Abstract
Functional MRI (fMRI) can be applied to study the functional connectivity of the human brain. It has been suggested that fluctuations in the blood oxygenation level-dependent (BOLD) signal during rest reflect the neuronal baseline activity of the brain, representing the state of the human brain in the absence of goal-directed neuronal action and external input, and that these slow fluctuations correspond to functionally relevant resting-state networks. Several studies on resting fMRI have been conducted, reporting an apparent similarity between the identified patterns. The spatial consistency of these resting patterns, however, has not yet been evaluated and quantified. In this study, we apply a data analysis approach called tensor probabilistic independent component analysis to resting-state fMRI data to find coherencies that are consistent across subjects and sessions. We characterize and quantify the consistency of these effects by using a bootstrapping approach, and we estimate the BOLD amplitude modulation as well as the voxel-wise cross-subject variation. The analysis found 10 patterns with potential functional relevance, consisting of regions known to be involved in motor function, visual processing, executive functioning, auditory processing, memory, and the so-called default-mode network, each with BOLD signal changes up to 3%. In general, areas with a high mean percentage BOLD signal are consistent and show the least variation around the mean. These findings show that the baseline activity of the brain is consistent across subjects exhibiting significant temporal dynamics, with percentage BOLD signal change comparable with the signal changes found in task-related experiments.
Keywords: functional connectivity, functional MRI, resting fluctuations
Typical functional MRI (fMRI) research focuses on the change in blood oxygenation level-dependent (BOLD) signal caused by the neural response to an externally controlled stimulus/task. The fMRI signal during “on” periods is contrasted with recordings during a baseline or control condition, resulting in the relative signal change because of the specific process being studied. Recently, increased attention has been directed at investigating the features of the baseline state of the brain. Of particular interest are low-frequency fluctuations (≈0.01–0.1 Hz) observed in the BOLD signal, which have been found to display spatial structure comparable to task-related activation (1–3). There is an ongoing discussion as to whether these fluctuations in the BOLD signal predominantly reflect changes of the underlying brain physiology independent of neuronal function (4–6), or instead reflect the neuronal baseline activity of the brain when goal-directed neuronal action and external input are absent (7, 8). The view that coherencies in resting fluctuations represent functional resting-state networks linked to underlying neuronal modulations is consistent with the appearance of these coherencies within cortical gray matter areas of known functional relevance. For example, one of the first studies of resting fluctuations identified the motor network (9). More recent studies have identified associated fluctuations in brain regions involved in visual, motor, language, and auditory processing (10–17). Brain regions that show greater BOLD signal during rest than during any one of a broad range of experimental tasks have also received attention. The default-mode network has been hypothesized to be active during rest and suspended/deactivated when specific goal-directed behavior is needed, as demonstrated in a task-related positron-emission tomography study (18). Connectivity has been reported between regions in the brain that form the default-mode network during resting states as well as inverse correlations among prefrontal regions (which show increased activity during a cognitive task) and the posterior cingulate cortex (an area within the default-mode network) (16, 17). However, although the observation that these resting fluctuations are located in gray matter is consistent with the notion of their representing neuronal modulations, a recent study shows a correlation between changes in respiration and BOLD signal also located in gray matter areas (6).
In this article, we focus on the following questions: (i) How many coherent spatiotemporal patterns can we distinguish? (ii) How strong are these fluctuations? and (iii) How consistent are these fluctuations across subjects and sessions? To infer these signal coherencies, most studies apply a region-of-interest cross-correlation analysis approach (9–13), where the spatial characteristics of these resting fluctuations are estimated by using correlation analysis against a reference time course derived from secondary recordings (19) or the resting data itself (seed-voxel-based correlation analysis) (9). More recently, some studies employed a model-free analysis by using independent component analysis (ICA) (14, 15, 20) instead of time-course regression. Such decompositions are of particular importance because they allow for a simultaneous separation into individual maps. These decompositions can simultaneously extract a variety of different coherent resting networks and separate such effects from other signal modulations such as those induced by head motion or physiological confounds, such as the cardiac pulsation or the respiratory cycle (13, 15).
