Abstract
Aminoacylation of RNA minihelices is speculated to be a key step in the transition from the putative RNA world to the theater of proteins. This reaction affords the opportunity to make chiral selection of an l- or d-amino acid and thus determine the ultimate chirality that is incorporated into proteins. Previous work showed chiral preference of aminoacylation with a nonprotein, nonribozyme, RNA-directed aminoacylation system. This preference was, in turn, determined by the preexisting chirality of the RNA. The α-amino group attached to the asymmetric α-carbon of the amino acid was an obvious candidate to play a role in chiral selectivity through interactions with the RNA. Also not clear was whether a simple manipulation could change the chiral selectivity, thereby giving insight into the basis of chiral selection in the first place. Here we show, surprisingly, no role for the free α-amino group in chiral selection. However, by a sequence manipulation, chiral preference was suppressed and partly reversed. This result and those with further RNA constructs support the idea that the chiral preference for an l-amino acid in these constructs depends on avoiding a sugar-pucker-sensitive steric clash between a pendant group of a base with the amino acid side chain.
Keywords: chirality, RNA world, origin of aminoacylation
Natural proteins throughout evolution are made up of l-amino acids. According to the RNA world hypothesis, proteins emerged after the establishment of RNAs that had the capacity for many chemical transformations with assorted molecules, including amino acids (1, 2). Recent work supported the idea that l-amino acid homochirality of proteins could be determined by the homochirality of the RNA, which has a d-ribose configuration (3). Because aminoacylation of transfer RNA is the first step of protein synthesis and establishes the genetic code (4), chiral selectivity could plausibly have originated in an early version of that reaction. The plausibility of chiral selectivity during aminoacylation was supported by the recent observation that nonprotein, nonribozyme, RNA-directed aminoacylation of an RNA minihelix [that recapitulates the amino acid acceptor domain of, and is thought to be the progenitor of, modern transfer RNAs (ref. 5)] is chiral-selective, with three different amino acids (alanine, leucine, and phenylalanine) (3). The observed selectivity was 4-fold. Reiterated many times under selective pressure, a 4-fold effect, although small, can lead to an overwhelming preference for an l-amino acid in a biological system.
In previous work that characterized more of the features of the RNA-based aminoacylation with a minihelix based on Escherichia coli tRNAAla, the 3′ terminal A was replaced by either 2′-dA or 3′-dA. In this instance, the 2′-dA substrate was charged, and the 3′-dA derivative was not (3). Thus, aminoacylation was specific for the 3′-OH. We also synthesized RNA components having the opposite chirality, and we tested both l- and d-amino acids. Because natural RNAs are constructed from d-ribose, the aminoacyl phosphate oligonucleotide, minihelixAla, and oligonucleotide bridge were synthesized with l-ribose. With these components of opposite chirality, the ratio of l-[14C]Ala- to d-[14C]Ala-minihelixAla was 1:3.6, namely, approximately the reverse of that determined when the RNA components were made up of d-ribose (3). Our experiments pointed to the idea that the homochirality of modern polypeptides was established during aminoacylation and that this chirality, in turn, was determined by the preexisting chirality of the RNA.
Although these results suggested a plausible explanation for the origin of the homochirality of contemporary proteins, the mechanistic side of the results needed further investigation. Specifically, we wondered whether the free amino group attached to the asymmetric α-carbon of the amino acid played a role in the chiral selectivity of aminoacylation. This question is of particular interest because of the capacity of a free amino group (whether or not protonated) to interact with a phosphate oxygen or a free ribose hydroxyl, for example, during the transition state of the reaction. Second, we wanted to explore whether a simple manipulation of sequence could change the chiral selectivity. If so, the specific atomic groups involved in influencing a change in selectivity could offer insight into mechanism. Last, although we had preliminary data supporting the hypothesis that chiral selectivity was determined in the aminoacyl transfer step of the reaction, further work was needed to establish this point. Pursuant to addressing these issues, we approached the first two questions by making RNA constructs that specifically addressed each, of them, whereas the last issue required more extensive kinetic studies.
