Abstract
Adrenomedullin (AM) is a multifunctional peptide vasodilator that is essential for life. Plasma AM expression dramatically increases during pregnancy, and alterations in its levels are associated with complications of pregnancy including fetal growth restriction (FGR) and preeclampsia. Using AM+/– female mice with genetically reduced AM expression, we demonstrate that fetal growth and placental development are seriously compromised by this modest decrease in expression. AM+/– female mice had reduced fertility characterized by FGR. The incidence of FGR was also influenced by the genotype of the embryo, since AM–/– embryos were more often affected than either AM+/– or AM+/+ embryos. We demonstrate that fetal trophoblast cells and the maternal uterine wall have coordinated and localized increases in AM gene expression at the time of implantation. Placentas from growth-restricted embryos showed defects in trophoblast cell invasion, similar to defects that underlie human preeclampsia and placenta accreta. Our data provide a genetic in vivo model to implicate both maternal and, to a lesser extent, embryonic levels of AM in the processes of implantation, placentation, and subsequent fetal growth. This study provides the first genetic evidence to our knowledge to suggest that a modest reduction in human AM expression during pregnancy may have an unfavorable impact on reproduction.
Introduction
Normal mammalian fetal growth and survival depend on the proper development and function of the placenta, which serves to maintain a maternal-fetal interface for the exchange of blood gases, nutrients, and waste. Reductions in placental blood flow due to placental abruption (placental/uterine detachment), placenta previa (low placental/uterine attachment), or local uterine infection are known causes of fetal growth restriction (FGR) in humans (reviewed in ref. 1). Likewise, physiological disruption of placental blood flow in sheep (2), guinea pigs (3), and rats (4–6) also leads to FGR. Most recently, as reviewed by Rossant and Cross (7), the increasing number of genetically modified mouse models with fetal developmental defects caused by abnormal placental development or function underscores the importance of a fully functional placenta for normal fetal growth and survival.
The earliest stages of placental development in humans and mice occur during implantation, when mural trophectoderm cells from the fetal blastocyst attach and invade the receptive maternal uterine wall. Although there are morphological differences between humans and mice in the type of implantation that occurs (humans undergo interstitial implantation, while mice have eccentric implantation), the eventual fate and function of the trophectoderm — to bring fetal placental tissue into contact with maternal blood — are conserved (8). Consequently, the trophectoderm cells of the blastocyst differentiate into polyploid trophoblast cells (called trophoblast giant cells [TGCs] in mice and extravillous cytotrophoblasts in humans), which invade deeply into the receptive uterine lining, providing an endothelium-like barrier to support a low-resistance, hemochorial placental network. The process through which these specialized cells become invasive likely involves a complex dialogue between the embryo and uterus comprised of numerous biological processes such as vasodilation, angiogenesis, cellular growth and differentiation, and tissue remodeling (9). Each of these processes in turn is mediated by a group of diverse gene products — an increasing number of which are still being identified — that form a “local cocktail” of factors essential for the appropriate recognition, attachment, and invasion of the blastocyst to the uterine wall (10). The diversity of factors required for normal implantation is aptly reflected by the wide variety of mutant gene products in genetically engineered mice that affect normal placental development (10, 11). Moreover, defects in the ability of trophectoderm cells to fully invade the maternal uterine wall and vessels are thought to underlie many serious reproductive conditions in humans, such as preeclampsia, spontaneous abortion, and FGR (12, 13). Conversely, in placenta accreta, an overgrowth of fetal placental tissue beyond the maternal decidua causes abnormal anchoring of the placenta to the uterine myometrium (14, 15). Thus, there is a growing interest in identifying the cohort of genes and factors that are expressed and secreted by maternal and/or fetal tissues within the local implantation site and determining how these factors come together to control normal implantation, placentation, and fetal growth.
Adrenomedullin (AM) is a multifunctional peptide that incorporates many of the biological functions associated with normal implantation. It is now recognized that this 52-amino-acid peptide, identified over a decade ago as a potent vasodilator (16), can also act as an angiogenic factor, growth factor, natriuretic factor, immune modulator, and antimicrobial agent, to name a few functions (17). AM, its G protein–coupled receptor (calcitonin receptor–like receptor [CLR]), and its associated receptor activity–modifying proteins (RAMPs) are expressed by numerous reproductive tissues including the uterine endometrium (18), fetal membranes (19, 20), placenta (21), and trophoblast cells (19, 22–26), suggesting an important role for AM signaling in either fetal development or maternal adaptation to pregnancy.
Plasma levels of AM generally increase approximately 2-fold in response to a variety of cardiovascular conditions such as essential hypertension, renal failure, and congestive heart failure (27, 28). Yet one of the greatest increases in plasma AM levels occurs during the course of a normal pregnancy, when plasma levels of the peptide, secreted by the placenta, are 4- to 5-fold higher by the third trimester (21, 29–32) and rapidly return to pre-pregnancy values after delivery (21). As recently reviewed by Wilson et al. and Di Iorio et al., numerous clinical studies have attempted to correlate altered AM levels with a variety of pregnancy complications, including preeclampsia, spontaneous abortion, gestational diabetes, and FGR (33, 34). However, the findings are inconsistent among groups and are significantly affected by sampling time and the types of tissue that can be reasonably obtained from human pregnancies.
Our previous work using knockout mice has demonstrated the importance of fetal AM for survival, since AM–/– embryos that completely lack endogenous AM die at mid-gestation (E14.5) from extreme hydrops fetalis and cardiovascular defects (35). However, the effects of genetically reduced maternal and/or fetal AM levels on fertility and reproduction have not been described. Therefore, in this article we have used our genetically engineered AM+/– mice to ask whether a modest change in the level of expression of the AM gene affects pregnancy, placentation, and fetal growth.
Results
Reduced litter size in AM+/– matings that is independent of AM–/– embryonic lethality.
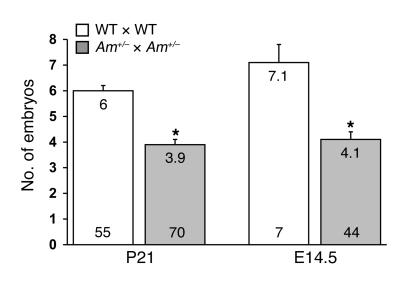
During the course of our studies to characterize the embryonic lethal phenotype of AM–/–embryos that occurs at E14.5 (35), we noticed a small but consistent decrease in the number of weaned offspring (~2.1 pups per litter) resulting from AM+/–heterozygote intercrosses compared with wild-type matings (Figure 1), which appeared greater than would be expected from the Mendelian in utero loss of AM–/–embryos alone (expected loss of 1.5 pups per litter). Therefore, to determine whether AM+/–matings resulted in a greater reduction in litter sizes than that expected from AM–/–embryonic lethality, we established timed pregnancies and counted the total number of viable, healthy embryos at E14.5, including in our assessment AM–/–embryos but excluding embryonic resorptions. We still found a significantly smaller litter size (4.1 pups per litter) compared with that resulting from similar timed pregnancies in wild-type 129S6/SvEv intercrosses (7.1 pups per litter) (Figure 1). Similar results were obtained when the litter sizes of wild-type and AM+/– females born to wild-type dams were compared (AM+/–, 4.2, n = 25 litters versus wild-type, 7.2, n = 6, at E14.5; P < 0.004), thereby ruling out any adverse effects of inherited epigenetic factors on the AM+/–phenotype. These data demonstrate that AM+/–intercrosses result in reduced fertility that occurs before the embryonic lethal loss of AM–/–embryos at E14.5.
Figure 1. Reduced litter size in AM+/– matings.
The average number of viable offspring at postnatal day 21 and the average number of embryos at gestational day E14.5 were determined for wild-type matings (white bars) and AM+/– intercrosses (gray bars). The actual average number is recorded at the top of each bar, while the number at the bottom of the bar represents the number of litters examined for each cross. *P < 0.01 compared with wild-type crosses.
Reduced litter size correlates with maternal AM+/– genotype.