ICA-based studies have identified components that resemble several functionally relevant cortical networks such as visual and auditory cortical areas as well as the default-mode network. Different studies have identified qualitatively similar areas of functional coherence across subjects, but the extent to which these fluctuations are consistent within a population has not previously been quantified. To characterize this level of consistency between and within subjects, it is necessary to employ techniques designed for the analysis of multisession/multisubject fMRI data. A promising method for the investigation of coherent signals at a group level is the recently described tensor probabilistic ICA (PICA) (21). This analysis simultaneously decomposes group fMRI data into modes describing variations across space, time, and subjects. It has been demonstrated that the tensor-PICA approach can provide useful representations of group fMRI data in task-related fMRI experiments and that it also seems capable of analyzing resting-state studies (21). In this study, we apply tensor-PICA to resting-state fMRI data with the aim of identifying resting coherencies that are consistent across subjects and sessions. To quantify the consistency of such patterns, we used bootstrapping. Based on 20 resting data sets, we generated 100 surrogate multisubject data sets from which we estimated these fluctuations. Using these separate estimates, we quantified common networks in terms of their expected percentage BOLD signal change, which provides a measure of the dynamics of these fluctuations. In addition, we calculated the amount of typical percentage variation around the expected percentage BOLD signal change at each voxel's location, which encodes the uncertainty of estimating any given voxel as part of an associated coherent network.
Results
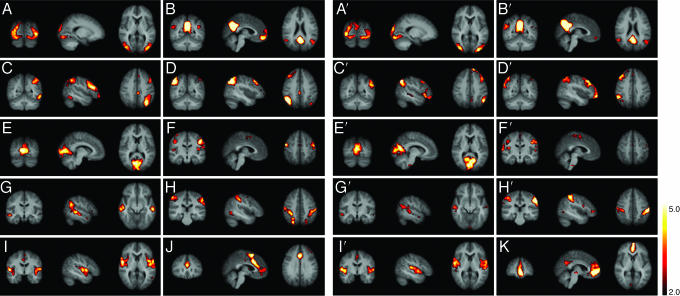
In the initial analyses, two data sets (10 subjects scanned at rest, and the same set of subjects scanned on a separate occasion) were analyzed separately. These analyses resulted in 10 components per data set, showing variations in the frequency, spatial, and subject domains. Spatial maps of the coherent resting fluctuations of both analyses are shown in Fig. 1. The 10 components showed low-frequency variations in time (mean peak frequency: 0.015 Hz; range 0.005–0.030 Hz) and can be described as follows. Fig. 1 A and A′ shows a pattern that consists predominantly of the peristriate area, and lateral and superior occipital gyrus [Brodmann area (BA) 19], which are areas recognized as part of the visual cortex. Fig. 1 B and B′ shows a cluster consisting of the prefrontal (BA 11), anterior cingulate (BA 32), posterior cingulate (BA 23/31), the inferior temporal gyrus (BA 20/37), and the superior parietal region (BA 7), known as the default-mode network as described by Raichle et al. (18) and Greicius et al. (17). Hippocampal involvement in this component, as described by Greicius et al. (22), is not found. Fig. 1 C, C′, D, and D′ shows components that are predominantly in the left (C and C′) and right (D and D′) hemispheres, the middle frontal and orbital (BA 6/9/10), superior parietal (BA 7/40), middle temporal gyrus (BA 21), and the posterior cingulate (BA 23/31; C and C′ only). These are the only components that show strong lateralization and are areas known to be involved in memory function. Fig. 1 E and E′ encompasses part of the striate and parastriate (BA 17/18). The visual cortex is apparent in two separate components. The more lateral visual areas are in Fig. 1 A and A′, and the more medial visual areas are in this figure. Fig. 1 F and F′ shows the pre- and postcentral gyri (BA 1/2/3/4) in one component, representing the motor and sensory network. Fig. 1 G and G′ shows the superior temporal (BA 22) area as the main element of this spatial map. Involvement of the cingulate (BA 23) and superior frontal (BA 9/10) areas is also seen. This cluster of brain regions bears a strong resemblance to the occipitotemporal pathway (ventral stream). Fig. 1 H and H′ involves mainly the superior parietal cortex (BA 7) with additional involvement in the occipitotemporal (BA 37) and precentral (BA 4) areas. Fig. 1 I and I′ involves the superior temporal (BA 22) and insular and postcentral cortex (BA 1/2), which are areas acknowledged to represent the auditory cortex.
