Abstract
The accumulation of α-1,4-polyglucans is an important strategy to cope with transient starvation conditions in the environment. In bacteria and plants, the synthesis of glycogen and starch occurs by utilizing ADP-glucose as the glucosyl donor for elongation of the α-1,4-glucosidic chain. The main regulatory step takes place at the level of ADP-glucose synthesis, a reaction catalyzed by ADP-Glc pyrophosphorylase (PPase). Most of the ADP-Glc PPases are allosterically regulated by intermediates of the major carbon assimilatory pathway in the organism. Based on specificity for activator and inhibitor, classification of ADP-Glc PPases has been expanded into nine distinctive classes. According to predictions of the secondary structure of the ADP-Glc PPases, they seem to have a folding pattern common to other sugar nucleotide pyrophosphorylases. All the ADP-Glc PPases as well as other sugar nucleotide pyrophosphorylases appear to have evolved from a common ancestor, and later, ADP-Glc PPases developed specific regulatory properties, probably by addition of extra domains. Studies of different domains by construction of chimeric ADP-Glc PPases support this hypothesis. In addition to previous chemical modification experiments, the latest random and site-directed mutagenesis experiments with conserved amino acids revealed residues important for catalysis and regulation.
INTRODUCTION: FUNCTION AND REGULATION OF ADP-Glc PPase IN BACTERIA AND IN PLANTS
Many organisms, including plants, accumulate carbon and energy reserves to cope with starvation conditions temporarily present in the environment (42, 72, 73, 78, 88). The biosynthesis of α-1,4-polyglucans is a main strategy for such metabolic storage. One outstanding advantage in using polysaccharides as reserve compounds is that after their high molecular weights and other physical properties, they have little effect on the internal osmotic pressure in the cell. The metabolic routes for polyglucan accumulation were elucidated after the discovery of nucleoside diphosphate sugars by Luis F. Leloir and coworkers in the 1950s (51). The seminal work of Leloir's group clearly established that biosynthesis and degradation of glycogen occur by different pathways, the former involving the use of an activated form of glucose, specifically UDP-Glc in cells from mammals, fungi, and eukaryotic heterotrophic microorganisms and ADP-Glc in bacteria and photosynthetic eukaryotes (88).
The precise role that the glycogen may play in bacteria is still not clear; however, it was suggested that the accumulation of glycogen by bacteria may give advantages during starvation periods, providing a stored source of energy and carbon surplus (95). In bacteria such as Bacillus subtilis and Streptomyces coelicolor, glycogen synthesis has been associated with sporulation and the supply of resources necessary to drive differentiation (43, 56, 68), whereas in Mycobacterium smegmatis, recycling of the polysaccharide during exponential phase was shown to be essential for growth (7). In Streptococcus mutans, it has been shown that a glycogen-like intracellular polysaccharide plays a central role in cariogenesis (92). Also, a relationship between glycogen synthesis, biofilm formation, and virulence has been reported in Salmonella enteritidis (9).
The process for the synthesis of storage polysaccharides in bacteria and plants, namely glycogen and starch, respectively, occurs by utilizing ADP-Glc as the glucosyl donor for the elongation of the α-1,4-glucosidic chain (42, 72, 73, 77, 78, 88). Moreover, in these organisms the main regulatory step of the metabolism takes place at the level of ADP-Glc synthesis, a reaction catalyzed by ADP-Glc pyrophosphorylase (ATP:α-d-glucose-1-phosphate adenylyltransferase; EC 2.7.7.27; ADP-Glc PPase): ATP + Glc-1-phosphate ⇔ ADP-Glc + inorganic pyrophosphate. This reaction was first described in soybean (16) and was subsequently found in many bacterial extracts and plant tissues (42, 70, 72, 73, 76-78, 88). The enzymatic reaction takes place in the presence of a divalent metal ion, Mg2+, and it is freely reversible in vitro, with an equilibrium close to 1. The hydrolysis of inorganic pyrophosphate by inorganic pyrophosphatase and the use of the sugar nucleotide for polysaccharide synthesis causes the ADP-Glc synthesis reaction to be essentially irreversible in vivo (42).
