Abstract
Retroviruses selectively package two copies of their RNA genomes in the context of a large excess of nongenomic RNA. Specific packaging of genomic RNA is achieved, in part, by recognizing RNAs that form a poorly understood dimeric structure at their 5′ ends. We identify, quantify the stability of, and use extensive experimental constraints to calculate a 3D model for a tertiary structure domain that mediates specific interactions between RNA genomes in a gamma retrovirus. In an initial interaction, two stem–loop structures from one RNA form highly stringent cross-strand loop–loop base pairs with the same structures on a second genomic RNA. Upon subsequent folding to the final dimer state, these intergenomic RNA interactions convert to a high affinity and compact tertiary structure, stabilized by interdigitated interactions between U-shaped RNA units. This retroviral conformational switch model illustrates how two-step formation of an RNA tertiary structure yields a stringent molecular recognition event at early assembly steps that can be converted to the stable RNA architecture likely packaged into nascent virions.
Keywords: retroviral RNA dimer, RNA folding, Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) chemistry, site-directed cleavage
Retroviral genomes usually consist of two sense-strand RNAs that are noncovalently linked near their 5′ ends to form a dimeric structure (1–3). Recognition of this dimeric state ensures that exactly two RNA genomes are packaged into each nascent virion (1, 2). Mature retroviral virions contain almost exclusively retroviral genomic RNA plus a few select cellular RNAs (4, 5). Many other cellular RNAs, including mRNAs (6–8), are accessible to the retroviral packaging process. Specific recognition of retroviral genomic RNA against a large background of cellular RNA thus represents a striking example of molecular recognition in biology.
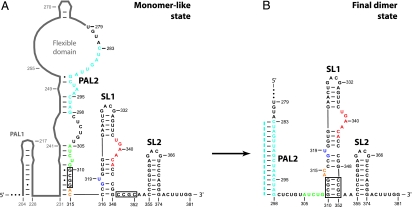
We have recently identified a minimal dimerization active sequence (MiDAS) (9) for a representative gamma retrovirus, the Moloney murine sarcoma virus (MuSV; Fig. 1A). The MiDAS domain correlates closely with retroviral genomic sequences sufficient to package heterologous RNAs into virions (6, 8, 11, 12), as dimers (8). The MiDAS domain also includes conserved sequence elements previously proposed to specify the noncovalent interactions that mediate RNA dimerization.
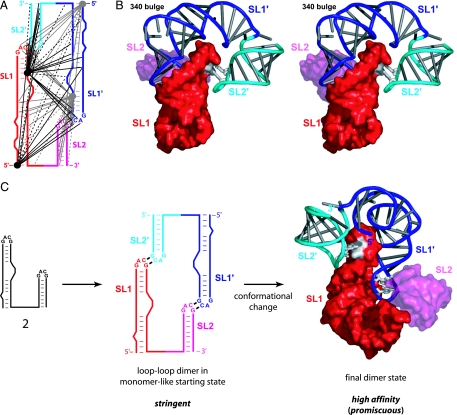
Fig. 1.
Conformational switch in the SL1-SL2 domain during retroviral RNA dimerization, defined by RNA SHAPE chemistry. (A) Structure of the MiDAS domain for the Moloney murine sarcoma virus in the monomeric starting state (9). RNA sequences that contribute to the SL1-SL2 conformational change are shown explicitly; other MiDAS structures are represented in gray. (B) SL1-SL2 domain conformation in the final dimer state. Regions in the RNA that undergo significant structural changes are emphasized in color. Dashed line (light blue) illustrates cross-strand base pairing in PAL2. Multiple systems are in use for naming structural features in gamma retroviral dimerization domains; we use the system introduced with the initial characterization of these elements (10).