Fig. 1.
Tensor-PICA estimated resting patterns of the first (A–J) and second (A′–I′ and K) multisubject data sets: coronal, sagittal, and axial view of spatial map for each component. A–I and A′–I′ show components found in both data sets. J and K components are unique to their data sets. Images are z statistics overlaid on the average high-resolution scan transformed into standard (MNI152) space. Black to yellow are z values, ranging from 2.0 to 5.0. The left hemisphere of the brain corresponds to the right side of the image.
All of the spatial maps mentioned above are found in both data sets. Each separate analysis, however, also yielded a component that was not present in the other. In the analysis of the first scan, the following component was found: Fig. 1J shows a cluster consisting of the frontopolar area (BA 10), prefrontal cortex (BA 11), dorsal anterior cingulate (BA 32), and superior parietal cortex (BA 7). This predominantly frontal spatial map has been found in earlier research and is proposed to be involved in executive control and working memory function (14, 23). In the analysis of the second scan, another component was found: Fig. 1K shows the prefrontal (BA 11), dorsal anterior cingulate (BA 32), and posterior cingulate (BA 23/31) regions, which seem to be part of the default-mode network shown in Fig. 1B. A combination of the map in Fig. 1K and the map in Fig. 1B′ resembles the map in Fig. 1B.
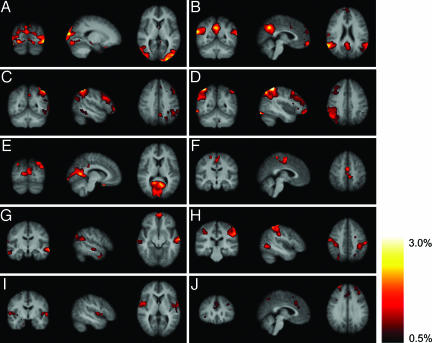
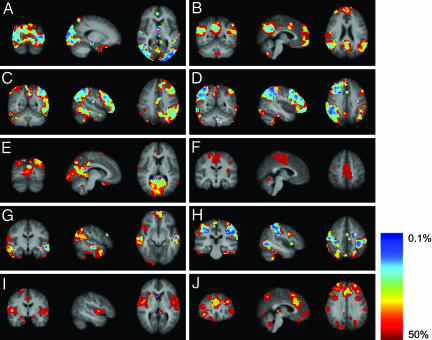
The results shown in Fig. 1 suggest that 9 of the 10 components are qualitatively spatially consistent. To quantify the consistency, the tensor-PICA analysis was repeated 100 times on surrogate data sets (see Data Analysis). The outcome was averaged, resulting in 10 mean maps of coherent resting fluctuations, presented here in terms of percentage BOLD signal change (Fig. 2). All components except K are reproducible in this analysis. In these maps of coherent resting fluctuations, the average percentage BOLD signal fluctuation reaches values of up to 3%. Map B, for example, shows that in the default-mode network, >2% signal fluctuation can be found in the posterior cingulate and superior parietal areas, whereas in map D, ≈3% BOLD signal fluctuation can be seen in the superior parietal region. Fig. 3 shows the mean variability (across different surrogate data sets) per component on a scale from 0.1% to 50% around this estimated percentage BOLD signal change. Variation of these coherent resting fluctuations differs among and within components. For example, map A shows less variation overall than map F. Also, as shown in map B, the posterior areas are more consistent than the anterior areas. When comparing Figs. 2 and 3, it is noticeable that, in general, areas with a relatively high mean percentage BOLD signal change are also the areas that are the most consistent, i.e., show the least variation around this mean.
Fig. 2.
Mean (across 100 surrogate multisubject data sets) tensor-PICA estimated resting patterns: coronal, sagittal, and axial view of spatial map for each component. Images are percentage BOLD signal change, overlaid on the average high-resolution scan transformed into standard (MNI152) space. Black to yellow is percentage signal change, ranging from 0.5% to 3.0%.
Fig. 3.