Results
Experimental Setup and Mechanistic Questions.
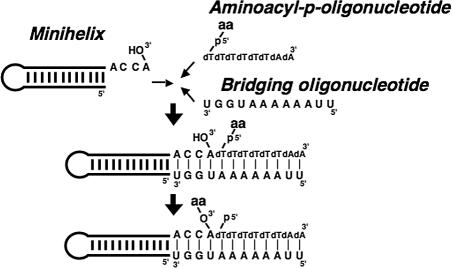
The system that we used comprised a minihelix (minihelixAla, based on the minihelix domain of E. coli tRNAAla) that was hybridized through its single-stranded 3′ end to a 5′-aminoacyl phosphate oligonucleotide. [The aminoacyl phosphate oligonucleotide resembles contemporary systems that use 5′-aminoacyl phosphate mononucleotides (adenylates) (refs. 3, 6, 7).] This hybridization was achieved through a bridging oligonucleotide (template) that rendered the reaction template-dependent (Fig. 1). Aminoacylation was strictly specific for the 3′-OH (3) most likely because, in the extended Watson–Crick helix of the hybridization complex, the 3′-OH is closer (than the 2′-OH) to the 5′-aminoacyl moiety of the aminoacyl phosphate oligonucleotide. Here, the feature that might be important in determining the chiral selectivity of aminoacylation is a hydrogen bond between the amino group of Ala and the phosphate oxygen of the terminal adenosine of minihelixAla. The hydrogen bonding might endow the constraint that is critical for determining the chiral selectivity because the approach of a nucleophile to a carbonyl carbon is not random, but it is instead described by the Bürgi–Dunitz angle (8).
Fig. 1.
Scheme of aminoacylation reaction used in this work. Minihelix, bridging oligonucleotide, and 5′-aminoacyl-p-(dT)6(dA)2 were mixed together.
Role of the α-Amino Group.
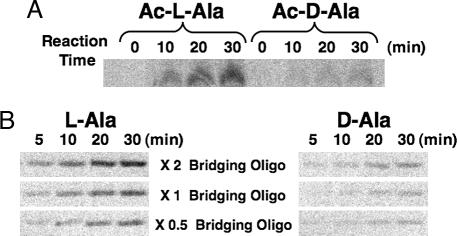
To investigate this role, the amino group of Ala was acetylated, and N-acetyl-l- or d-Ala-aminoacyl oligonucleotide was used for the aminoacylation reaction. Acetylation of Ala was performed by the method of Cardillo et al. (9), with >70% of both l-[14C]Ala and d-[14C]Ala being acetylated. The aminoacylation reaction was performed with a 100 μM concentration each of minihelixAla, bridging oligonucleotide, and 5′-N-acetyl-[14C]Ala-p-(dT)6(dA)2. The ratio of the product formed (calculated from the band intensities of products resolved by gel electrophoresis) after 30 min at 0°C was 3.7:1 (Ac-l-Ala:Ac-d-Ala) (Fig. 2A). Strikingly, this selectivity was similar to that for nonacetylated Ala (3). These results suggest that, at least during the rate-determining step, the amino group of Ala does not make any hydrogen bonds with the phosphate oxygen of the terminal adenosine of the minihelix or with another H-bond acceptor located elsewhere in the hybridization complex.
Fig. 2.
Aminoacylation of minihelix. (A) PAGE analysis of aminoacylation in both N-acetyl-l-Ala and N-acetyl-d-Ala. (B) PAGE analysis of aminoacylation with different concentrations of the bridging oligonucleotide.
Chiral Suppression.