Conserved sequence elements include self-complementary (palindromic) sequences (PAL1 and PAL2) and stem–loop structures 1 and 2 (SL1 and SL2) (10, 13–16). SL1 and SL2 contain GACG tetraloops that form stable loop–loop interactions with a second RNA molecule. Loop–loop interactions are mediated by canonical intermolecular C-G base pairing and additional stacking and intra- and intermolecular hydrogen bonds (17) (see Fig. 6, which is published as supporting information on the PNAS web site). In addition, the self-complementary PAL1 and PAL2 sequences form extended heteroduplexes involving both strands in the dimer (refs. 13–16; C.S.B. and K.M.W., unpublished data). However, the kinetic process of duplex formation typically exhibits poor sequence selectivity because duplexes containing mismatches form as fast as the thermodynamically more stable perfect duplex (18, 19). What nucleic acid structures then contribute to the process that allows retroviral RNA genomes to dimerize and be packaged into nascent virions with such exquisite selectivity?
Results
The SL1-SL2 Domain Undergoes a Conformational Switch upon RNA Dimerization.
The structure of the monomeric RNA starting state for the MuSV MiDAS was determined by using RNA Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) chemistry (ref. 9; Fig. 1A). In a SHAPE analysis, local nucleotide flexibility is monitored at every position in an RNA via structure-selective reaction between an anhydride reagent (N-methylisatoic anhydride; NMIA) and the ribose 2′-hydroxyl group. Flexible and single-stranded nucleotides react efficiently, whereas nucleotides constrained by base pairing or tertiary interactions are unreactive (20, 21).
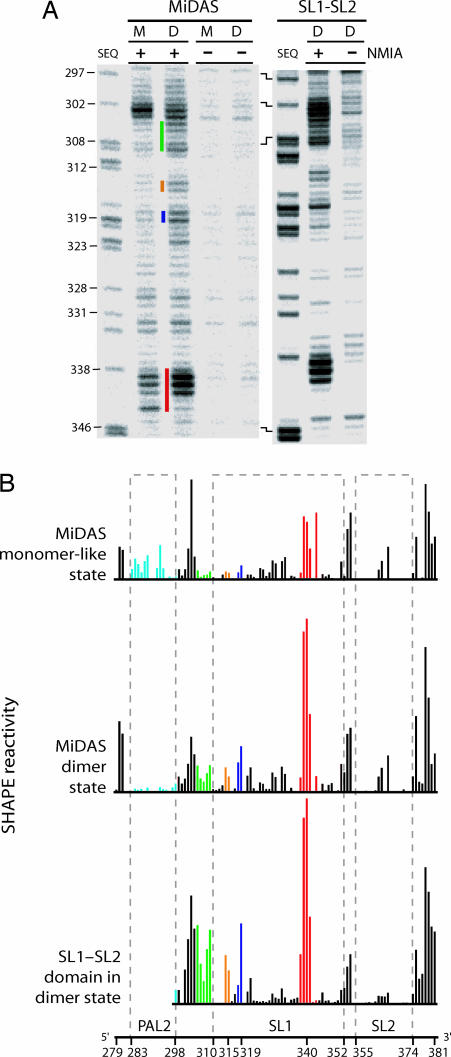
We monitored RNA structure in the final dimer state and, for comparison, in the monomer-like starting state by using SHAPE chemistry and the resulting 2′-O-adducts were identified by primer extension and sequencing gel analysis (Fig. 2A). The initial monomer can be converted to a well defined dimer state by incubating the RNA at 60°C for 30 min (in 200 mM potassium acetate/5 mM MgCl2, pH 7.5). Multiple regions in the MiDAS domain undergo a conformational change as the initial monomer (M) folds to the final dimer (D) state (Fig. 2A, colored bars). Structural changes are highlighted by using a consistent coloring scheme in Figs. 1 and 2.
Fig. 2.
SHAPE analysis of the MiDAS RNA in starting monomer-like (M) and final dimer (D) conformations and a simplified SL1-SL2 domain RNA in the final dimer state (D). (A) 2′-O-Adduct formation upon addition of NMIA (+) detected by primer extension. −, reactions omitting NMIA. Sequencing lanes (SEQ) were generated by dideoxy cytosine nucleotide incorporation; nucleotide positions are labeled with respect to NMIA lanes. (B) Quantitative histograms for NMIA reactivity. Columns are colored using the scheme shown in Fig. 1. Column heights report band intensities in the (+) NMIA reactions minus background.