Maps of coefficient of variation: coronal, sagittal, and axial view of spatial map for each component. Images are percentage variation around the mean percentage BOLD signal change, overlaid on the average high-resolution scan transformed into standard (MNI152) space. Red denotes much variation around the mean; blue denotes little variation.
Conclusion and Discussion
In this article, we show a variety of coherent low-frequency fluctuations in the BOLD signal identified across subjects. In addition to being consistent, these coherent fluctuations also seem to be very dynamic. The percentage BOLD signal change in these areas can reach levels as high as 2–3%, and areas with high mean percentage BOLD signal change also show high levels of consistency.
The coherent resting fluctuations presented in this article include functionally relevant regions involved in motor function, visual processing, executive functioning, auditory processing, memory, and the default-mode network (encompassing attention and/or even consciousness). This finding is in line with previous research, which has consistently shown coherent fluctuations in the BOLD signal within specific neuroanatomical systems (9, 12, 14, 15, 17). As in previous studies, we denote these regional coherent fluctuations as representing a “network.” Whether these fluctuations are of neuronal origin or reflect changes in local physiology remains unclear. The most direct evidence of an association between some of these fluctuations and neuronal activity comes from the observed correlation between the BOLD signal and cortical electrical activity in studies employing simultaneous fMRI and electroencephalograms (19, 24) and the observation of change in these networks resulting from neurological disease (22). However, other studies suggest that these fluctuations in the BOLD signal primarily reflect changes in underlying brain physiology independent of neuronal function (4–6). A number of the networks presented in this study were already recognized in earlier connectivity studies aimed at those specific regions, for example, the motor cortex (9) and the visual cortex (12, 19). Besides areas involved in sensory-related processing such as the auditory and visual cortex, regions involved in higher cognitive functioning are also represented by separate networks. Some networks involve groups of areas known to activate together in task-related fMRI. For example, components C and D encompass areas such as the dorsolateral–prefrontal cortex and superior parietal cortex, i.e., areas that have repeatedly been found to be active during memory tasks (25–27). In this analysis, these networks show very little spatial variation around the mean percentage BOLD signal change (see Fig. 3). An interesting aspect of components C and D is that these are the only ones to show a similar activation pattern in opposite hemispheres in separate components. This result could potentially reflect hemispheric specialization in memory function. Map B, which includes the cingulate cortices, inferior parietal, and medial prefrontal regions, has consistently demonstrated deactivation in spatial maps of task-related activation (17, 28–31). This component is now commonly referred to as the default-mode network because it seems to be active during rest and suspended when specific goal-directed behavior is needed (7, 18). The existence of this network during resting state has already been validated in several studies (14, 15, 17), and it shows great resemblance to the brain areas found to be involved in random episodic silent thinking, as described by Andreasen et al. (32). The appearance is consistent across samples, but within the network the posterior parts show less variation than the anterior parts (see Fig. 3). This result possibly explains the separation of this component in the tensor-PICA analysis of the second original data set in components B′ and K in Fig. 1.
Ten different and reproducible components are presented in this article. This division into separate systems potentially reflects the organization of the human brain, although we do not suggest that the brain consists of disconnected functional resting networks. In this article, components are defined as spatially independent areas showing coherence in low-frequency fluctuations. This by no means excludes the notion that there are interactions possible between different resting patterns and that the human brain demonstrates properties of a small-world network, as described by Achard et al. (3). The number of identifiable patterns is to some extent determined by the limitations of current imaging and analysis sensitivity and specificity. Additional improvements in modeling and/or data acquisition might result in a more fine-grained characterization of such functional connectivity.
In this article, we apply a technique that identifies multiple, separate, coherent resting fluctuations in group fMRI data. Compared with single time course correlation approaches, an ICA approach does not require predefined regions of interest or the identification of a seed voxel location. One of the main advantages of an ICA approach over simple regression approaches is that multiple regression techniques like ICA can separate resting fluctuations from other structured noise-related signal variations, such as those induced by head motion or cardiac and respiratory pulsations. The fact that the decomposition in this case is determined by the spatial signal characteristics (optimization for spatially independent components) also implies that these resting fluctuations can be separated from cardiac or respiratory physiology despite the fact that the typical echo-planar image sampling (2.85 s in the present case) renders the latter effects to be aliased. For example, refs. 14 and 16 provide experimental evidence that estimated resting fluctuations are not contaminated by aliased physiological noise such as respiration and the cardiac cycle.
