. 2002;184:1827–1831. doi: 10.1128/JB.184.7.1827-1831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark KC, Azzam Yasseen A, Phillips SM, Mehregany M. Sensors 2002 Proc IEEE. 2002;2:1751–1756. [Google Scholar]
- 25.Xi J, Schmidt JJ, Montemagno CD. Nat Mater. 2005;4:180–184. doi: 10.1038/nmat1308. [DOI] [PubMed] [Google Scholar]
- 26.Darnton N, Turner L, Breuer K, Berg HC. Biophys J. 2004;86:1863–1870. doi: 10.1016/S0006-3495(04)74253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weibel DB, Garstecki P, Ryan D, DiLuzio WR, Mayer M, Seto JE, Whitesides GM. Proc Natl Acad Sci USA. 2005;102:11963–11967. doi: 10.1073/pnas.0505481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razin S, Yogev D, Naot Y. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aluotto BB, Wittler RG, Williams CO, Faber JE. Int J Syst Bacteriol. 1970;20:35–58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.