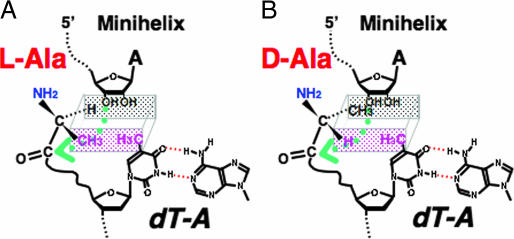
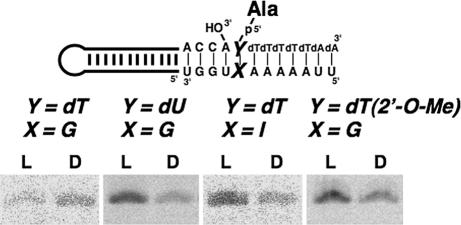
In studying steric models of the base pairing at the position closest to the amino acid attachment site, a potential clash of the CH3 of dT with the CH3 of l-Ala was seen to be avoided in the Watson–Crick dT·A pair (Fig. 3). (Whereas the CH3 of l-Ala is distal to the 3′-OH of A, the CH3 of d-Ala crowds this 3′-OH.) With these considerations in mind, a dT·G pair was introduced to create a potential clash of the CH3 of l-Ala with that of dT (Fig. 3). This substitution sharply reduced the yield of l-Ala-minihelixAla without altering production of d-Ala-minihelixAla (Fig. 4). As a consequence, chiral-selective aminoacylation was suppressed and even somewhat reversed in favor of the d-Ala-minihelixAla product. Consistent with these results and the idea of a clash between the ring and l-amino acid methyl groups (Fig. 3), ablation of the ring CH3 through substituting at dU·G for a dT·G pair gave a construct in which chiral preference for l-Ala was retained (Fig. 4).
Fig. 3.
Schematic representation of the positioning of l-Ala (A) and d-Ala (B), related to 3′-OH of minihelix and CH3 of thymidine. The rectangular parallelpiped shows the depth of the space. The green arrow indicates the nucleophilic attack of 3′-O of the minihelix.
Fig. 4.
PAGE analysis of the aminoacylation using wobble base pairing at the position closest to the amino acid attachment site, dT·G, dU·G, dT·I, or dT(2′-O-Me)·G. Reaction time was 30 min.
Role of Sugar-Pucker-Sensitive Steric Clash.
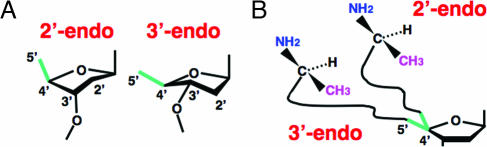
In the Watson–Crick dT·A pair, the potential clash of the CH3 of dT with the CH3 of l-Ala could be avoided through a 3′-endo pucker of dT (Fig. 5) (10). In contrast, conversion to the 2′-pucker could bring the CH3 of dT close to the CH3 of l-Ala (Fig. 5) (10). The NMR structure of a dT·dG base pair established a 2′-endo preference for the deoxyribose of dT (11). The distance between the 3′-O of dT and the 2-amino group of dG is 7.2 Å, a distance that could be bridged by a water molecule. A 3′-endo pucker preference can be created by making a ribose 2′-O-CH3 substitution (12). Accordingly, a dT(2′-O-Me)·G pair was introduced, and chiral-selective aminoacylation in favor of the l-Ala product was restored (Fig. 4). Also, to remove the capacity for making a water bridge between the 3′-O and a base that would stabilize the 2′-endo conformation, a dT·I pair (I, inosine) was also tested. This construct also showed the chiral preference for l-Ala (Fig. 4). These results support the idea that the chiral preference for l-Ala in these constructs depends on avoiding a sugar-pucker-sensitive steric clash between a pendant group of a base with the CH3 of l-Ala.
Fig. 5.
Differences dependent on sugar puckering. (A) Schematic representation of the pucker of the ribose ring (10). (B) Possible differences of the spatial positioning of l-Ala based on the pucker differences of the ribose.
Evidence That the Rate-Determining Step Occurs During the Aminoacyl Transfer Step.