Most of the ADP-Glc PPases so far characterized are allosterically regulated by small effector molecules. Although the major activators vary according to the source, they share the characteristic of being intermediates in the major carbon assimilatory pathway in the organism (72, 73, 75, 78, 88). For instance, many of the enzymes from heterotrophic bacteria are activated by metabolites of glycolytic pathways (either the classical Embden-Meyerhof or the Entner-Doudoroff metabolism), such as fructose 6-phosphate, fructose 1,6-bisphosphate, or pyruvate, and inhibited by AMP, ADP, and/or Pi (73, 75, 78). In general, ADP-Glc PPase activators are key metabolites that represent signals of high carbon and energy contents within the cell. The opposite occurs for inhibitors of the enzyme, which are intermediates of low metabolic energy levels. These regulatory properties of ADP-Glc PPase, together with the fact that ATP is one of the substrates of the enzyme, have the rationale that synthesis of storage polysaccharides in bacteria and plants will be maximal when cellular carbon and energy are in excess, and vice versa (39, 78, 88).
Cross talk between activators and inhibitors in the ADP-Glc PPase from different sources that renders an amplified response to small changes in concentration of the activator has been described. This response, defined as ultrasensitive behavior (45), is observed because the inhibitor, at higher concentrations, increases the sigmoidicity of the activation curves in spite of decreasing the activity of the enzyme. Detailed studies performed in cyanobacteria have shown that the inhibitor Pi elicits an ultrasensitive response of the enzyme towards 3-phosphoglycerate (3-PGA) activation, which is operative within the cell (27, 28) and allows the enzyme to respond efficiently to minimal changes in 3-PGA levels despite a background of high Pi that may be present inside the cyanobacterial cell (28). Early characterization of the enzyme purified from Escherichia coli also showed the interaction between the activator and inhibitors (24).
PHYSIOLOGICAL ROLE
There is strong experimental evidence to support the view that ADP-Glc PPase is a regulatory enzyme on the pathway for bacterial glycogen and plant starch biosynthesis. In E. coli and Salmonella enterica serovar Typhimurium, several mutants affected in the ability to accumulate glycogen were isolated after chemical mutagenesis, and their ADP-Glc PPases displayed altered regulatory properties (31, 74, 94). It was shown that there was a direct relationship between the affinity of the enzyme for the activator, fructose-1,6-bisphosphate, and the ability of the mutant to accumulate glycogen (79). Similar results were obtained with oxygenic photosynthetic organisms, in which the activator is 3-PGA and the inhibitor is Pi. In the unicellular green alga Chlamydomonas reinhardtii, starch-deficient mutants were isolated and shown to have ADP-Glc PPases that could not be activated by 3-PGA (2). Comparable experimental data support the physiological importance of ADP-Glc PPase allosteric regulation in both photosynthetic and nonphotosynthetic tissues from higher plants (26, 53-55, 90, 93). Thus, regulation of ADP-Glc synthesis in bacteria and plants agrees with the generalization that a biosynthetic pathway is effectively regulated at its first unique step.
REGULATORY PROPERTIES AND QUATERNARY STRUCTURE OF ADP-Glc PPases FROM DIFFERENT SOURCES
Based on specificity for activator and inhibitor, ADP-Glc PPases have been grouped into different classes (42, 70, 72, 78, 88). The former classifications can be updated to include nine distinctive classes of ADP-Glc PPases (Table 1) to include recent reports on the properties of the enzymes from gram-positive bacteria (96) and from endosperm tissues of higher plants (30). Also reported in Table 1 is the quaternary structure of the enzymes from different prokaryotic and eukaryotic organisms.
TABLE 1.