Previous work on group resting fMRI analysis using ICA (14, 15) simply concatenated individual time-series data from different subjects. By concatenating individual data, the subject-to-subject variance is not modeled as a separate variance component. The tensor-PICA approach used in this study differs in that the analysis simultaneously decomposes all data in three domains: space, subject, and frequency. The advantage of this method is that the between-subject variation is estimated directly as part of the decomposition, enabling consistent between-group as well as within-group analysis to quantify the consistency of these networks.
In this study, transformation of fMRI time courses into the frequency domain was used because strong similarity in the temporal domain across subjects cannot be assumed for any of these resting-state fluctuations. Estimating ICA components from the power spectra effectively discounts possible phase variations between components. The decomposition does not impose sparseness in the spectral domain, and it also does not imply that isolated frequencies are associated with an individual component. Indeed, the estimated power spectra do not show isolated peak frequencies for any of these effects, but instead all show distribution of strong power at low frequency across a range of frequencies, consistent with the majority of resting-fMRI studies (9, 10, 14, 15). The estimated power spectra did not show isolated frequencies, and the separation of signal into different components, therefore, is largely driven by the difference in the spatial characteristics. This finding is consistent with results from tensor-PICA decompositions on task fMRI data (see ref. 21 for details) where, for example, the decomposition successfully separates task-related activation from task-related deactivation and stimulus-correlated head motion, despite the fact that all three effects are similar in the temporal and frequency domain.
There are, however, limitations to the method presented here. First, as with cross-correlation analysis, the analysis is based on a linear decomposition of the data. As such, it is possible that this linear decomposition misses interesting functional connectivity between brain regions with highly nonlinear relationships in the data (33). Note, however, that the first-order (linear) part of any possible functional relationship between different voxels will be reflected in the current set of components. Second, functional connectivity at an individual subject level cannot easily be inferred from these findings because we present results at a mixed-effects level. In this study, the aim was to perform a group analysis, i.e., characterize the expected average level of resting fluctuation at the population level. In clinical studies, however, it might be more appropriate to investigate the individual subject level instead. Third, whereas the trilinear decomposition gives interpretable discrimination across the spatial modes, the estimates across the frequency domain appear less informative. To better understand the temporal dynamics of these effects, the combination of fMRI with other imaging techniques such as electroencephalograms (19, 24) shows great promise. Finally, note that bootstrapping methods like those used here can underestimate variances. Note, however, that each surrogate data set was limited to a rather small group size of six, which in turn will give a larger estimate of the within-group variation at the mixed-effects level. Analysis of a larger pool of scans and subjects would generate a more reliable estimation of the consistency of these effects.
Our findings confirm the idea that the baseline state of the brain is by no means an inactive state. Instead, the brain appears to be very dynamic, showing coherent slow fluctuations in the BOLD signal of a magnitude comparable to task-induced signals. These coherent, slow fluctuations are grouped in separate anatomically and functionally plausible networks. The sensory cortices as well as areas involved in higher cognitive functioning, e.g., memory function, constitute separate networks. The observed magnitude of these fluctuations in the BOLD signal at rest is consistent with previous findings (9, 19), and this magnitude at rest puts the magnitude of “activations” usually found in task-related experiments into perspective. An important issue is the implication of these large fluctuations at rest on the interpretation of activation found in task-related fMRI studies. In these studies, the fMRI signal during “on” periods is contrasted with recordings during a baseline or control condition. Previous studies have demonstrated that the default-mode network is still detectable during such a control condition (16, 17), and it is highly likely that this is the case as well for other resting patterns we found in this study. If the fluctuations in the BOLD signal within these brain areas are correlated to the design used, it will be difficult to interpret the origin of the significant results because in some brain areas it will be hard to distinguish whether the activation is because of the task or the control condition. In cases where these fluctuations are uncorrelated to the task, they will be treated as noise and thus reduce the signal-to-noise ratio and statistical power of the fMRI experiment. This phenomenon has been described by Fox et al. (34), who demonstrate that coherent spontaneous fluctuations indeed account for a significant fraction of the variability in measured event-related BOLD responses.