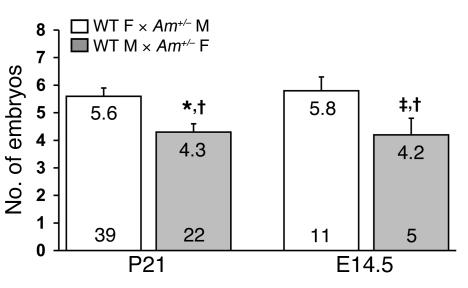
To determine whether the reduced fertility among AM+/–intercrosses was associated with the maternal and/or paternal AM+/–genotype, reciprocal matings were established in which only 1 parent was heterozygous for the targeted AM allele. The numbers of viable offspring at weaning and at E14.5 were determined and compared with those of either the opposite reciprocal cross or wild-type matings. We found a statistically significant drop in litter size in female AM+/–× male wild-type matings examined at either postnatal day 21 or E14.5 compared with that in either male AM+/– × female 129S6/SvEv or 129S6/SvEv intercrosses (Figure 2; P < 0.05 and P < 0.001, respectively). The number of viable offspring from male AM+/– × female 129S6/SvEv crosses did not differ significantly from that from female AM+/–× male wild-type matings or 129S6/SvEv intercrosses at weaning or at E14.5. Incidentally, the wild-type/heterozygote ratio of offspring from both reciprocal crosses was the expected Mendelian ratio of 54%:46% (male AM+/– × female wild-type) and 45%:55% (female AM+/–× male wild-type), demonstrating that neither the paternal nor the maternal AM allele is silenced by imprinting. Results from these reciprocal breeding studies establish that fertility defects observed in AM+/–× 129S6/SvEv crosses are caused largely by the heterozygous genotype of the mother.
Figure 2. Reduced litter size correlates with maternal AM+/– genotype.
Reciprocal heterozygote × wild-type matings were established, and the number of viable offspring at postnatal day 21 and the average number of embryos at gestational day 14.5 were determined. The gray bars represent wild-type males crossed with AM+/–females and the white bars represent wild-type females crossed with AM+/–males. The actual average number for each cross is recorded at the top of each bar, while the number at the bottom of the bar represents the number of litters examined for each cross. *P < 0.01, ‡P < 0.05 compared with the reciprocal cross; †P < 0.01 compared with wild-type crosses (Figure 1).
Abnormal implantation spacing in AM+/– females.
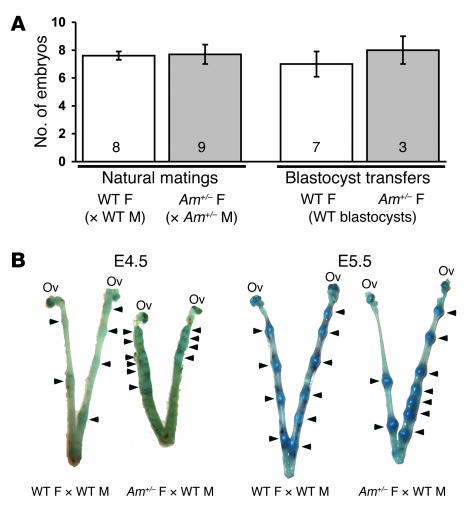
To determine the gestational stage at which embryonic loss occurred in AM+/–females, AM+/–intercrosses were established, and litters were dissected at various days between E5.5 and E8.5. Assessing litter size and embryonic viability during this gestational window provided information about the ability and efficiency of AM+/–female mice to ovulate, mate, and undergo fertilization and implantation. We found no significant difference in the total number or viability of embryos from natural AM+/–intercrosses compared with 129S6/SvEv wild-type natural matings (Figure 3A). Furthermore, transfer of wild-type blastocysts into receptive wild-type or AM+/–females also resulted in an equivalent number of implantations for AM+/–females compared with wild-type females (Figure 3A). Evans blue dye injection at E4.5 or E5.5 did not reveal any significant differences in the size of the conceptuses between AM+/–and wild-type females, suggesting that the timing of implantation in AM+/–female mice was appropriate (Figure 3B). However, the implantation staining revealed overcrowded conceptuses and abnormal spacing in AM+/–females compared with WT littermates (Figure 3B, arrowheads). Taken together, these results demonstrate that ovulation, fertilization, success rate and timing of implantation, and embryonic development up to E8.5 are not significantly different when the mother is heterozygous for AM. However, AM+/–females display abnormal spacing of and overcrowded conceptuses within the uterine horns.
Figure 3. Abnormal implantation spacing in AM+/–females.
(A) Timed natural matings or blastocyst transfers were performed in wild-type (white bars) and AM+/– female mice (gray bars). The number of implanted embryos was determined between gestational stages E5.5 and E8.5. The number at the bottom of the bars represents the number of litters examined for each cross. (B) Evans blue dye injections of pregnant wild-type and AM+/– female mice at gestational stages E4.5 and E5.5 revealed comparable conceptus sizes, demonstrating that timing of implantation was conserved in AM+/– female mice. However, AM+/– female mice displayed abnormal spacing of and overcrowded conceptuses in the uterine horn (arrowheads) compared with wild-type control females.
Embryonic loss in AM+/– matings is concurrent with placental development.
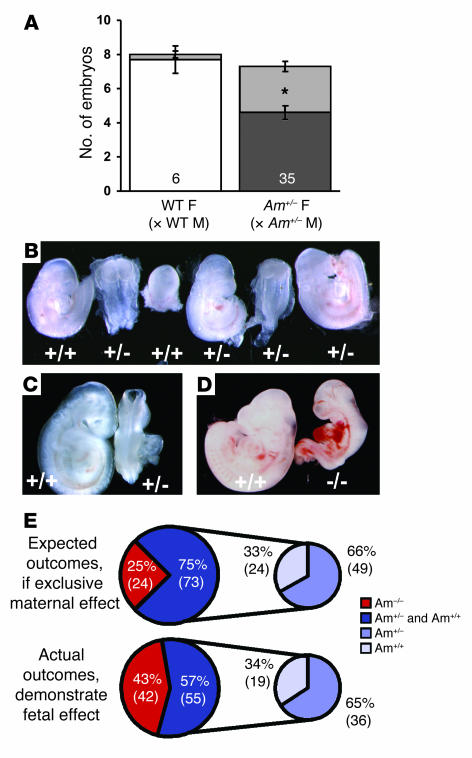
Using AM+/–intercrosses and 129S6/SvEv wild-type matings, we next examined litters between gestational days E9.5 and E12.5, which correlates with the development and maturation of the functional placenta in the mouse. Among the 6 litters of wild-type matings that were examined between E9.5 and E12.5, we found 2 of 48 total embryos were either resorbed or severely malformed (4% affected, represented as light gray in Figure 4A, left). In contrast, among the 35 dissected litters from AM+/–intercrosses, we found that 97 of 257 total embryos were either resorbed or severely malformed (38% affected, represented as light gray in Figure 4A, right). Importantly, this high percentage of embryonic loss is consistent with the assessment of reduced litter sizes at E14.5 and postnatal day 21 (42% and 35%, compared with 129S6/SvEv wild-type matings; shown in Figure 1). We conclude that the reduced fertility observed in the female AM+/– mice is due to embryonic loss that occurs between E9.5 and E12.5, a time that precedes the loss of AM–/–embryos and is simultaneous with the development of a mature and functional placenta.
Figure 4. FGR and developmental defects correlate with placentation and fetal AM genotype.
(A) Timed matings were established for wild-type (white bar) and AM+/– crosses (dark gray bar), and viable embryos were counted for gestational stages E9.5–E12.5. Severely growth restricted or resorbed embryos were included in the total litter size and are represented by the light gray area of the bars. * P < 0.001, viable embryos from AM+/– intercrosses compared with viable embryos from wild-type crosses at the same gestational stage. (B) Representative E9.5 litter from AM+/– intercross. The genotype of each embryo is indicated below. Note that growth restriction occurred in AM+/– and AM+/+ embryos. (C) Severe growth restriction and failure of neural tube closure in an AM+/– embryo (right) compared with an AM+/+ littermate (left) isolated at E9.5. (D) Moderate growth restriction and severe heart malformations in an AM–/– embryo (right) compared with an AM+/+ littermate (left) isolated at E9.5. (E) Expected outcomes and actual outcomes for genotypes of growth-restricted or abnormal embryos expressed as a percentage of total affected embryos. The smaller pie charts only include AM+/– and AM+/+affected embryos. Data are representative of 35 AM+/– litters examined at E9.5, which included 97 growth-restricted or malformed embryos.