Relationships between carbon metabolism and regulatory and structural properties of ADP-Glc PPase from different organisms
| Organism | Main carbon utilization | Major reserve polyglucan | ADP-Glc PPase
|
|||
|---|---|---|---|---|---|---|
| Class | Allosteric regulatorsa
|
Quaternary structure | ||||
| Activator(s) | Inhibitor(s) | |||||
| Prokaryotes | ||||||
| Escherichia coli | Embden-Meyerhof pathway (glycolysis) | Glycogen | I | Fru 1,6-bisP | AMP | Homotetramer (α4) |
| Salmonella enterica serovar Typhimurium | ||||||
| Enterobacter aerogenes | ||||||
| Aeromonas formicans | Glycolysis | Glycogen | II | Fru 1,6-bisP, Fru 6-P | AMP, ADP | |
| Micrococcus luteus | ||||||
| Mycobacterium smegmatis | ||||||
| Serratia marcescens | Glycolysis | Glycogen | III | None | AMP | |
| Enterobacter hafniae | ||||||
| Clostridium pasteurianum | ||||||
| Agrobacterium tumefaciens | Entner-Doudoroff pathway | Glycogen | IV | Pyruvate, Fru 6-P | AMP, ADP | Homotetramer (α4) |
| Arthrobacter viscosus | ||||||
| Chromatium vinosum | ||||||
| Rhodobacter capsulata | ||||||
| Rhodomicrobium vannielii | ||||||
| Rhodobacter gelatinosa | Glycolysis and Entner-Doudoroff pathways | Glycogen | V | Pyruvate, Fru 6-P, Fru 1,6-bisP | AMP, Pi | Homotetramer (α4) |
| Rhodobacter globiformis | ||||||
| Rhodobacter sphaeroides | ||||||
| Rhodocyclus purpureus | ||||||
| Rhodospipillum rubrum | Tricarboxylic acid cycle | Glycogen | VI | Pyruvate | None | |
| Rhodospirillum tenue | Reductive carboxylic acid cycle | |||||
| Bacillus subtilis | Tricarboxylic acid cycle during sporulation | Glycogen | VII | None | None | Heterotetramer (α2β2) |
| Bacillus stearothermophillus | ||||||
| Cyanobacteria | ||||||
| Synechococcus sp. strain PCC 6301 | Oxygen evolving photosynthesis | Glycogen | VIII | 3-PGA | Pi | Homotetramer (α4) |
| Synechocystis sp. strain PCC 6803 | ||||||
| Anabaena sp. strain PCC 7120 | Calvin cycle | |||||
| Eukaryotes | ||||||
| Green algae | ||||||
| Chlorella fusca | Oxygen evolving photosynthesis | Starch | VIII | 3-PGA | Pi | Heterotetramer (α2β2) |
| Chlorella vulgaris | ||||||
| Chlamydomonas reinhardtii | Calvin cycle | |||||
| Higher plants | ||||||
| Photosynthetic tissues | ||||||
| Leaves of spinach, wheat | Oxygen evolving photosynthesis | Starch | VIII | 3-PGA | Pi | Heterotetramer (α2β2) |
| Arabidopsis, maize, rice | Calvin cycle | |||||
| Nonphotosynthetic tissues | Catabolism of sucrose imported from photosynthetic tissues | Starch | VIII | 3-PGA | Pi | Heterotetramer (α2β2) |
| Potato tubers | ||||||
| Endosperm of maize, barley and wheat | Starch | IX | None directly, 3-PGA and Fru 6-P reverse inhibitor's effect | Pi, ADP, Fru 1,6-bisP | Heterotetramer (α2β2) | |
Fru, fructose; P, phosphate; bisP, bisphosphate; Pi, inorganic phosphate.