Absolute 2′-hydroxyl SHAPE chemistry reactivities were quantified for every position in both the starting monomer-like and final dimer states (Fig. 2B). Nucleotides in PAL2 are reactive in the monomeric starting state (Figs. 1A and 2B Top, light blue). Upon dimerization, reactivity in PAL2 decreases dramatically, consistent with a conformational change in which PAL2 lies in a flexible domain in the monomer state and then folds to form a stable 16-bp intermolecular duplex in the dimer state (Figs. 1 and 2B).
SHAPE analysis also identifies changes in several other MiDAS RNA structures. NMIA reactivity increases in the stem that anchors the flexible domain at nucleotides 305–309, in the AC bulge (nucleotides 314 and 315), and at the U319 bulge (Fig. 2B Top and Middle, green, orange, and dark blue symbols). These changes in SHAPE reactivity are consistent with a register shift in which the SL1 stem–loop extends by four base pairs during dimerization (Fig. 1, boxed nucleotides). We term these two conformations the monomer-like and final dimer states.
To confirm this secondary structure assignment, we analyzed the secondary structure of an RNA spanning nts 295–381. This simplified RNA is only capable of forming the SL1-SL2 domain structure found in the final dimer state (Fig. 1B). SHAPE reactivity in the simplified SL1-SL2 domain construct is essentially identical to this region in the complete MiDAS domain in the final dimer state (compare Fig. 2B Middle and Bottom). Notably, the structural similarity includes the fine-scale reactivity patterns characteristic of the 298–310, 314–319, and 328–361 regions. Thus, SHAPE analyses of the monomer, final dimer, and SL1-SL2 domain RNAs indicate that the SL1-SL2 domain changes conformation upon dimerization (Fig. 1).
The SL1-SL2 Domain Is an Autonomous Dimerization Motif.
If GACG tetraloops form loop–loop interactions (ref. 17; Fig. 6) in the context of the SL1-SL2 domain, then there are at least two distinct dimer states: a loop–loop dimer state in which the RNA is in the starting monomer-like conformation (Fig. 3A) and a loop–loop state for the final dimer conformation (Fig. 3D).
Fig. 3.
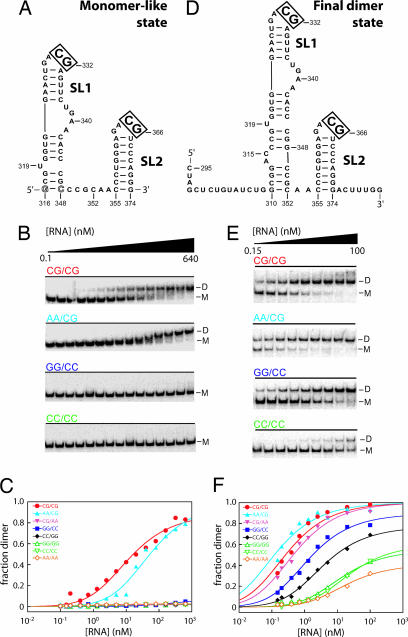
Dimerization specificity of the SL1-SL2 domain in monomer-like versus final dimer states. (A and D) Monomer-like and final dimer states for the SL1-SL2 domain. Monomer contains a single inverted base pair to facilitate transcription (open letters). Control experiments show this base-pair change does not affect dimerization (data not shown). (B and E) Native gel analysis of RNA dimerization for wild type (CG/CG) and representative mutant sequences in monomer-like and final dimer conformations. M, monomer; D, dimer. (C and F) Binding curves for RNA dimerization in the monomer-like and final dimer states.
To understand the contribution of these loop–loop interactions for the monomer-to-dimer conformational change, we measured dimerization affinities for RNAs forced to be in either the monomer-like or final dimer states by incubating these RNAs at 25°C and resolving monomer and dimer species in nondenaturing gels (Fig. 3 B and E). We evaluated dimerization affinities for native sequence RNAs and also for mutants containing changes in the CG sequence that mediates intermolecular base pairs in the loop–loop interaction (Fig. 6). RNA constructs are identified by their sequences at the cross-strand base-pairing positions. Thus, the native sequence RNAs are termed CG/CG and the mutants are GG/GG, GG/CC, CC/GG, CC/CC, AA/CG, CG/AA, and AA/AA, respectively.