Morphological characterization of embryos from AM+/–intercrosses at E9.5 revealed a high incidence of FGR and abnormal embryonic development. In a representative litter from an AM+/–intercross at E9.5, 3 of 6 embryos were severely developmentally delayed by as much as 2–4 days compared with normal E9.5 littermates (Figure 4B). We also observed a high incidence of morphological and developmental abnormalities that included failure of neural tube closure (Figure 4C), torqued body symmetry, and severe heart malformations (Figure 4D), which together are indicative of placental insufficiency.
FGR and developmental defects occur more often in AM–/– embryos.
Genotyping of the growth-restricted and malformed embryos revealed that homozygous loss of AM in the embryo is associated with a greater incidence of FGR and demise. As illustrated in Figure 4E, if the observed FGR were solely caused by maternal reduction in AM expression, then one would expect that the incidence of FGR should be random with respect to genotype and thus follow Mendelian distribution (top diagram of Figure 4E). However, our genotyping analysis revealed that 43% of the growth-restricted embryos were AM–/–, nearly twice the expected random incidence of 25% (bottom diagram of Figure 4E). More than half (57%) of the growth-restricted embryos were either AM+/+ or AM+/–, yet their relative distribution to each other remained in the expected 2:1 ratio (19 AM+/+:36 AM+/–; small pie charts). These genotyping analyses allow us to conclude that while heterozygous loss of embryonic AM does not significantly contribute to the growth restriction phenotype, complete loss of AM in the embryo is significantly detrimental to the process of early fetal growth and development.
High incidence of morphological placental defects in AM+/– intercrosses.
During the process of implantation, fetal mural trophectoderm cells lining the blastocyst differentiate into invasive TGCs, which burrow into the maternal uterine wall and demarcate the transition from fetal placental tissue to maternal decidual tissue. Figure 5A shows the gross morphology of a wild-type conceptus unit in which the fetus and fetal membranes have been removed by dissection. Histological sections show that by E9.5, the wild-type placenta lies firmly anchored to the central portion of the underlying maternal decidua (Figure 5B), and the morphologically and functionally distinct layers of the placenta can be easily distinguished (Figure 5C). Placentas from normal, non–growth-restricted fetuses of all genotypes from AM+/–intercrosses were morphologically and histologically indistinguishable from those of wild-type placentas at all gestational stages examined (E8.5–E14.5; data not shown).
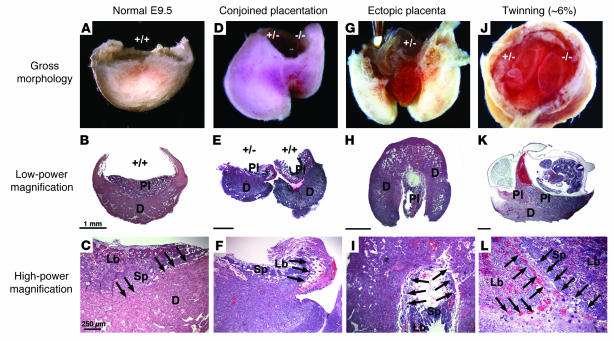
Figure 5. High incidence of morphological placental defects in AM+/– intercrosses at E9.
. (A) Saggital image of WT placental/decidual unit from E9.5 wild-type cross. Embryo and fetal membranes were removed. (B) H&E section of the sample in A, revealing fetal placental and maternal decidual tissues. (C) Normal histology of placental layers and maternal-fetal interface. (D) Saggital image of 2 placental/decidual units from E9.5 AM+/– intercross. Note the juxtaposition of implantations, with fusion of decidua and smaller size of right placental/decidual unit. The right embryo was growth restricted. (E) H&E section of another conjoined placenta unit embedded in the same juxtaposed manner as in uterus. The right sample has detached maternal-fetal interface, with invasion of the ectopic placenta from the left embryo. (F) Abnormal invasion of TGCs (arrows), small labyrinth layer, significantly reduced spongiotrophoblast layer, and almost no maternal-fetal interface are present. (G) Saggital image of placental/decidual unit with ectopic placenta that has overgrown the decidual tissue. (H and I) H&E sections through distal pole of the sample in G. Note the absence of placental layers and maternal-fetal interface and the presence of edema. (J) Rostral image of twinned placental/decidual unit from E9.5 AM+/– intercross. Note 2 fetal membranes within 1 decidua. Neither embryo appeared growth restricted. (K) H&E section through another twinned conceptus unit. Note shared decidua, 2 placentas, pool of maternal blood, and growth-restricted embryo. Genotypes were not determined. (L) Maternal-fetal interface of the sample in K. Arrows show 2 populations of TGCs invading at the same place. Pl, placenta; D, decidua; Lb, labyrinth; Sp, spongiotrophoblast. Arrows show TGCs. Scale bars: 1 mm (B, E, H, and K); 250 μm (C, F, I, and L).
However, most of the growth-restricted embryos from AM+/–intercrosses also had marked morphological placental defects (Figure 5, D–L). Most commonly observed were closely spaced, conjoined placentas, which at the macroscopic level appeared as 2 interlocked deciduas within 1 conceptus unit (Figure 5D). Histology of the conjoined placentas showed fetal placental tissue from one conceptus inappropriately invading the adjoining maternal-fetal interface (Figure 5E). In all cases, the leading edge of the invading placental tissue was lined with TGCs immersed in large pools of maternal blood (Figure 5F). We also observed a high incidence of ectopic placentation, in which the fetal placental tissue hypertrophied and extended beyond the confines of the maternal decidua (Figure 5, G and H) and completely lacked the normal histological characteristics of a wild-type placenta. Finally, the spontaneous occurrence of twinning in mice is exceedingly rare and has previously been reported to be as low as 0.4% in an inbred strain (36). Remarkably, we found a high incidence of dizygotic twinning among AM+/–intercrosses (approximately 6%), which we interpret as the most severe manifestation of uterine overcrowding, placental sharing, and conjoined placentation (Figure 5J). Histological analysis of twinned conceptus units revealed significant FGR (Figure 5K), with 2 TGC-lined placentas sharing a single maternal decidua (Figure 5L). In all instances of morphologically abnormal placentation, we found evidence of fragile or partial maternal-fetal interfaces and misplaced TGCs. Taken together, these observations imply that the early processes of uterine spacing and TGC invasion during implantation are disrupted in AM+/–intercrosses, thus leading to poor maternal-fetal exchange and eventual FGR.
Fetal AM gene expression is upregulated at implantation.
Since our fertility and developmental studies of AM+/–intercrosses demonstrated a significant effect of fetal AM–/–genotype on the incidence of FGR and abnormal placental development, we sought to characterize the relative levels of AM gene expression by fetal-derived preimplanting trophectoderm cells and differentiated TGCs. Using an in vivo enhanced GFP (EGFP) biomarker that was inserted at the initiator methionine of the AM coding sequence during homologous recombination (35), we found robust levels of EGFP fluorescence (as a biomarker of AM gene expression) in the mural trophectoderm cells of a preimplanting blastocyst (E3.5) (Figure 6A). The relative level of fluorescence in the trophectoderm lineage was slightly higher than the levels observed in the inner cell mass. Based on the fact that AM–/– blastocysts carry 2 copies of the EGFP gene, we found that, as expected, they displayed approximately twice the intensity of reporter gene fluorescence as AM+/– blastocysts with only 1 copy of EGFP (Figure 6A), demonstrating an appropriate dose of the AM gene in preimplanting trophectoderm.