Class I comprises ADP-Glc PPases from bacteria that perform glycolysis (typically enterobacteria: E. coli, S. enterica serovar Typhimurium), mainly regulated by fructose bisphosphate, the activator, and AMP, the inhibitor (76). The enzyme from class I is encoded by a single gene, giving rise to a native homotetrameric structure (α4) with a molecular mass of about 200 kDa (Table 1) (42, 73, 75, 88). Other bacteria that perform glycolysis contain ADP-Glc PPases that are allosterically activated by fructose bisphosphate and fructose 6-phosphate and inhibited by AMP and ADP (class II) or exhibit no sensitivity to activator and are inhibited by AMP (class III) (Table 1) (42, 71-73, 88). The enzymes included in class IV are those from bacteria that mainly utilize the Entner-Doudoroff glycolytic pathway, which are distinctively activated by fructose 6-phosphate and pyruvate, with ADP, AMP, and Pi behaving as inhibitors (15, 97). Interestingly, ADP-Glc PPases from organisms using both the Embden-Meyerhoff and the Entner-Doudoroff pathways are activated by the three main effectors: fructose 1,6-bisphosphate, fructose 6-phosphate, and pyruvate (class V, Table 1) (32, 36). As also specified in Table 1, ADP-Glc PPases from Agrobacterium tumefaciens (97) and Rhodobacter sphaeroides (36) have been characterized as tetramers composed of a single subunit with a molecular mass of about 50 kDa.
Class VI includes ADP-Glc PPases from anaerobic bacteria that are capable of growth under heterotrophic conditions in the dark or being autotrophic in the light and performing anoxygenic photosynthesis (Table 1). These organisms cannot catabolize glucose but grow very well on pyruvate and tricarboxylic acid cycle intermediates as carbon sources and photosynthetic electron donors. Enzymes from class VI are specifically regulated by pyruvate (Table 1) (22, 101).
ADP-Glc PPases grouped as class VII include the enzymes from sporulating bacteria of the genus Bacillus (Table 1). These microorganisms accumulate glycogen only during sporulation and in the presence of a carbon source that does not interfere with such a process for survival in hostile environments (96). Under these conditions, the main pathway for carbon utilization is the tricarboxylic acid cycle, which fully metabolizes the by-products of glycolysis (57). It has been determined that in Bacillus subtilis and Bacillus stearothermophilus, the genes for glycogen synthesis are clustered in one operon, glgBCDAP (43, 96). A comparative analysis of the gene clusters showed that glgC and glgD encode proteins homologous to ADP-Glc PPases from prokaryotes. Thus, the putative GlgC protein from B. stearothermophilus has 387 amino acids, with a predicted molecular mass of 43.3 kDa and showing 42 to 70% identity with bacterial ADP-Glc PPases. The GlgD product is a shorter protein (343 amino acids and a predicted molecular mass of 38.9 kDa) with a lower homology to ADP-Glc PPase (20 to 30% identity) (96).
Expression of the glgC gene from B. stearothermophilus rendered an active recombinant enzyme; whereas GlgD exhibited negligible activity. However, when the glgC and glgD genes were expressed together, the resulting GlgCD protein exhibited higher affinity for substrates and twofold higher Vmax in catalyzing ADP-Glc synthesis than GlgC by itself. The different recombinant enzymes from B. stearothermophilus were insensitive to regulation by different metabolites typically affecting the activity of other bacterial ADP-Glc PPases (96). Thus, the enzymes grouped in class VII in Table 1 are very distinct from other ADP-Glc PPases, as they are apparently unregulated enzymes, being the only bacterial ADP-Glc PPases that exhibit a heterotetrameric structure of the type α2β2.
The last group of bacterial ADP-Glc PPases are those from cyanobacteria, prokaryotes that perform an oxygenic photosynthetic process similar to that occurring in plants (class VIII, Table 1). These enzymes have 3-PGA and Pi as the main activator and inhibitor, respectively (13, 40). Remarkably, the specificity for allosteric regulators of the cyanobacterial ADP-Glc PPase is identical to that found in eukaryotic photosynthesizers, such as green algae and higher plants, which are also grouped in class VIII (Table 1) (40, 42). All these photosynthetic organisms utilize the reductive pentose phosphate pathway or Calvin cycle to photoassimilate atmospheric CO2, rendering 3-PGA as the first intermediate product. Pi under light conditions is utilized to regenerate ATP through photophosphorylation (41). Thus, class VIII ADP-Glc PPases are typically regulated by the 3-PGA/Pi ratio under physiological conditions (41, 42, 73, 78, 88).