Strikingly, in the monomer-like conformation, the SL1-SL2 domain forms a high-affinity complex, resolved as a well defined species by nondenaturing gel electrophoresis (Kdimer = 11 nM; Fig. 3B, CG/CG; Fig. 3C, red symbols). This tight binding observed for constructs containing two GACG loop motifs (equal to –11 kcal/mol) is broadly consistent with the prior measurement (made under slightly different conditions) that a single loop–loop interaction has a favorable free energy increment of –6.5 kcal/mol (17). Dimerization by the AA/CG variant is weakened by 5-fold (Kdimer = 51 nM; Fig. 3 B and C). This mutant also is unable to form heterodimers with the native sequence RNA, indicating that this sequence fails to form key interactions that stabilize the native structure (data not shown). Mutating the loops in SL1 and SL2 to all other tested sequences completely eliminates detectable dimerization. Our observation that variants like CC/GG and GG/CC that, in principle, maintain Watson–Crick-type base-paring interactions do not form stable dimers is consistent with disruption of noncanonical hydrogen-bonding interactions that can form only in the context of the native sequence (17). Together, these data demonstrate that, in the monomer-like state, dimerization strongly discriminates against nonnative sequences.
We performed comparable experiments by using a SL1-SL2 domain RNA that folds to the conformation characteristic of the final dimer state (Figs. 2B and 3D). The native (CG/CG) sequence dimerizes with high affinity (Kdimer = 0.3 nM) or ≈40-fold more tightly than in the monomer state. Control experiments showed that the 5′ and 3′ flanking sequences in our dimer state construct (nucleotides 295–309 and 375–381; see Fig. 3D) have no effect on Kdimer as compared with a sequence spanning nucleotides 310–374 (data not shown). Most SL1-SL2 domain sequence variants dimerized with high affinities (Fig. 3 E and F). For example, the AA/CG and CG/AA variants have dimerization affinities comparable with wild type. Other sequence variants have lower, but still quite high, dimerization affinities (Fig. 3F; see also Table 1, which is published as supporting information on the PNAS web site).
Thus, the SL1-SL2 region functions as an autonomous domain that dimerizes in two distinct modes (Fig. 3 C and F). In the monomer-like conformation, loop–loop mediated dimerization is highly selective for the native sequence. Upon rearranging to the final dimer state, dimerization affinity increases but now shows low sequence specificity (compare Fig. 3 C with F).
Architecture of SL1-SL2 Domain.
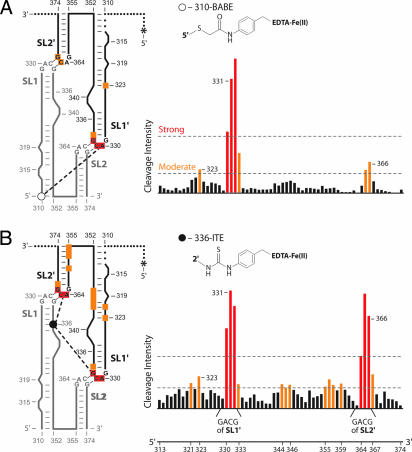
Because the SL1-SL2 domain contains two GACG RNA tetraloops, additional experiments are required to establish the interaction partners for SL1 and SL2 in one RNA with their tetraloop partners from the second RNA in the dimer (Fig. 6). We used site-directed hydroxyl radical cleavage experiments (22) (i) to identify the orientation and pairing partners for the SL1 and SL2 loops and (ii) to obtain constraints for the global architecture of the SL1-SL2 domain in the monomer-like and final dimer states. We engineered two 65-nt RNAs (spanning nucleotides 310–374), confined to be in the final dimer state, and derivatized these RNAs with Fe(II)-EDTA groups at defined positions (Fig. 4, gray). The 310-bromoacetamidobenzyl-EDTA-Fe(II) (BABE) construct contained a BABE adduct at the 5′ end of the RNA. The 336-isothiocyanobenzyl-EDTA-Fe(II) (ITE) construct was derivatized with ITE via a 2′ amino nucleotide at position 336. In the presence of hydrogen peroxide and ascorbic acid, highly reactive hydroxyl radicals capable of cleaving the RNA backbone are generated at the Fe(II)-EDTA moiety.