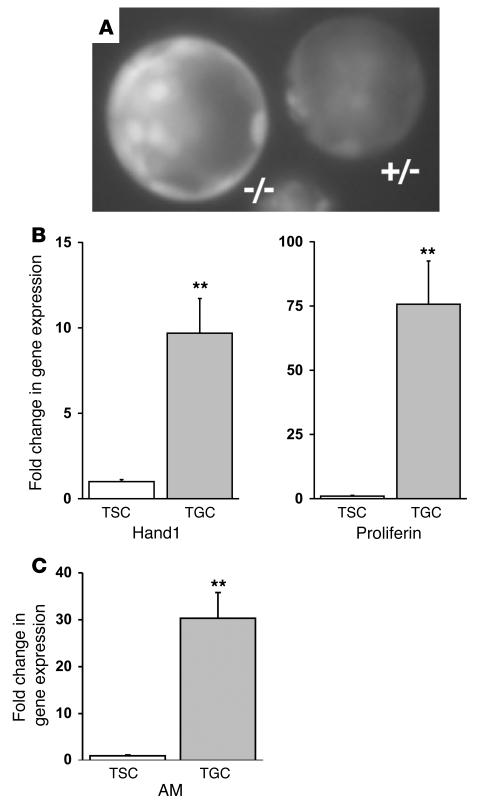
Figure 6. Fetal AM gene expression is upregulated in invasive TGCs.
(A) Fluorescence microscopy of preimplanting blastocysts generated from an AM+/– intercross and flushed from the uterine horn of an AM+/– female at E3.5. Note the strong fluorescence present in both mural and polar trophectoderm cells and a fainter signal in the inner cell mass. The left blastocyst is AM–/– and has approximately twice the intensity of fluorescence as the right AM+/– blastocyst. (B) Confirmation of TGC differentiation in a TS cell line using Hand1 and proliferin gene expression markers. Results from the quantitative RT-PCR experiment show the fold change in gene expression in differentiated TGCs compared with that in the undifferentiated TS cell culture, which was arbitrarily set at 1. (C) Quantitative measurement of AM gene expression in differentiated TGCs compared with that in the undifferentiated TS cell culture. All data were normalized to a GAPDH internal control. **P < 0.001 compared with undifferentiated TS cells.
To determine whether AM continued to be expressed by invading TGCs, we used a cell culture–based system in which murine trophoblast stem cells (TS cells; analogous to preimplanting trophectoderm cells) are differentiated into invading TGCs by removal of FGF from the growth media (37). To confirm that our cell culture and differentiation conditions resulted in a population of cells enriched for the TGC lineage, we measured the relative gene expression of 2 TGC-specific markers, Hand1 and proliferin, using real-time quantitative RT-PCR. As shown in Figure 6B, we found that the differentiated cell population expressed nearly 10-fold higher levels of Hand1 and 75-fold higher levels of proliferin compared with the undifferentiated TS cell population. Consistent with these results, we also observed by microscopy an increase in extremely large, nucleated cells resembling TGCs (data not shown). Quantitative RT-PCR revealed dramatic (30-fold) upregulation of AM gene expression in differentiated TGCs compared with the undifferentiated TS cells. Taken together, these in vitro results indicate that the expression of AM is dynamically and markedly increased during the transition from mural trophectoderm cells to invading TGCs, a process that occurs during implantation in vivo.
Maternal uterine expression of AM is localized to implantation sites.
Because the reproductive and developmental phenotypes we observed occurred only in AM+/–females, we sought to characterize the pattern and level of maternal AM expression in the receptive uterus at implantation. In situ hybridization for AM was performed on serial sections from individual implantation sites collected from wild-type 129S6/SvEv females at E5. As shown in the consecutive images in Figure 7, A–E, uterine AM expression is strictly localized and concentrated around the implanting and developing embryo. Specifically, the luminal epithelium and several surrounding subepithelial cell layers of the stroma express high levels of AM (Figure 7F), which rapidly dissipate away laterally from the implanted embryo (Figure 7, A and E). The robust expression of AM in the maternal decidua, compared with weak expression in the fetal placenta, persists throughout development (Figure 7, G and H). These results demonstrate that maternal AM expression is tightly coordinated, localized to implantation sites, and persistently robust throughout development.
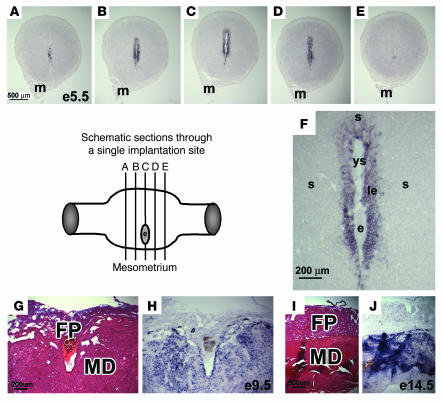
Figure 7. Maternal uterine expression of AM is localized to implantation sites.
(A–E) In situ hybridizations for AM gene expression in consecutive serial sections of an implantation site isolated from a wild-type cross at E5. The schematic diagram depicts the approximate location of the series of sections shown in A–E, which were each approximately 150 μm apart. Note the gradual increase in maternal uterine AM gene expression that peaked at the center of the implantation site in C next to the developing embryo. (F) High-power magnification of the implantation site shown in C. Note the presence of the embryo (e) and yolk sac cavity (ys). The fetal tissue is surrounded by a thin line of intense, punctuate staining, likely representative of mural TGCs. Maternal expression of AM can be visualized in the luminal epithelium (le) and surrounding stroma (s). H&E staining of wild-type placentas at E9.5 (G) and E14.5 (I), with corresponding in situ hybridizations of serial sections (H and J, respectively) showing robust AM expression in the maternal decidua (MD) compared with weak expression in the fetal placenta (FP). m, mesometrium.
Discussion
The process of implantation consists of characteristic increases in angiogenesis and vascular permeability around the blastocyst, which aid in the delivery of substances that are required for the establishment of the placental maternal-fetal circulation. Together these substances (including vasodilators, angiogenic factors, growth factors, tissue remodeling factors, and immune modulators) contribute to a localized environment that supports decidualization, invasion of fetal trophoblasts into the maternal tissue, and eventual placental development and fetal growth. The characterization of numerous genetically engineered mouse models with either maternal-derived or fetal-derived defects in implantation make it clear that biological signals from both mother and fetus are essential for maintaining the complex “maternal-fetal dialogue” that mediates these processes (8, 10). However, in nearly all of these models, the phenotypes only occur on a homozygous-null background. To our knowledge, this is the first report of reproductive defects that occur on a maternal heterozygote background in which AM expression is modestly reduced, and so the implications of variation in AM peptide levels for human reproductive health are likely significant.
As summarized in Figure 8, our data show that AM peptide is highly expressed by both the maternal uterine luminal epithelium and the fetal trophectoderm at the time and site of implantation. When female mice were wild-type for AM, we found normal fertility and no appreciable FGR, even when they were mated to AM+/–males. In contrast, a modest 50% reduction in uterine AM in AM+/– female mice caused significantly reduced fertility and FGR even in wild-type and AM+/–embryos, demonstrating a critical role for the dose of maternal AM on fetal growth. Genotype analysis from AM+/– intercrosses and results from reciprocal crosses using wild-type females mated to AM+/–males did not reveal a significant dose effect of heterozygous loss of fetal AM on fertility or fetal growth. However, the incidence of FGR was significantly exacerbated when the implanting blastocyst was null for AM. These data suggest that fetal expression of AM also contributes significantly to the early stages of embryonic development. Taken together, our data provide a genetic in vivo model that implicates both maternal and, to a lesser extent, fetal sources of AM peptide in mediating the complex process of implantation and early fetal growth.
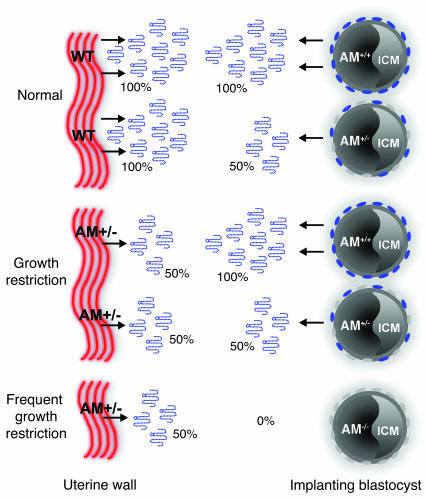
Figure 8. Effects of maternal and fetal depletion of AM in pregnancy.