Concerning ADP-Glc PPases from nonphotosynthetic tissues of higher plants, two different types can be distinguished (Table 1). The potato tuber enzyme is the best-characterized ADP-Glc PPase from reserve tissues that are typically activated by 3-PGA and inhibited by Pi and thus grouped as class VIII (Table 1) (5, 37). In addition, the potato enzyme is subject to regulation by a redox mechanism involving Cys-12, with the thioredoxin-mediated reduction of an intermolecular disulfide bridge resulting in activation of the enzyme (3, 20). This was proposed to be operative in different tissues of higher plants (leaves, tuber, fruit, and cotelydons, except in endosperms from monocots). In contrast, the ADP-Glc PPases from bacteria lack a Cys-12 homologous residue (3).
ADP-Glc PPases from reserve tissues of cereals have been reported to exhibit distinctive regulatory properties, mainly related to a lower sensitivity to activators (30, 35, 44, 69, 82, 98). Recently (30), a complete characterization of the ADP-Glc PPase purified from wheat endosperm showed that the enzyme is subject to regulation by the coordinate action of a series of metabolites. The wheat endosperm enzyme is allosterically inhibited by Pi, ADP, and fructose 1,6-bisphosphate. In all cases, inhibition can be reversed by 3-PGA and fructose 6-phosphate, which individually (in the absence of the inhibitors) have no effect on enzyme activity (30). Thus, rather than being an unregulated PPase, this enzyme seems to have distinctive regulatory properties accounting for a class IX group of ADP-Glc PPases (Table 1) that have Pi inhibition as a key signal, as shown in genetically modified plants (90).
Cyanobacterial ADP-Glc PPase occupies a central position with respect to structure/regulation relationships, as its properties are intermediate between those of the bacterial and plant enzymes. Thus, cyanobacterial PPase is homotetrameric in structure, as observed for the protein from other bacteria (Table 1), but it is regulated like and is immunologically more related to the plant enzyme (12, 40). A main difference between the cyanobacterial and plant ADP-Glc PPases is the quaternary structure (38).
SUBUNIT STRUCTURE OF PLANT ENZYMES
Early studies on the spinach leaf ADP-Glc PPase showed the existence of two distinct subunits (62). Other immunological studies in maize endosperm suggested that in both nonphotosynthetic and photosynthetic tissues, the ADP-Glc PPase comprised two subunits that are the products of two genes (77). ADP-Glc PPases from all the eukaryotes characterized so far (starting with the green alga proteins; see Table 1) is composed of α and β subunits to form a heterotetrameric structure (38, 42, 73, 77, 78, 88). In the recombinant potato tuber ADP-Glc PPase, it was shown by N terminus sequencing that the structure is α2β2 (17). For convenience, these subunits were named the small (α subunit, 50 to 54 kDa) and large (β subunit, 51 to 60 kDa) subunits, even though the difference in mass between them in some cases is not more than 1 kDa (63, 64). The small subunit of the higher plant ADP-Glc PPase is highly conserved (85 to 95% identity), whereas the large subunit is less conserved (50 to 60% identity) (91). Nevertheless, both subunits seem to derive from the same ancestor, based on the homology of conserved regions.
IDENTIFICATION OF IMPORTANT AMINO ACID RESIDUES
Chemical modification has been used to identify important amino acids in the ADP-Glc PPases, and site-directed mutagenesis was employed to confirm their roles. Photoaffinity analogs of ATP and ADP-Glc, 8-azido-ATP and 8-azido-ADP-Glc, respectively, were used to identify a residue at the substrate-binding site. When UV light at 257 nm is used to irradiate azido compounds, a nitrene radical is formed, which reacts with electron-rich residues. In the E. coli enzyme, it was shown after covalent labeling of these analogs, tryptic digestion, separation, and isolation of the peptides by high-pressure liquid chromatography and subsequent amino acid sequencing that Tyr114 was modified (49, 50). Site-directed mutagenesis of this residue showed a marked decrease in affinity for ATP, but it did not seem to be specific only for ATP, since the affinity for Glc 1-phosphate and the activator fructose 1,6-bisphosphate also decreased (49). This residue must be close to the adenine ring of ATP or ADP-Glc but probably also near the Glc 1-phosphate and the fructose 1,6-bisphosphate regulatory sites.