Fig. 4.
Architecture of the SL1–SL2 interaction in the final dimer conformation mapped by site-directed hydroxyl radical footprinting. (A) Fe(II)-BABE (open circle) mediated cleavage from nucleotide 310. (B) Fe(II)-ITE (filled circle) cleavage from nucleotide 336. Small spheres indicate RNA regions that were not monitored; the position of 5′ radiolabel on second RNA strand is indicated by an asterisk.
The 310-BABE or 336-ITE constructs were allowed to form loop–loop dimers with a second 5′ 32P-labeled SL1-SL2 domain RNA (radiolabels are indicated with asterisks in Fig. 4). By this scheme, only intermolecular cross-strand RNA cleavages are detected. Dimerization affinities for the Fe(II)–EDTA-containing complexes were ≈0.4 nM or identical, within error, to underivatized complexes. Cross-strand RNA cleavage products were resolved on sequencing gels. Cleavages greater than three or greater than five times background were judged to be moderate and strong, respectively (Fig. 4).
For the 310-BABE construct, strong cleavage was obtained only at the apex of SL1′ (Fig. 4A, red symbols). With the 336-ITE construct, strong cleavage was observed in both SL1′ and SL2′ (Fig. 4B). These cleavage data strongly support a secondary structure arrangement in which SL1 forms a loop–loop interaction with SL2′ and SL2 interacts with SL1′ (Fig. 4).
Finally, to evaluate structural changes in the SL1-SL2 domain that occur during dimerization, we performed site-directed cleavage experiments by using our RNA construct that imitates the SL1-SL2 domain structure in the initial monomer-like starting state (Fig. 3A). This RNA was derivatized with BABE at its 5′ end at nucleotide 316 (Fig. 7, which is published as supporting information on the PNAS web site). For this RNA, no strong cross-strand cleavages were detected (Fig. 7). These experiments support a model for the monomer-like starting state in which loop–loop interactions still form but in which the structure is less intimately folded overall than in the final dimer state.
We then used solvent-based hydroxyl radical footprinting to map tertiary contacts in the SL1-SL2 domain in the final dimer state. Under conditions that support loop–loop interactions, multiple regions in the SL1-SL2 domain are strongly protected from hydroxyl radical cleavage mediated by untethered Fe(II)-EDTA (Fig. 8, which is published as supporting information on the PNAS web site). Reassuringly, solvent accessible regions are compatible with the site-directed cleavage information obtained by using the 310-BABE and 336-ITE constructs because strong and moderate site-directed cleavage occurs in regions that are accessible to the untethered Fe(II)-EDTA probe (compare Figs. 4 and 8). Together, the site-directed and solvent-based chemical probing experiments emphasize that SL1 and SL2 function as a true tertiary structure domain.
Distance Constraints Refinement of a 3D Structure for the SL1-SL2 Domain.
RNA SHAPE chemistry (Fig. 2) and site-directed cleavage (Fig. 4) experiments provide extensive information that strongly constrain allowed 3D structures for the SL1-SL2 domain in the final high-affinity dimer state. We used this experimental information to refine structures for the final dimer state by using distance constraints-based algorithms (23–25). Canonical hydrogen bonding and planarity constraints were imposed for predicted (26) base pairs, as constrained by experimental SHAPE reactivities (Figs. 1B and 2). Base pairing also was enforced between C-G pairs in the GACG tetraloops, as demonstrated in an independent NMR study (17). The site-directed hydroxyl radical cleavage experiments yielded 24 long-range intermolecular constraints between the two RNA strands that comprise the dimer (Fig. 4). In addition, seven positions that were both unreactive by site-directed cleavage and also solvent-accessible as judged by cleavage with free Fe(II)-EDTA, were constrained to be distant from the tethered Fe(II)-EDTA groups. Because the SL1-SL2 domain is a symmetrical dimer, the number of constraints used to constrain the refinement is doubled to 62 (Fig. 5A).