Last, although previous work suggested that the chiral preference was derived during the process of aminoacyl transfer from the 5′-phosphate of the oligonucleotide to the minihelix (3), additionally or alternatively, it could come from a difference in template (bridging oligonucleotide) hybridization efficiency between the l-amino acid- and d-amino acid-p-oligonucleotides, rather than from a reaction within the ternary complex itself. To investigate these possibilities with one set of experiments, we explored the consequences of varying the concentration of the bridging oligonucleotide to determine whether the rate of aminoacylation was sensitive to its concentration and, at the same time, affected the ratio of l- to d-products. Incubation of a 100 μM concentration each of minihelixAla, bridging oligonucleotide, and 5′-[14C]Ala-p-(dT)6(dA)2 was used at starting point (3), and, in further experiments, the concentration of bridging oligonucleotide was increased or decreased by 2-fold to give an overall concentration range of 4-fold. Significantly, the amount of aminoacylation measured at four different time points was sensitive to the concentration of bridging oligonucleotide (Fig. 2B). However, at all time points and at all concentrations of the bridging oligonucleotide, the ≈4-fold preference of l-Ala-minihelixAla to d-Ala-minihelixAla was observed. Thus, although the higher concentrations of the bridging oligonucleotide raised the rate of aminoacylation, presumably because more of the reacting partners were sequestered together in the tripartite complex, the chiral selectivity was retained. This result further supports the idea that chiral selectivity comes during aminoacyl transfer in the tripartite complex.
Discussion
We were surprised that acetylation of the α-amino group of alanine did not block or suppress chiral-specific aminoacylation. As stated above, this result eliminates a role for the free amino group of alanine in forming some sort of H-bonding complex during the rate-determining step. The most stable isomer of Gly·Na+ contains the metal ion complexed between the carbonyl and amine groups in a bidentate mode (13). Bidentate coordination of Na+ between the two carbonyl oxygen atoms of N-acetyl-Gly has also been shown (13). From this perspective and because high concentrations of NaCl are contained in our reaction mixture, Ala and N-acetyl-Ala (being attached to the oligonucleotide portion through a phosphate group) may have the same conformation. The similar conformations are consistent with the same chiral selectivity seen for both Ala- and N-acetyl-Ala oligonucleotide substrates. In these situations, the amino group of Ala could be located on the outside of the helix (Fig. 3).
The positioning of the 3′-O just before the nucleophilic attack should determine the efficiency of the transition intermediate formation. Although the structures shown in Fig. 3 are not intended to correspond to the transition state, the nucleophilic attack of the 3′-O at the carbonyl carbon (shown with green arrow in Fig. 3) is thought to be controlled by the Bürgi–Dunitz angle (8). Our results suggest that this angle is determined not by direct interactions of the α-amino group, but rather by more subtle sugar-puckering-dependent steric factors coming from both the base and ribose moieties.
Making further progress on understanding these subtle parameters is inherently limited by the small energetic differences needed to switch chiral preference from one side to the other. For example, an energetic difference of <1 kcal·mol−1 in the rate-determining step of the transition state is sufficient to give the observed 4-fold preference for l- vs. d-specific aminoacylation. Energy differences of this small magnitude cannot reliably be calculated from energy-minimization programs applied to different models for the transition state. On the other hand, that small energetic differences could have such a profound effect on the development of the chirality of proteins further illustrates how living systems are dependent on subtle “tipping points.”
Materials and Methods
Synthesis and Preparation of Substrates.