Pyridoxal 5-phosphate (PLP) is a reagent that is able to react with lysine residues to form Schiff bases that can be covalently bonded after reduction with NaBH4. Since PLP may be considered a structural analog of fructose 1,6-bisphosphate and 3-PGA (it activates the ADP-Glc PPases from E. coli, Anabaena sp., and spinach leaf) (71), it was used to find lysine residues located in those activator sites. In the enzyme from spinach leaf, PLP bound at Lys440 very close to the C terminus of the small subunit and also to three other Lys residues in the large subunit. Binding to these sites was prevented by the allosteric effector 3-PGA, which indicated that they are close to or directly involved in the binding of this activator (1, 61). Similar results were obtained with the ADP-Glc PPase from the Anabaena sp. In this case, the modified residues were identified as Lys419, which is homologous to Lys440 and Lys441 in the small subunits of the spinach and potato tuber enzymes, respectively, and Lys382 is analogous to Lys404 of the potato tuber small subunit. Identification of these residues as regulatory binding sites was confirmed by site-directed mutagenesis of the Anabaena ADP-Glc PPase (12, 85).
Mutation of these Lys residues in the potato tuber ADP-Glc PPase revealed that they are also part of the 3-PGA site in heterotetrameric enzymes and that the contribution of these residues to the binding of 3-PGA is additive (4). However, mutation of the small subunits yielded enzymes with less affinity for 3-PGA than homologous mutants of the large subunit. These data indicate that Lys404 and Lys441 on the potato tuber small subunit are more important than their homologous counterparts on the large subunit, suggesting that the large subunit does not modify the regulatory properties of the small subunit, providing more effective allosteric sites but making the 3-PGA activator sites which are already present in the small subunit more efficient (4).
Chemical modification studies on the E. coli enzyme showed that it was covalently modified with [3H]PLP by reduction with NaBH4. It was demonstrated that the PLP could bind to two different lysine residues. Allosteric activators protected binding to Lys39, and substrate ADP-Glc protected binding to Lys195 (66, 67). Site-directed mutagenesis of Lys39 showed that this residue is important for the interaction of the activator fructose 1,6-bisphosphate with the enzyme (23). Interestingly, PLP, as an analog of the activator, was reactive with lysine in the N terminus of the E. coli enzyme rather than to the C terminus, as in enzymes activated by 3-PGA.
In E. coli ADP-Glc PPase, site-directed mutagenesis of Lys195 produced enzymes whose Km for Glc 1-phosphate was 100- to 10,000-fold greater than that of the wild type (34). On the other hand, kinetic constants for ATP, Mg2+, and fructose 1,6-bisphosphate were similar to those of the wild-type enzyme, suggesting that this Lys is specifically involved in the binding of Glc 1-phosphate. Furthermore, the kcat for the glutamine mutant was similar to that of the wild type, ruling out the participation of this residue in the catalytic reaction (34). Site-directed mutagenesis was used to determine the role of this conserved residue in the small (Lys198) and large (Lys213) subunits of the potato tuber ADP-Glc PPase (21). Mutation of Lys198 of the small subunit to Arg, Ala, or Glu had little effect on kinetic constants for ATP, Mg2+, activator (3-PGA), and inhibitor (Pi), but the apparent affinity for Glc 1-phosphate decreased 135- to 550-fold. However, similar mutations on Lys213 of the large subunit had little effect on the affinity for Glc 1-phosphate. These results indicate that Lys198 in the small subunit is directly involved in the binding of Glc 1-phosphate and that the homologous counterpart in the large subunit it is not (21). This is in good agreement with the idea that the large subunit does not have a catalytic role but only a modulatory one (20).
Arginine residues in ADP-Glc PPases were found to be functionally important, as shown by chemical modification with phenylglyoxal (39, 86). Alanine scanning mutagenesis of ADP-glucose pyrophosphorylase from Anabaena sp. strain PCC 7120 indicated that Arg294 plays a role in inhibition by orthophosphate (86). Recently, it was shown that replacement of this residue with Ala or Gln reversed the pattern of inhibitor specificity; the main inhibitor was NADPH rather than Pi (18). All of these results suggest that the positive charge of Arg294 may not be specifically involved in orthophosphate binding but that it plays a role in determining inhibitor selectivity.