In wild-type mice (Normal), AM gene expression is highly and coordinately upregulated in the maternal uterine luminal epithelium and surrounding stroma as well as fetal trophectoderm cells at the time of implantation. Due to effective gene targeting of the AM gene, AM peptide secretion is reduced by 50% in the uterine wall of heterozygote females. This modest genetic reduction results in abnormal implantation spacing, morphological defects in placentation, FGR, and subsequent reduced fertility. Implanting blastocysts from AM+/– intercrosses can be of 3 genotypes, AM+/+, AM+/–, or AM–/–, and thus they contribute different amounts of AM peptide to the implantation site: 100%, 50%, or 0% of normal, respectively. Thus, the combined effects of maternal and fetal depletion of AM result in reduced and varying concentrations of AM peptide at the implantation site. While heterozygous loss of fetal AM does not significantly affect the incidence of FGR, a greater incidence of FGR is significantly associated with AM–/–embryos, suggesting an essential function for the peptide during early embryonic development. ICM, inner cell mass.
The hallmark function of AM is mediation of vasodilation (38), and thus the most likely role for AM in the maternal uterine tissue is to promote blood flow to the implantation site. However, it is worth noting that AM is also an angiogenic factor (39–41) and has been shown to regulate vascular permeability in vitro (42), so that its contributions to maternal vascular remodeling and permeability that occur during implantation are likely several fold. In support of this notion, maternal AM expression has been localized to both the epithelium and endothelium of receptive uterine tissue (18) as well as stromal macrophages (43). The occurrence of conjoined placentation and our histological observation of “mis-guided” TGCs imply that AM, secreted either from maternal or fetal tissues, may act as a migratory factor for these cells during invasion. In support of this possibility, AM can be an effective chemoattractant and migratory factor for a variety of cell types (44, 45), including cultured choriocarcinoma JAr cells and first-trimester cytotrophoblast cells (46). Therefore, it is quite likely that maternally derived AM at the implantation site plays numerous roles in the establishment and maintenance of a normal pregnancy. Finally, it is worth considering that AM may mediate these processes through interactions with other molecular factors previously shown to be important for postimplantation development (for example, growth factors, vasoactive mediators, cytokines, adhesion and matrix remodeling factors, and/or transcription and cell cycle factors; reviewed in ref. 10).
Our previous studies showed an essential role for the AM gene for embryonic survival (35). The data presented in this study demonstrate that absence of AM in fetal tissues is also correlated with an increase in the incidence of FGR during the early stages of gestation (Figure 4). In agreement with our findings, others have shown that injection of an AM peptide inhibitor, AM22-52, in the uterus of pregnant rats during early and late gestation caused significant FGR and demise (47, 48). However, it is not possible from these experiments to determine whether the effects of the inhibitor on fetal growth were due to inhibition of AM in the fetus, in the maternal uterine tissue or circulation, or in both. Significantly, Di Iorio and colleagues found that fetoplacental levels of AM peptide were increased in human patients with intrauterine growth restriction (49), while another group found no significant differences in fetal or maternal AM levels between normal pregnancies and pregnancies with FGR (50). Several other human studies have shown altered AM levels in the fetoplacental tissues associated with other types of pregnancy complications, such as spontaneous abortion (34, 51), gestational diabetes (52), and preeclampsia (53–57). Thus, the effects of fetal-derived AM on the outcome of pregnancy in humans have been difficult to define. These studies with mice establish a critical role for fetal-derived AM peptide in the early process of implantation and normal fetal growth and survival.
According to previous studies in mice, the earliest expression of AM in fetal tissues occurs at E8 in the heart (24) and at E6.5 in the TGCs and ectoplacental cone (25). Interestingly, Yotsumoto et al. also showed that the expression of AM in TGCs peaked at E9.5, the same gestational stage at which we observe FGR and demise (25). Using an in vivo EGFP reporter gene, we now extend these findings by showing that fetal AM is expressed in the trophectoderm as early as E3.5 in preimplanting blastocysts. Using cultured TS cells, we also found that AM gene expression is profoundly increased during the differentiation of these cells into an invasive, TGC lineage. Similar studies using purified trophoblast cells from term human placentas have shown that these cells actively secrete AM into the culture media for 48 hours and subsequently reduce their production of AM by 70% as the cells spontaneously differentiate into noninvasive syncytiotrophoblast cells (34). Taken together, these data show that expression and secretion of AM peptide from trophoblast cells is tightly and dynamically regulated during the early stages of implantation and support our current model, according to which the gene significantly affects fetal growth and development.
More clues about the function of AM in early gestation can be gathered from the expression profiles of other genes that are required for AM signaling. The AM signaling system consists of CLR and its associated RAMPs (RAMP2 and RAMP3). Association of CLR with either RAMP2 or RAMP3 confers specificity for AM binding, while association with RAMP1 changes the ligand binding specificity to another related vasodilator called calcitonin gene–related peptide (CGRP) (58, 59). Consequently, in this novel paradigm of signaling, the spatial and temporal expression of the RAMPs with the CLR receptor is what dictates tissue responsiveness to AM peptide. Using Northern blot analysis, Yotsumoto et al. found high levels of CLR expression in late-gestation mouse embryonic samples but little to no expression in E7 samples that contained both embryonic and extraembryonic tissues (25). The authors speculated that the lack of CLR in the same fetal tissues that highly secrete AM peptide signifies a paracrine mode of action (19, 60). In contrast, Tsatsaris et al. used RT-PCR to show high expression of CLR in extravillous trophoblast cells isolated from first-trimester human placentas, suggesting the possibility of an autocrine, fetal function for AM (26). In preliminary breedings of mice heterozygous for deletion of the Calcrl gene (61), we also find reduced fertility that is independent of homozygous embryonic lethality, suggesting an overlapping role for the AM receptor gene in reproductive outcomes (R.T. Dackor and K.M. Caron, unpublished observations). Perhaps most compelling have been studies that show an increased expression of the RAMPs in pregnant versus nonpregnant rat uterus (62) and a remarkable 30- to 45-fold increase in murine uterine RAMP3 expression with estradiol treatment (63). Results from these expression studies suggest a signaling mechanism, in part regulated by sex steroids (64, 65), that is capable of directing the dynamic modulation of AM responsiveness during implantation, fetal development, and pregnancy.
In general, the abnormal placental phenotypes we observed imply that AM peptide at the implantation site is involved in the early stages of trophectoderm invasion and guidance of developing placental tissue through the uterine luminal epithelium. Specifically, we found shallow placentation, fragile maternal-fetal interfaces, and ectopic growth of placental tissue beyond the decidual boundary (Figure 5). Similar defects in trophoblast invasion and placental development are thought to underlie several reproductive diseases in humans. Most notably, shallow invasion of spiral arteries leading to a poor maternal-fetal placental interface is associated with preeclampsia. In contrast, placenta accreta is a condition in which overgrowth of the placenta beyond the boundary of the decidua basalis causes adherent placental tissue to abnormally attach to the uterine wall. Both preeclampsia and placenta accreta are associated with significant maternal mortality and fetal complications, yet the etiological or genetic factors leading to these conditions remain unclear. Genetic polymorphisms in the human AM gene have been associated with alterations in basal blood pressure and diabetic nephropathy (66, 67). Thus, our current studies with heterozygote mice that have a genetic reduction in AM expression not only provide insights into the role AM has in the processes of implantation and fetal growth but further raise the intriguing possibility that modest alterations in AM gene expression in the human population may contribute significantly to overall reproductive health.
Methods
Mice.