Fig. 5.
Refined model and two-step assembly of the SL1-SL2 domain. (A) Summary of 62 long-range intermolecular distance constraints used for structure refinement. Adjacent and repelling constraints are shown with solid and dashed lines, respectively. (B) Stereo image of the SL1-SL2 domain in the final dimer state. One monomer (red and magenta) is shown in a surface representation. The second monomer (blue and cyan) is illustrated as a backbone cartoon; bases are shown as cylinders. Cross-strand G-C pairs in the tetraloops are white. (C) Assembly of the high-affinity SL1-SL2 dimer via a stringent loop–loop intermediate.
Refined structure models converged to an average rmsd of ≈4.1 Å over 128 phosphate positions (Fig. 9, which is published as supporting information on the PNAS web site). The high degree of convergence reflects that the 62 constraints provide dense information with which to position the five independent helical elements in each SL1-SL2 domain monomer (Fig. 5A).
The SL1-SL2 domain forms a tightly packed tertiary structure in which SL1 is bent at the 340 bulge, such that each SL1-SL2 element is roughly U-shaped (Figs. 5B and 9). Two U-shaped RNAs then form an interdigitated structure in which the apex of SL1 from one RNA forms extensive interfaces with both SL1′ and SL2′ from the second monomer. The minor groove side of SL1 faces the minor groove of SL2′, whereas the major groove of SL1 fits snugly against the major groove side of SL1′ in the second RNA (Fig. 5B).
Discussion
Our experiments support a model for retroviral genomic dimerization in MuSV in which the SL1-SL2 domain plays a major role. This conclusion is strongly corroborated by recent experiments showing that the SL1-SL2 domain (in the absence of PAL2 and most of PAL1) is sufficient to direct modest packaging of a heterologous RNA, in a dimeric state, into virions (8).
Conformational changes involving the dimerization domain have been proposed to expose single-stranded RNA regions that are recognized by the retroviral Gag protein (27, 28). SHAPE analysis indicates that two potential UCUG nucleocapsid-binding sites lie in the single-stranded linker between PAL2 and SL1 in the final dimer state (Fig. 1B). Selective packaging of retroviral genomes is likely to involve a mechanism significantly more complex than simple protein recognition of short RNA sequences. The RNA conformation itself is likely both to govern the stringency of dimerization (Fig. 3) and to present protein-binding elements in the context of a specific 3D architecture that enables selective recognition by the viral Gag protein (Fig. 5).
We propose that intergenomic interactions involving the SL1-SL2 domain form in two steps. Initially, SL1–SL2 domain interactions in the monomer-like conformation are mediated primarily by relatively simple cross-strand tertiary interactions involving the GACG tetraloops (Fig. 5C Center). This complex has a relatively loose tertiary structure as judged by site-directed hydroxyl radical cleavage (Fig. 7). Dimerization in this state can be defined as stringent because only the native sequence forms a high-affinity dimer.
During the dimerization reaction from the initial loop–loop pair, two PAL2 sequences eventually form an extended intermolecular duplex (Fig. 1B; refs. 13 and 14; C.S.B. and K.M.W., unpublished data). Coincident with PAL2 duplex formation, the SL1-SL2 domain changes conformation such that the 231–241/305–315 anchoring helix in the monomer-like state breaks and SL1 extends by 4 bp (Fig. 1). This conformation change allows extensive stabilizing tertiary interactions to form in the SL1-SL2 domain (Fig. 5C Right). An important consequence is that, in the final dimer state, RNAs with nonnative sequences in the GACG tetraloops also are able to form high-affinity complexes (Fig. 3). Thus, dimer formation is more stable for both native and nonnative sequences and, therefore, is less stringent (Fig. 5C).