Alanine scanning mutagenesis of the arginine residues located in the N terminus of the enzyme from Agrobacterium tumefaciens demonstrated the presence of separate subsites for the activators fructose 6-phosphate and pyruvate (29). The R32A mutant enzyme had reduced affinity for fructose 6-phosphate (11.5-fold) and behavior identical to the wild-type enzyme with respect to pyruvate activation. Both the R33A and R45A mutant enzymes had higher activity than the wild-type enzyme in the absence of activators and no response to fructose 6-phosphate, but partial activation by pyruvate and desensitization to phosphate inhibition (29).
Random mutagenesis experiments were performed on the potato tuber ADP-Glc PPase to find residues that are important for the enzyme. Even though several residues were found, some of them did not show a very big decrease in activity or a very specific effect. The most interesting finding was that Asp403 (in the article it is described as Asp413) in the small subunit is important for activation by 3-PGA (33). This residue is adjacent to the lysine that is responsible for PLP binding and 3-PGA activation. Mutation of residue Asp253 on the small subunit showed a specific effect on the apparent affinity for Glc 1-phosphate, but the Km only increased 10-fold (48). Interestingly, this residue is conserved in the sugar nucleotide pyrophosphorylases that have been crystallized and whose structure has been solved when an alignment is made according the secondary-structure elements (19). This residue seems to be close the substrate site without a direct interaction with Glc 1-phosphate.
PREDICTION OF THE STRUCTURE OF ADP-Glc PPases
Information about the three-dimensional structure of any ADP-Glc PPase would be tremendously helpful for structure-function relationship studies. Unfortunately, it is not currently available. For that reason, several methods to predict the structure have been applied (19, 81). A modified hydrophobic cluster analysis (52) was applied to several ADP-Glc PPases from different sources representing different classes according to homology of subunits and tissue, i.e., E. coli, Anabaena, Chlamydomonas, potato (Solanum tuberosum L.) tuber small subunit and different large subunits from maize embryo, maize shrunken 2, and Arabidopsis thaliana. Hydrophobic cluster analysis showed that the ADP-Glc PPases were extremely similar in the distribution and pattern of the clusters, even between bacterial and plant enzymes. This strongly suggests that the ADP-Glc PPases have a common folding pattern despite a different quaternary structure (α2β2 in plants and α4 in bacteria) and specificity for the activator.
If the ADP-Glc PPases from different sources have a similar three-dimensional structures, their secondary-structure predictions should be similar. All the sequences mentioned above, and also those from A. tumefaciens, Bacillus stearothermophilus, and Rhodobacter sphaeroides, were analyzed with the PHD program to predict the secondary structure (81). The alignment helped to establish a structure for regions where the predictions were not conclusive for one of the enzymes but very clear for the rest (19). A similar alignment of representative bacterial ADP-Glc PPases from each class is shown in Fig. 1. The small-subunit sequences of the enzymes from C. reinhardtii and potato tuber were also included for comparison with the proteins from uni- and pluricellular eukaryotes, respectively (Fig. 1). From these analyses, a general structure that fits all of these proteins was postulated (Fig. 2). There are also biochemical data that support the model (19).
FIG. 1.
Alignment of ADP-Glc PPases from different classes. Amino acid alignment was performed with the program PILEUP from the Wisconsin package (http://www.gcg.com). The alignment was fine tuned manually based on the secondary structure of each enzyme as predicted by the PHD program (81). Residues in blue and red were predicted to be β-sheets and α-helixes, respectively; pale shades indicate a lower level of confidence. Green residues were predicted to be neither of these (loops). In black are residues for which the PHD program could not make a prediction. Insertions and deletions were introduced to maximize the alignment of both primary and secondary structure. a, sequence of the small (catalytic) subunit; b, sequence of the subunit encoded by glgC (catalytic).