Generation of mice with a targeted, homozygous deletion of the AM gene has been previously described (35). The AM+/– line is maintained as a heterozygote breeding colony on an isogenic 129S6/SvEv background. Unless otherwise stated, experimental and control animals were strain- and age-matched and born from AM+/–intercrosses. For timed pregnancies, heterozygote AM+/–or wild-type 129S6/SvEv breedings were established, and the day when the vaginal plug was detected was considered E0.5. Genotyping was performed by a 3-primer, PCR-based strategy: primer 1: 5′-CAGTGAGGAATGCTAGCCTC-3′; primer 2: 5′-GCTTCCTCTTGCAAAACCACA-3′; primer 3: 5′-TCGAGCTTCCAAGGAAGACCAGG-3′. Primers 1 and 3 amplify a 1.8-kb wild-type product, while primers 2 and 3 amplified a 1.3-kb targeted product. All experiments were approved by the Institutional Animal Care and Use Committee of The University of North Carolina at Chapel Hill (UNC-CH).
Blastocyst transfer.
Blastocyst transfer experiments were performed as previously described (68). Briefly, 3.5-day-old blastocysts were collected from uteri of superovulated C57BL/6 donor female mice purchased from Harlan. Fourteen to 16 blastocysts were transferred into the uteri of 8- to 12-week-old AM+/–females or wild-type recipient females anesthetized with avertin (0.5 mg/g body weight) at pseudopregnant day E3.5. In breedings with vasectomized males, the morning of the vaginal plug was designated as day E0.5. Recipient females were euthanized 5 days later, and the number of implanted blastocysts in the pregnant mice was calculated.
Histology.
Placentas were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Five-micrometer sections were stained with H&E.
TS cell culture and differentiation.
TS cells (1 × 105) were seeded on a 60-mm dish and grown at 37°C under 95% humidity and 5% CO2. TS cell medium contained 20% FBS, 1 mM sodium pyruvate, 50 μg/ml penicillin/streptomycin, 100 μM β-mercaptoethanol, 25 ng/ml FGF4 (F8424; Sigma-Aldrich), and 1 μg/ml heparin (H3149; Sigma-Aldrich) in RPMI 1640 (Invitrogen), with 70% of the medium being preconditioned by incubation with mouse embryonic fibroblasts for 72 hours. To differentiate TS cells into TGCs, the TS cells were grown in TS medium that was not preconditioned and lacked FGF4 and heparin. After 5 days of culture, TS cells and differentiated TG cells were washed with PBS and collected for RNA extraction.
Gene expression analysis.
Hand1, proliferin, AM, and Calcrl gene expression were analyzed by quantitative RT-PCR with the Mx3000P from Stratagene. Primers for Hand1 amplification were 5′-GGATGCACAAGCAGGTGAC-3′ and 5′-CTCGAGAAGGCATCAGGGTACT-3′. The probe sequence for Hand1 detection was 5′-FAM TCGTCCCCTTCTTGGCTCTGCG-TAMRA-3′. Primers for proliferin amplification were 5′-CAAGCCAGGCTCACACACTA-3′ and 5′-GATCGTCCAGAGGGCTTTCC-3′. The probe sequence for proliferin detection was 5′-FAM-TGAAATCAGGAGCCATGATTTTGGATGC-TAMRA-3′. Primers for AM amplification were 5′-GAGCGAAGCCCACATTCGT-3′ and 5′-GAAGCGGCATCCATTGCT-3′. The probe sequence for AM detection was 5′-FAM-CTACCGCCAGAGCATGAACCAGGG-TAMRA-3′. GAPDH served as an internal control, and the reagents were purchased from Applied Biosystems (part number 4308313).
RNA was isolated from TS cell cultures and differentiated TGC cultures with an RNeasy Kit (74104; Qiagen) and subsequently DNase treated (79254; Qiagen). Two hundred nanograms of total RNA was used in each reaction. The ΔΔCt method was used to determine the relative levels of gene expression, and values were expressed as the fold change over TS cell expression, normalized to whole placenta RNA and arbitrarily set at 1. Assays were repeated 3 times, each with triplicates.
In situ hybridization.
Wild-type 129S6/SvEv female mice at E4.5 were euthanized 5 minutes after tail vein injection of 1 ml 0.5% Evans blue dye. Uteri were fixed in 4% paraformaldehyde overnight and 30% sucrose in diethylpyrocarbonate-PBS (DEPC-PBS) overnight. Blue implantations were serially cryosectioned (20 μm). Sense and anti-sense RNA probes were synthesized from a plasmid containing 524 bp cDNA sequence spanning the 3 coding exons of the AM gene. Nonradioactive in situ hybridization reagents were used (1745816, 1585762, 1093274; Roche Diagnostics). Following prehybridization, sections were hybridized to digoxigenin-labeled AM sense or antisense RNA probes for 12–16 hours at 60°C. After washing, the sections were incubated with anti-digoxigenin Fab-AP diluted 1:2,000 in blocking buffer at room temperature for 2 hours. After washing, the sections were developed in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) solution in the dark overnight, then rinsed several times in PBS at room temperature and mounted. Sections hybridized with the sense probe served as negative controls and showed no positive signal.
Statistics.
Statistical analyses were performed with JMP software (SAS). One-way ANOVA was used to determine significance at P < 0.05. In all figures, error bars represent SEM.
Acknowledgments
This work was supported by grants from the Burroughs Wellcome Fund and the NIH (HD046970 to K.M. Caron and HL49277 to O. Smithies). The authors gratefully acknowledge James Cross for his expert advice and discussions about mouse placental biology and Steve Young for his discussions about human reproductive disorders. We also thank Robert Bagnell, Sundeep Kalantry, Kathleen S. Christine, Kimberly Fritz-Six, and members of the UNC-CH Neuroscience Center Expression Localization Core Facility (NIH grant NS031768) for helpful advice and technical assistance.
Footnotes
Nonstandard abbreviations used: AM, adrenomedullin; CLR, calcitonin receptor–like receptor; EGFP, enhanced GFP; FGR, fetal growth restriction; RAMP, receptor activity–modifying protein; TGC, trophoblast giant cell; TS cell, trophoblast stem cell.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:2653–2662 (2006). doi:10.1172/JCI28462
References
- 1.Baschat A.A. Pathophysiology of fetal growth restriction: implications for diagnosis and surveillance. Obstet. Gynecol. Surv. 2004;59:617–627. doi: 10.1097/01.ogx.0000133943.54530.76. [DOI] [PubMed] [Google Scholar]
- 2.Anthony R.V., Scheaffer A.N., Wright C.D., Regnault T.R. Ruminant models of prenatal growth restriction. Reprod. Suppl. 2003;61:183–194. [PubMed] [Google Scholar]
- 3.Scheffen I., et al. Alterations of the fetal capillary bed in the guinea pig placenta following long-term hypoxia. Adv. Exp. Med. Biol. 1990;277:779–790. doi: 10.1007/978-1-4684-8181-5_89. [DOI] [PubMed] [Google Scholar]
- 4.Ogata E.S., Swanson S.L., Collins J.W., Finley S.L. Intrauterine growth retardation: altered hepatic energy and redox states in the fetal rat. Pediatr. Res. 1990;27:56–63. doi: 10.1203/00006450-199001000-00017. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz J.E., Kovach A., Meyer J., McConnell C., Iwamoto H.S. Brief, intermittent hypoxia restricts fetal growth in Sprague-Dawley rats. Biol. Neonate. 1998;73:313–319. doi: 10.1159/000013990. [DOI] [PubMed] [Google Scholar]