In this study, 10 consistent resting patterns with relatively large coherent fluctuations in the BOLD signal are presented. These results show very plausible networks that are in line with findings in previous research (9, 12, 14, 15, 17, 20). The coherent fluctuations have been identified in resting fMRI data of healthy subjects. It is of great interest to investigate whether these patterns are present to the same extent under different conditions. For example, it has already been shown that one or more of these patterns are affected by disease (22, 35, 36). Using resting-state fMRI to investigate the influence of disease and/or medication on the brain has apparent clinical advantages because no complicated experimental design is required, and no task needs to be practiced beforehand. Not having to use a task is a significant benefit, especially when studying patients who may have difficulties performing a task, for example, patients with Alzheimer's disease. Understanding these resting fluctuations in terms of their spatial and temporal characteristics, their magnitude and consistency, and their origin is, therefore, of prime importance to taking advantage of the benefits of resting fMRI.
Materials and Methods
Subjects.
Ten healthy right-handed subjects (age 22.8 ± 2.3, five male/five female) participated in this study after giving written informed consent in accordance with the VU University Medical Center Medical Ethical Committee. All subjects underwent an fMRI session of ≈40 min on two different occasions. The time between the first and second fMRI session averaged 8.7 days (5–14 days). For the resting-state scan, subjects were instructed to lie with their eyes closed, think of nothing in particular, and not fall asleep.
Imaging Methods.
Two hundred whole-brain, T2*-weighted echo planar images were acquired on a 1.5-T Sonata scanner (Siemens, Erlangen, Germany) (sequence parameters: repetition time = 2,850 ms; echo time = 60 ms; flip angle = 90°). Thirty-six axial slices (3.3 mm isotropic) were acquired. For registration purposes, a high-resolution T2*-weighted echo planar image and a high-resolution T1-weighted image were also acquired.
Data Analysis.
Preprocessing.
The image preprocessing was carried out by using the Oxford Centre for Functional Magnetic Resonance Imaging of the Brain Software Library (FMRIB, Oxford U.K.; FSL version 3.2; ref. 37). The following prestatistics processing was applied: motion correction (38), removal of nonbrain structures from the echo planar imaging volumes (39), spatial smoothing by using a Gaussian kernel of 6 mm FWHM, mean-based intensity normalization of all volumes by the same factor (i.e., 4D grand-mean), high-pass temporal filtering (Gaussian-weighted least-squares straight line fitting) (FWHM = 150 s), and Gaussian low-pass temporal filtering (FWHM = 5.6 s). After preprocessing, the functional scans were registered to the MNI152 standard space (average T1 brain image constructed from 152 normal subjects at the Montreal Neurological Institute, Montreal, QC, Canada) by using affine registration (FLIRT; ref. 38). From the resulting affine transformation matrices, a midspace was defined as the transformation that approximates the average size and shape of the individual subjects' spaces by calculating the geometric mean of the affine transformation matrices. Within this midspace, the data were kept at the original echo-planar imaging resolutions, reducing the computational burden of a simultaneous decomposition. Finally, the individual time-series data were converted to voxel-wise power spectra. Because there is no task in resting-state data to restrict what subjects are doing during a specific time course, we cannot assume a consistent temporal response for individual components between subjects. However, in the frequency domain, these resting fluctuations of interest are typically associated with strong power in the range of 0.01–0.1 Hz. This preprocessing resulted in 20 data sets, with data obtained from two different resting fMRI sessions for each of the 10 subjects.
Statistical analyses.