Dimerization of retroviral genomes appears to occur in the cytoplasm and before encapsidation into the immature viral particle (29). Dimerization therefore must exclude noncognate interactions with the enormous background (30, 31) of both retroviral and general cellular mRNAs (6–8). We suggest that the two-step mechanism (Fig. 5C) functions to facilitate specific dimerization of native sequence genomic RNAs. At early stages of new viral RNA production in an infected cell, dimerization via loop–loop interactions in the SL1-SL2 domain is highly specific for the native sequence, likely contributing to accurate dimerization and packaging. In subsequent steps, the SL1-SL2 domain and PAL2, and perhaps other RNA structures, undergo a conformational rearrangement to the final dimer state. Formation of the high-affinity final dimer is likely to be important for preventing dissociation and separation of the two genomic RNA strands during packaging, reverse transcription, or other viral processes. Analogous, multistep formation of dimeric tertiary structures from simpler initial base pairing interactions may be broadly used to ensure stringent dimerization and packaging of retroviral genomes.
Methods
Retroviral RNA Constructs.
In addition to the full-length MiDAS sequence, two types of simplified RNA constructs were used in this work that constrained the SL1-SL2 domain to be in either the monomer-like or final dimer states (Fig. 3 A and D). For some experiments, dimer state RNAs contained 5′ and 3′ tails of different lengths; control experiments demonstrated that these extensions had no affect on dimerization affinity. RNAs were generated by T7 RNA polymerase-mediated transcription by using PCR-generated templates and purified by denaturing gel electrophoresis (9). For additional information, see Supporting Methods,which is published as supporting information on the PNAS web site.
SHAPE Analysis of MiDAS and SL1-SL2 RNAs.
NMIA modification, primer extension, and band quantification steps were performed exactly as described in ref. 9. MiDAS and SL1-SL2 constructs contained a nonviral RNA cassette at their 3′ ends to facilitate analysis of the entire sequence by primer extension (20). SHAPE reactivity data were normalized to nucleotide 393 in the UUCG tetraloop of the nonviral RNA cassette.
Concentration-Dependent Dimerization of SL1-SL2.
Internally 32P-labeled RNA (≈0.1 nM) was combined with identical unlabeled RNA (at 0.15–640 nM in 8 μl). RNAs were denatured at 90°C, snap-cooled on ice for 30 sec, treated with 2 μl 5× dimer buffer [1 M potassium acetate/25 mM MgCl2/250 mM Hepes (pH 7.5)], incubated at 25°C for 50 min, and placed on ice. Samples (3 μl) were mixed with 1 μl of loading dye (30% glycerol/5 mM MgCl2/0.01% xylene cyanol/bromophenol blue) and resolved on nondenaturing gels [12% polyacrylamide/5 mM MgCl2/1× TBE (90 mM Tris/90 mM boric acid/2 mM EDTA, pH 8.3)] at a gel temperature of ≈20°C for 1 h by using a running buffer that also contained 5 mM MgCl2. Gels were prerun for 15 min before loading; the running buffer was reequilibrated every 20 min to maintain a uniform [MgCl2]. Monomer and dimer species were visualized by phosphorimaging, and the dimerization dissociation constant (Kdimer) was obtained by fitting to:
where A is the fraction dimer at saturating RNA concentrations and CT is the total concentration of RNA.
Site-Directed Hydroxyl Radical Cleavage.
5′ 32P-labeled SL1-SL2 RNA (0.1 pmol) (Fig. 4) was mixed with 1 pmol unlabeled 310-BABE or 336-ITE RNA in 5.25 μl of water, heated for 3 min at 90°C, snap-cooled on ice, brought to 1× dimer buffer (in 7 μl), and incubated for 30 min at 25°C. Hydroxyl radical cleavage was initiated by adding hydrogen peroxide and ascorbic acid (in 3 μl) to final concentrations of 0.1% and 2.5 mM, respectively. Reactions (4 min at 25°C) were quenched by adding 11 μl of stop solution [0.1 M thiourea/73% (vol/vol) formamide/81 mM Tris-borate/1.8 mM EDTA/marker dyes]. Sites of cleavage were resolved by denaturing gel electrophoresis (15% polyacrylamide/7 M urea) and intensities for individual bands were obtained by integration (32). Background was established in control experiments performed with the 310-BABE and 336-ITE constructs, in which reagents or tethering components were omitted.
RNA Dimer Structure Refinement.