- 6.Van Geijn H.P., Kaylor W.M., Nicola K.R., Zuspan F.P. Induction of severe intrauterine growth retardation in the Sprague-Dawley rat. Am. J. Obstet. Gynecol. 1980;137:43–47. doi: 10.1016/0002-9378(80)90384-1. [DOI] [PubMed] [Google Scholar]
- 7.Rossant J., Cross J.C. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 8.Lee K.Y., DeMayo F.J. Animal models of implantation. Reproduction. 2004;128:679–695. doi: 10.1530/rep.1.00340. [DOI] [PubMed] [Google Scholar]
- 9.Simmons D.G., Cross J.C. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 2005;284:12–24. doi: 10.1016/j.ydbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Dey S.K., et al. Molecular cues to implantation. Endocr. Rev. 2004;25:341–373. doi: 10.1210/er.2003-0020. [DOI] [PubMed] [Google Scholar]
- 11.Watson E.D., Cross J.C. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda). 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- 12.Chaddha V., Viero S., Huppertz B., Kingdom J. Developmental biology of the placenta and the origins of placental insufficiency. Semin. Fetal Neonatal Med. 2004;9:357–369. doi: 10.1016/j.siny.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Lala P.K., Chakraborty C. Factors regulating trophoblast migration and invasiveness: possible derangements contributing to pre-eclampsia and fetal injury. Placenta. 2003;24:575–587. doi: 10.1016/s0143-4004(03)00063-8. [DOI] [PubMed] [Google Scholar]
- 14.Kazandi M. Placenta percreta: report of two cases and review of the literature. Clin. Exp. Obstet. Gynecol. 2003;30:70–72. [PubMed] [Google Scholar]
- 15.Comstock C.H. Antenatal diagnosis of placenta accreta: a review. Ultrasound Obstet. Gynecol. 2005;26:89–96. doi: 10.1002/uog.1926. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura K., et al. Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma. Biochem. Biophys. Res. Commun. 1993;192:553–560. doi: 10.1006/bbrc.1993.1451. [DOI] [PubMed] [Google Scholar]
- 17.Beltowski J., Jamroz A. Adrenomedullin — what do we know 10 years since its discovery? Pol. J. Pharmacol. 2004;56:5–27. [PubMed] [Google Scholar]
- 18.Hague S., et al. Expression of the hypoxically regulated angiogenic factor adrenomedullin correlates with uterine leiomyoma vascular density. Clin. Cancer Res. 2000;6:2808–2814. [PubMed] [Google Scholar]
- 19.Nikitenko L.L., et al. Differential and cell-specific expression of calcitonin receptor-like receptor and receptor activity modifying proteins in the human uterus. Mol. Hum. Reprod. 2001;7:655–664. doi: 10.1093/molehr/7.7.655. [DOI] [PubMed] [Google Scholar]
- 20.Trollmann R., et al. Adrenomedullin gene expression in human placental tissue and leukocytes: a potential marker of severe tissue hypoxia in neonates with birth asphyxia. Eur. J. Endocrinol. 2002;147:711–716. doi: 10.1530/eje.0.1470711. [DOI] [PubMed] [Google Scholar]
- 21.Minegishi T., et al. Adrenomedullin and atrial natriuretic peptide concentrations in normal pregnancy and pre-eclampsia. Mol. Hum. Reprod. 1999;5:767–770. doi: 10.1093/molehr/5.8.767. [DOI] [PubMed] [Google Scholar]
- 22.Gratton R.J., Gluszynski M., Mazzuca D.M., Nygard K., Han V.K. Adrenomedullin messenger ribonucleic acid expression in the placentae of normal and preeclamptic pregnancies. . J. Clin. Endocrinol. Metab. 2003;88:6048–6055. doi: 10.1210/jc.2003-030323. [DOI] [PubMed] [Google Scholar]
- 23.Marinoni E., et al. Immunoreactive adrenomedullin in human fetoplacental tissues. Am. J. Obstet. Gynecol. 1998;179:784–787. doi: 10.1016/s0002-9378(98)70083-3. [DOI] [PubMed] [Google Scholar]
- 24.Montuenga L.M., Martinez A., Miller M.J., Unsworth E.J., Cuttitta F. Expression of adrenomedullin and its receptor during embryogenesis suggests autocrine or paracrine modes of action. Endocrinology. 1997;138:440–451. doi: 10.1210/endo.138.1.4881. [DOI] [PubMed] [Google Scholar]
- 25.Yotsumoto S., et al. Expression of adrenomedullin, a hypotensive peptide, in the trophoblast giant cells at the embryo implantation site in mouse. Dev. Biol. 1998;203:264–275. doi: 10.1006/dbio.1998.9073. [DOI] [PubMed] [Google Scholar]
- 26.Tsatsaris V., et al. Calcitonin gene-related peptide (CGRP) and CGRP receptor expression at the human implantation site. J. Clin. Endocrinol. Metab. 2002;87:4383–4390. doi: 10.1210/jc.2002-020138. [DOI] [PubMed] [Google Scholar]
- 27.Eto T., Kitamura K., Kato J. Biological and clinical roles of adrenomedullin in circulation control and cardiovascular diseases. Clin. Exp. Pharmacol. Physiol. 1999;26:371–380. doi: 10.1046/j.1440-1681.1999.03047.x. [DOI] [PubMed] [Google Scholar]
- 28.Jougasaki M., Burnett J.C. Adrenomedullin: potential in physiology and pathophysiology. Life Sci. 2000;66:855–872. doi: 10.1016/s0024-3205(99)00358-6. [DOI] [PubMed] [Google Scholar]
- 29.Di Iorio R., Marinoni E., Scavo D., Letizia C., Cosmi E.V. Adrenomedullin in pregnancy. Lancet. 1997;349:328. doi: 10.1016/s0140-6736(05)62827-9. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi K., et al. Immunoreactive adrenomedullin (AM) concentration in maternal plasma during human pregnancy and AM expression in placenta. Eur. J. Endocrinol. 2000;142:683–687. doi: 10.1530/eje.0.1420683. [DOI] [PubMed] [Google Scholar]
- 31.Hoshimoto K., Hayashi M., Ohkura T. Mature adrenomedullin concentrations in plasma during pregnancy. J. Matern. Fetal Neonatal Med. 2002;11:126–129. doi: 10.1080/jmf.11.2.126.129. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi Y., Ueyama H., Mashimo T., Kangawa K., Minamino N. Circulating mature adrenomedullin is related to blood volume in full-term pregnancy. Anesth. Analg. 2005;101:1816–1820. doi: 10.1213/01.ANE.0000182329.02880.83. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C., Nikitenko L.L., Sargent I.L., Rees M.C. Adrenomedullin: multiple functions in human pregnancy. Angiogenesis. 2004;7:203–212. doi: 10.1007/s10456-004-4183-5. [DOI] [PubMed] [Google Scholar]
- 34.Di Iorio R., Marinoni E., Letizia C., Cosmi E.V. Adrenomedullin in perinatal medicine. Regul. Pept. 2003;112:103–113. doi: 10.1016/s0167-0115(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 35.Caron K.M., Smithies O. Extreme hydrops fetalis and cardiovascular abnormalities in mice lacking a functional Adrenomedullin gene. Proc. Natl. Acad. Sci. U. S. A. 2001;98:615–619. doi: 10.1073/pnas.021548898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan Y.J., Wu T.C., Damjanov I. Twinning and conjoined placentation in mice. J. Exp. Zool. 1982;221:81–86. doi: 10.1002/jez.1402210110. [DOI] [PubMed] [Google Scholar]
- 37.Hughes M., et al. The Hand1, Stra13 and Gcm1 transcription factors override FGF signaling to promote terminal differentiation of trophoblast stem cells. Dev. Biol. 2004;271:26–37. doi: 10.1016/j.ydbio.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura M., et al. Potent and long-lasting vasodilatory effects of adrenomedullin in humans. Comparisons between normal subjects and patients with chronic heart failure. Circulation. 1997;95:1214–1221. doi: 10.1161/01.cir.95.5.1214. [DOI] [PubMed] [Google Scholar]
- 39.Oehler M.K., Hague S., Rees M.C., Bicknell R. Adrenomedullin promotes formation of xenografted endometrial tumors by stimulation of autocrine growth and angiogenesis. Oncogene. 2002;21:2815–2821. doi: 10.1038/sj.onc.1205374. [DOI] [PubMed] [Google Scholar]