A modified version of the tensor-PICA approach was used for statistical analyses (21). The 20 individual data sets were divided into two group data sets: the first included the first resting-state scan of every subject; the second included the second scan of each. These two group data sets were decomposed separately into groups of vectors that characterize the structured signals in the spatial, frequency, and subject domain by using tensor PICA (21). Twenty-five components per data set were estimated, the number of which was determined from the estimated data covariance matrix of all 20 individual subject data sets by using the Laplace approximation of the model order for a probabilistic principal component analysis model (40). From these components, 10 were qualitatively selected by visual inspection as anatomically relevant areas across subjects, potentially depicting functionally relevant resting-state networks. Components excluded from further analysis show clearly interpretable distinct artifacts caused by, among other things, head motion, physiological noise, misregistration, or image fluctuations in cerebral spinal fluid (see refs. 14 and 21 for examples of the spatial patterns of head motion and physiological noise). Final maps were thresholded at a posterior probability threshold of P > 0.5 by using an alternative hypothesis-testing approach based on the fit of a Gaussian/Gamma mixture model to the histogram of intensity values in each map (40).
To quantify the spatial consistency of the fluctuations, the individual subjects' data sets were combined into surrogate group fMRI data sets by using a bootstrapping procedure. Each surrogate data set contained data from six randomly selected subjects and one of the two possible subjects' sessions. Overall, 100 of a possible 13,440 multisubject data sets were created, each containing 100 (frequencies) × 41,582 (voxels) × 6 (subjects) data points. These 100 group data sets were analyzed similarly to the 2 original data sets. To reduce computational burden, the analysis was performed by using a mask to exclude ventricular spaces. For each group data set, 25 components were extracted. Based on their spatial cross-correlations, the 2,500 components were automatically grouped into pairs of 100 maps, which successively explained less amount of residual variation among the set of all components. Sets were selected to represent >97% of the total variance. For each of these sets of components, voxel-wise mean and standard deviation maps were calculated. The corresponding mean maps for each independent component were converted to expected voxel-wise percentage BOLD signal change. These maps were combined with the estimated standard deviation across the pool of virtual populations into maps of voxel-wise coefficients of variation (i.e., by calculating CVj = σj/μj × 100 at each voxel j). These maps describe the amount of typical percentage variation around the expected percentage BOLD signal change at every voxel's location across the decompositions. For example, a value of CVj = 25 indicates that at voxel j, there is an expected ±25% deviation around the reported mean percentage BOLD signal change value μj. These maps were subsequently thresholded at <50% variation. Artefactual maps showing head motion or the effect of magnetic field B0 inhomogeneities were excluded from further analysis. Note that the maps of coefficients of variation show the variability between different analyses of the virtual populations, not the (average) variability within any one of the 100 analyses. As such, Figs. 2 and 3 present complementary pieces of information. For example, two areas can show identical expected percentage BOLD fluctuation with very different coefficients-of-variation.
Acknowledgments
This work was supported by Institute for the Study of Aging Grant 231002, Netherlands Organization for Scientific Research Grant 916.36.117, and the U.K. Engineering and Physical Sciences Research Council.
Abbreviations
- BA
Brodmann area
- BOLD
blood oxygenation level-dependent
- fMRI
functional MRI
- ICA
independent component analysis
- PICA
probabilistic ICA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The neuroimaging data have been deposited with the fMRI Data Center, www.fmridc.org (accession no. 2-2006-1226A).