Three-dimensional models were refined by using simulated annealing and molecular mechanics computations (23–25). Base pairing was enforced at the G-C pairs in the tetraloops (17); strong and medium site-directed cleavages (Fig. 5) were refined to optimal distances of 0–25 and 0–35 Å (from the 5′ OH or 2′ OH position to the appropriate C4′ atom), respectively, by using square-well potentials. Our analysis is based on the eight refined structures with clash scores (33, 34) <43. These eight structures superimpose with rmsd values of 3.5–5.0 (4.1 average) Å over 128 phosphate positions in the dimer (Fig. 9). Figures were composed by using PyMOL (DeLano Scientific LLC, South San Francisco, CA).
Supplementary Material
Acknowledgments
We thank Sam Butcher and Jared David for extensive advice with structure refinement and Cristina Gherghe, Alan Rein, and Andrew Kaplan for many helpful discussions. This work was supported by National Institutes of Health Grant GM64803 (to K.M.W. and A. Kaplan).
Abbreviations
- BABE
bromoacetamidobenzyl-EDTA-Fe(II)
- ITE
isothiocyanobenzyl-EDTA-Fe(II)
- NMIA
N-methylisatoic anhydride
- SHAPE
Selective 2′-Hydroxyl Acylation analyzed by Primer Extension.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Berkowitz R, Fisher J, Goff SP. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 2.Rein A. Nat Struct Mol Biol. 2004;11:1034–1035. doi: 10.1038/nsmb1104-1034. [DOI] [PubMed] [Google Scholar]
- 3.D'Souza V, Summers MF. Nat Rev Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 4.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Plainview, NY: Cold Spring Harbor Lab Press; 1997. [PubMed] [Google Scholar]
- 5.Onafuwa-Nuga AA, King SR, Telesnitsky A. J Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adam MA, Miller AD. J Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banks JD, Yeo A, Green K, Cepeda F, Linial ML. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibbert CS, Mirro J, Rein A. J Virol. 2004;78:10927–10938. doi: 10.1128/JVI.78.20.10927-10938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badorrek CS, Weeks KM. Nat Chem Biol. 2005;1:104–111. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- 10.Konings DA, Nash MA, Maizel JV, Arlinghaus RB. J Virol. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mougel M, Barklis E. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakuragi J, Shioda T, Panganiban AT. J Virol. 2001;75:2557–2565. doi: 10.1128/JVI.75.6.2557-2565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard PM, Bonnet-Mathoniere B, Muriaux D, Paoletti J. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 14.Paillart J, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 15.Oroudjev EM, Kang PCE, Kohlstaedt LA. J Mol Biol. 1999;291:603–613. doi: 10.1006/jmbi.1999.2984. [DOI] [PubMed] [Google Scholar]
- 16.Ly H, Parslow TG. J Virol. 2002;76:3135–3144. doi: 10.1128/JVI.76.7.3135-3144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim C, Tinoco I. Proc Natl Acad Sci USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Friedman AE, Kool ET. Biochemistry. 1995;34:9774–9784. doi: 10.1021/bi00030a015. [DOI] [PubMed] [Google Scholar]
- 19.Herschlag D. Proc Natl Acad Sci USA. 1991;88:6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson KA, Merino EJ, Weeks KM. J Am Chem Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 22.Joseph S, Noller HF. Methods Enzymol. 2000;318:175–190. doi: 10.1016/s0076-6879(00)18052-8. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 25.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. J Mol Biol. 2005;351:371–82. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 26.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza V, Summers MF. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 28.Dey A, York D, Smalls-Mantey A, Summers MF. Biochemistry. 2005;44:3735–3744. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 29.Pal BK, Scherer L, Zelby L, Bertrand E, Rossi JJ. J Virol. 1998;72:8349–8353. doi: 10.1128/jvi.72.10.8349-8353.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis AR, Nayak DP, Lofgren J. J Virol. 1978;26:603–614. doi: 10.1128/jvi.26.3.603-614.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.East JL, Chan JC, Bartlett RJ, Knesek JE. J Virol. 1979;29:818–824. doi: 10.1128/jvi.29.2.818-824.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. J Mol Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 34.Davis IW, Murray LW, Richardson JS, Richardson DC. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.