- 40.Kim W., et al. Angiogenic role of adrenomedullin through activation of Akt, mitogen-activated protein kinase, and focal adhesion kinase in endothelial cells. Faseb J. 2003;17:1937–1939. doi: 10.1096/fj.02-1209fje. [DOI] [PubMed] [Google Scholar]
- 41.Fernandez-Sauze S., et al. Effects of adrenomedullin on endothelial cells in the multistep process of angiogenesis: involvement of CRLR/RAMP2 and CRLR/RAMP3 receptors. Int. J. Cancer. 2004;108:797–804. doi: 10.1002/ijc.11663. [DOI] [PubMed] [Google Scholar]
- 42.Hippenstiel S., et al. Adrenomedullin reduces endothelial hyperpermeability. Circ. Res. 2002;91:618–625. doi: 10.1161/01.res.0000036603.61868.f9. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y., et al. PCR display identifies tamoxifen induction of the novel angiogenic factor adrenomedullin by a non estrogenic mechanism in the human endometrium. Oncogene. 1998;16:409–415. doi: 10.1038/sj.onc.1201768. [DOI] [PubMed] [Google Scholar]
- 44.Xia C.F., Yin H., Borlongan C.V., Chao J., Chao L. Adrenomedullin gene delivery protects against cerebral ischemic injury by promoting astrocyte migration and survival. Hum. Gene Ther. 2004;15:1243–1254. doi: 10.1089/hum.2004.15.1243. [DOI] [PubMed] [Google Scholar]
- 45.Miyashita K., et al. Adrenomedullin promotes proliferation and migration of cultured endothelial cells. Hypertens. Res. 2003;26(Suppl.):S93–S98. doi: 10.1291/hypres.26.s93. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Green K.E., Yallampalli C., Dong Y.L. Adrenomedullin enhances invasion by trophoblast cell lines. Biol. Reprod. 2005;73:619–626. doi: 10.1095/biolreprod.105.040436. [DOI] [PubMed] [Google Scholar]
- 47.Penchalaneni J., Wimalawansa S.J., Yallampalli C. Adrenomedullin antagonist treatment during early gestation in rats causes fetoplacental growth restriction through apoptosis. Biol. Reprod. 2004;71:1475–1483. doi: 10.1095/biolreprod.104.032086. [DOI] [PubMed] [Google Scholar]
- 48.Witlin A.G., et al. Placental and fetal growth and development in late rat gestation is dependent on adrenomedullin. Biol. Reprod. 2002;67:1025–1031. doi: 10.1095/biolreprod.101.002196. [DOI] [PubMed] [Google Scholar]
- 49.Di Iorio R., et al. Adrenomedullin is increased in the fetoplacental circulation in intrauterine growth restriction with abnormal umbilical artery waveforms. Am. J. Obstet. Gynecol. 2000;182:650–654. doi: 10.1067/mob.2000.103944. [DOI] [PubMed] [Google Scholar]
- 50.Yamashiro C., Hayashi K., Yanagihara T., Hata T. Plasma adrenomedullin levels in pregnancies with appropriate for gestational age and small for gestational age infants. J. Perinat. Med. 2001;29:513–518. doi: 10.1515/JPM.2001.071. [DOI] [PubMed] [Google Scholar]
- 51.Marinoni E., et al. Adrenomedullin and nitric oxide synthase at the maternal-decidual interface in early spontaneous abortion. J. Reprod. Med. 2004;49:153–161. [PubMed] [Google Scholar]
- 52.Di Iorio R., et al. Fetomaternal adrenomedullin levels in diabetic pregnancy. Horm. Metab. Res. 2001;33:486–490. doi: 10.1055/s-2001-16942. [DOI] [PubMed] [Google Scholar]
- 53.Di Iorio R., et al. Adrenomedullin, a new vasoactive peptide, is increased in preeclampsia. Hypertension. 1998;32:758–763. doi: 10.1161/01.hyp.32.4.758. [DOI] [PubMed] [Google Scholar]
- 54.Kanenishi K., Kuwabara H., Ueno M., Sakamoto H., Hata T. Immunohistochemical adrenomedullin expression is decreased in the placenta from pregnancies with pre-eclampsia. Pathol. Int. 2000;50:536–540. doi: 10.1046/j.1440-1827.2000.01085.x. [DOI] [PubMed] [Google Scholar]
- 55.Knerr I., et al. Adrenomedullin, calcitonin gene-related peptide and their receptors: evidence for a decreased placental mRNA content in preeclampsia and HELLP syndrome. Eur. J. Obstet. Gynecol. Reprod. Biol. 2002;101:47–53. doi: 10.1016/s0301-2115(01)00519-x. [DOI] [PubMed] [Google Scholar]
- 56.Makino Y., et al. Expression of adrenomedullin in feto-placental circulation of human normotensive pregnant women and pregnancy-induced hypertensive women. Endocrinology. 1999;140:5439–5442. doi: 10.1210/endo.140.11.7243. [DOI] [PubMed] [Google Scholar]
- 57.Li H., et al. Adrenomedullin is decreased in preeclampsia because of failed response to epidermal growth factor and impaired syncytialization. Hypertension. 2003;42:895–900. doi: 10.1161/01.HYP.0000095613.41961.6E. [DOI] [PubMed] [Google Scholar]
- 58.Conner A.C., et al. Heterodimers and family-B GPCRs: RAMPs, CGRP and adrenomedullin. Biochem. Soc. Trans. 2004;32:843–846. doi: 10.1042/BST0320843. [DOI] [PubMed] [Google Scholar]
- 59.Hay D.L., Poyner D.R., Sexton P.M. GPCR modulation by RAMPs. Pharmacol. Ther. 2006;109:173–197. doi: 10.1016/j.pharmthera.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 60.Nikitenko L.L., et al. Adrenomedullin and CGRP interact with endogenous calcitonin-receptor-like receptor in endothelial cells and induce its desensitisation by different mechanisms. J. Cell Sci. 2006;119:910–922. doi: 10.1242/jcs.02783. [DOI] [PubMed] [Google Scholar]
- 61.Dackor R., et al. Hydrops fetalis, cardiovascular defects and embryonic lethality in mice lacking the calcitonin receptor-like receptor gene. Mol. Cell. Biol. 2006;26:2511–2518. doi: 10.1128/MCB.26.7.2511-2518.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thota C., Yallampalli C. Progesterone upregulates calcitonin gene-related peptide and adrenomedullin receptor components and cyclic adenosine 3′5′-monophosphate generation in Eker rat uterine smooth muscle cell line. Biol. Reprod. 2005;72:416–422. doi: 10.1095/biolreprod.104.033779. [DOI] [PubMed] [Google Scholar]
- 63.Hewitt S.C., Collins J., Grissom S., Deroo B., Korach K.S. Global uterine genomics in vivo: microarray evaluation of the estrogen receptor alpha-growth factor cross-talk mechanism. Mol. Endocrinol. 2005;19:657–668. doi: 10.1210/me.2004-0142. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe H., et al. The estrogen-responsive adrenomedullin and receptor-modifying protein 3 gene identified by DNA microarray analysis are directly regulated by estrogen receptor. J. Mol. Endocrinol. 2006;36:81–89. doi: 10.1677/jme.1.01825. [DOI] [PubMed] [Google Scholar]
- 65.Ikeda K., Arao Y., Otsuka H., Kikuchi A., Kayama F. Estrogen and phytoestrogen regulate the mRNA expression of adrenomedullin and adrenomedullin receptor components in the rat uterus. Mol. Cell. Endocrinol. 2004;223:27–34. doi: 10.1016/j.mce.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 66.Ishimitsu T., Tsukada K., Minami J., Ono H., Matsuoka H. Variations of human adrenomedullin gene and its relation to cardiovascular diseases. Hypertens. Res. 2003;26(Suppl.):S129–S134. doi: 10.1291/hypres.26.s129. [DOI] [PubMed] [Google Scholar]
- 67.Li Y., et al. Blood pressure and urinary sodium excretion in relation to the A-1984G adrenomedullin polymorphism in a Chinese population. Kidney Int. 2006;69:1153–1158. doi: 10.1038/sj.ki.5000213. [DOI] [PubMed] [Google Scholar]
- 68. Nagy, A., Gertsenstein, M., Vintersten, K., and Behringer, R. 2003. Manipulating the mouse embryo, a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 764 pp. [Google Scholar]