References
- 1.Lowe MJ, Dzemidzic M, Lurito JT, Mathews VP, Phillips MD. NeuroImage. 2000;12:582–587. doi: 10.1006/nimg.2000.0654. [DOI] [PubMed] [Google Scholar]
- 2.Cordes D, Haughton VM, Arfanakis K, Carew JD, Turski PA, Moritz CH, Quigley MA, Meyerand ME. AJNR Am J Neuroradiol. 2001;22:1326–1333. [PMC free article] [PubMed] [Google Scholar]
- 3.Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obrig H, Neufang M, Wenzel R, Kohl M, Steinbrink J, Einhaupl K, Villringer A. NeuroImage. 2000;12:623–639. doi: 10.1006/nimg.2000.0657. [DOI] [PubMed] [Google Scholar]
- 5.Wise RG, Ide K, Poulin MJ, Tracey I. NeuroImage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Birn RM, Diamond JB, Smith MA, Bandettini PA. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 7.Gusnard DA, Raichle ME. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- 8.Peltier SJ. NeuroImage. 2002;16:985–992. doi: 10.1006/nimg.2002.1141. [DOI] [PubMed] [Google Scholar]
- 9.Biswal B. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 10.Cordes D, Haughton VM, Arfanakis K, Wendt GJ, Turski PA, Moritz CH, Quigley MA, Meyerand ME. AJNR Am J Neuroradiol. 2000;21:1636–1644. [PMC free article] [PubMed] [Google Scholar]
- 11.Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Hum Brain Mapp. 2002;15:247–262. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampson M, Olson IR, Leung HC, Skudlarski P, Gore JC. NeuroReport. 2004;15:1315–1319. doi: 10.1097/01.wnr.0000129997.95055.15. [DOI] [PubMed] [Google Scholar]
- 13.Lowe MJ, Mock BJ, Sorenson JA. NeuroImage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- 14.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Philos Trans R Soc London B. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. NeuroImage. 2005;29:1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greicius MD, Krasnow B, Reiss AL, Menon V. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman RI, Stern JM, Engel J, Jr, Cohen MS. NeuroReport. 2002;13:2487–2492. doi: 10.1097/01.wnr.0000047685.08940.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckmann CF, Smith SM. NeuroImage. 2005;25:294–311. doi: 10.1016/j.neuroimage.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 22.Greicius MD, Srivastava G, Reiss AL, Menon V. Proc Natl Acad Sci USA. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller EK, Cohen JD. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Montes E, Valdes-Sosa PA, Miwakeichi F, Goldman RI, Cohen MS. NeuroImage. 2004;22:1023–1034. doi: 10.1016/j.neuroimage.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 25.Buckner RL, Koutstaal W, Schacter DL, Wagner AD, Rosen BR. NeuroImage. 1998;7:151–162. doi: 10.1006/nimg.1998.0327. [DOI] [PubMed] [Google Scholar]
- 26.Achim AM, Lepage M. J Cognit Neurosci. 2005;17:652–667. doi: 10.1162/0898929053467578. [DOI] [PubMed] [Google Scholar]
- 27.Cabeza R, Nyberg L. Curr Opin Neurol. 2000;13:415–421. doi: 10.1097/00019052-200008000-00008. [DOI] [PubMed] [Google Scholar]
- 28.Gusnard DA, Akbudak E, Shulman GL, Raichle ME. Proc Natl Acad Sci USA. 2001;98:4259–4264. doi: 10.1073/pnas.071043098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazoyer B, Zago L, Mellet E, Bricogne S, Etard O, Houde O, Crivello F, Joliot M, Petit L, Tzourio-Mazoyer N. Brain Res Bull. 2001;54:287–298. doi: 10.1016/s0361-9230(00)00437-8. [DOI] [PubMed] [Google Scholar]
- 30.McKiernan KA, Kaufman JN, Kucera-Thompson J, Binder JR. J Cognit Neurosci. 2003;15:394–408. doi: 10.1162/089892903321593117. [DOI] [PubMed] [Google Scholar]
- 31.Shulman GL, Fiez J, Corbetta M, Buckner RL, Miezin F, Raichle ME, Peterson SE. J Cognit Neurosci. 1997;9:648–663. doi: 10.1162/jocn.1997.9.5.648. [DOI] [PubMed] [Google Scholar]
- 32.Andreasen NC, O'Leary DS, Cizadlo T, Arndt S, Rezai K, Watkins GL, Ponto LL, Hichwa RD. Am J Psychiatry. 1995;152:1576–1585. doi: 10.1176/ajp.152.11.1576. [DOI] [PubMed] [Google Scholar]
- 33.Friston KJ. Philos Trans R Soc London B. 2000;355:215–236. doi: 10.1098/rstb.2000.0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fox MD, Snyder AZ, Zacks JM, Raichle ME. Nat Neurosci. 2006;9:23–25. doi: 10.1038/nn1616. [DOI] [PubMed] [Google Scholar]
- 35.Gotman J, Grova C, Bagshaw A, Kobayashi E, Aghakhani Y, Dubeau F. Proc Natl Acad Sci USA. 2005;102:15236–15240. doi: 10.1073/pnas.0504935102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Cereb Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- 37.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, et al. NeuroImage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 38.Jenkinson M, Bannister P, Brady M, Smith S. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 39.Smith SM. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beckmann CF, Smith SM. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]