MinihelixAla, bridging oligonucleotides (5′-U2A6UG2U, 5′-U2A5GUG2U, and 5′-U2A5IUG2U) and 5′-p-dT(2′-O-Me)(dT)5(dA)2 were synthesized on an Expedite 8909 synthesizer (PE Biosystems, Foster City, CA). The chemicals for the synthesizer were purchased from ChemGenes (Wilmington, MA). The oligonucleotides 5′-p-(dT)6(dA)2 and 5′-p-dU(dT)5(dA)2 were synthesized by Invitrogen (Carlsbad, CA). All molecules were purified by denaturing PAGE. Aminoacyl phosphate oligonucleotides 5′-Ala-p- and 5′-N-acetyl-Ala-p-oligonucleotide were synthesized and purified by a published procedure with slight modifications (14, 15). N-acetylation of alanine [before coupling with p-(dT)6(dA)2] was performed by the method of Cardillo et al. (9). The acetylation was monitored by TLC developed with chloroform/methanol/acetic acid [85:10:5 (vol/vol/vol)] and analyzed on a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA).
Aminoacylation of Minihelix.
Aminoacylation reactions were carried out with a final reaction mixture that contained 50 mM Hepes/NaOH (pH 7.0), 1,000 mM NaCl, 100 μM minihelix, 100 μM [or 50 or 200 μM, only in the experiments with Ala-p-(dT)6(dA)2] bridging oligonucleotide, and 100 μM 5′-N-acetyl-[14C]Ala-p-oligonucleotide. Minihelix and bridging oligonucleotide were preincubated together for 1 h at 0°C in the same buffer condition, and then 5′-N-acetyl-[14C]Ala-p-oligonucleotide was added. After incubation at 0°C for the time described in the figures, 167 mM (final concentration) sodium citrate (pH 5.0) was added, and the products were applied to a denaturing PAGE in 22% polyacrylamide. Both the gel and electrophoresis buffer contained 25 mM sodium citrate (pH 5.0), and electrophoresis was performed at 4°C. The gel was dried and analyzed on a PhosphorImager screen.
Acknowledgments
We thank Prof. Stephen Benkovic of Pennsylvania State University for comments on the manuscript, and Drs. Tetsuo Kushiro of the University of Tokyo and Hirofumi Seike of Harvard University for helpful discussions. This work was supported by National Institutes of Health Grant GM15539 and by a fellowship from the National Foundation for Cancer Research (both to P.R.S.).
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rich A. In: Horizons in Biochemistry. Kasha M, Pullman B, editors. New York: Academic; 1962. pp. 103–126. [Google Scholar]
- 2.Gilbert W. Nature. 1986;319:618. [Google Scholar]
- 3.Tamura K, Schimmel P. Science. 2004;305:1253. doi: 10.1126/science.1099141. [DOI] [PubMed] [Google Scholar]
- 4.Schimmel P. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 5.Schimmel P, Giegé R, Moras D, Yokoyama S. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura K, Schimmel P. Proc Natl Acad Sci USA. 2003;100:8666–8669. doi: 10.1073/pnas.1432909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura K, Alexander RW. Cell Mol Life Sci. 2004;61:1317–1330. doi: 10.1007/s00018-004-3449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bürgi HB, Dunitz JD, Lehn JM, Wipff G. Tetrahedron. 1974;30:1563–1572. [Google Scholar]
- 9.Cardillo G, Gentilucci L, Tolomelli A, Tomasini C. Tetrahedron. 1999;55:6231–6242. [Google Scholar]
- 10.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. pp. 220–241. [Google Scholar]
- 11.Isaacs RJ, Rayens WS, Spielmann HP. J Mol Biol. 2002;319:191–207. doi: 10.1016/S0022-2836(02)00265-6. [DOI] [PubMed] [Google Scholar]
- 12.Venkateswarlu D, Lind KE, Mohan V, Manoharan M, Ferguson DM. Nucleic Acids Res. 1999;27:2189–2195. doi: 10.1093/nar/27.10.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerda BA, Hoyau S, Ohanessian G, Wesdemiotis C. J Am Chem Soc. 1998;120:2437–2448. [Google Scholar]
- 14.Berg P. J Biol Chem. 1958;233:608–611. [PubMed] [Google Scholar]
- 15.Illangasekare M, Sanchez G, Nickles T, Yarus M. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]