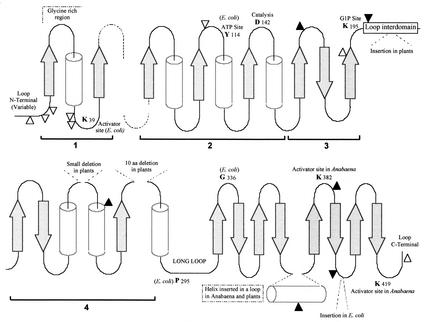
FIG. 2.
Prediction of secondary structure of ADP-glucose pyrophosphorylases. The secondary structure of various ADP-Glc PPases from bacteria as well as plants was predicted with the PHD program (81). The secondary structures align very well (19) with the sequences of UDP-N-acetylglucosamine pyrophosphorylase (11)and TDP-glucose pyrophosphorylase (8). Sequences shown as arrows are predicted to be β-pleated sheets, and sequences shown as cylinders are predicted to be α-helices. These structures are interconnected with amino acid sequences indicated as being neither α-helices or β-pleated sheets and are possibly random structures or loops. They are shown as lines. White triangles indicate areas where proteinase K hydrolyzes the E. coli enzyme (99). Black triangles indicate where the Anabaena ADP-Glc PPase is partially proteolyzed by trypsin, and gray triangles indicate partial hydrolysis of the E. coli enzyme by trypsin (unpublished data). The proteolysis results suggest that the areas sensitive to proteases are exposed random structures (loops). Residues K39, Y114, and K195 are the amino acids in the E. coli ADP-Glc PPase that bind the activator fructose 1,6-bisphosphate and the substrates ATP and Glc 1-phosphate, respectively. D142 is the amino acid shown to be a catalytic residue in the E. coli enzyme. P295 and G336 are amino acids that, when mutated, affect the allosteric properties of the ADP-Glc PPase (59, 60). Regions 1, 2, and 3 form the putative catalytic domain, and region 4 may also be part of the catalytic domain, as suggested by alignment with the crystal structures of UDP-N-acetylglucosamine pyrophosphorylase (11) and TDP-glucose pyrophosphorylase (8).
Controlled proteolysis experiments were in good agreement with the model. The exposed loops would be more sensitive to proteolytic cleavage, and the studies confirmed that the proteases analyzed cut in sites predicted to be loops (19). The only exception is the α-helix predicted near the C terminus on the Anabaena enzyme (Fig. 2). Since this is an insertion (20 amino acids) that is absent in the E. coli enzyme and is not predicted to be buried by the PHD program, it is most likely that this helix is not part of the core but part of a loop in a domain of eight β-sheets (Fig. 2).
Loops are prone to have insertions and deletions in homologous proteins that do not alter the structure. In our model, all the insertions and deletions observed fell in loops (Fig. 2). The conserved amino acids known to have specific roles in the binding of substrates (E. coli Tyr114 and Lys195) and activators (E. coli Lys39 and Anabaena Lys382 and Lys419) are located in loops. The residues Pro295 and Gly336, which seem to be located in a region important for the regulation of the E. coli enzyme, are also in loops (25, 59). The amino acid Asp142 in the E. coli enzyme was identified as a catalytic residue (19), and it is also present in a loop.
A structure usually observed in proteins that bind nucleotides is also predicted in this model. Region 1 has a Gly-rich loop after a β-sheet, which is similar to a P loop in protein kinases or nucleotide binding sites (84), and region 2 has three β-sheets and helices that are compatible with the Rossman fold (80). Thus, regions 1 and 2 comprise a putative domain or subdomain that binds ATP. Moreover, Tyr114, which was shown to be reactive to the azido analog of ATP (49, 50), is in this region.
α/β structures generally have a very particular topology regarding the loops. Some of them are “functional” because they carry residues important for binding and catalysis, and others are just “connectors” because they only connect one helix with the next sheet. It has been observed that functional loops are the ones that are located at the C-terminal end of the β-sheets (10). Supporting the model, those loops in regions 1, 2, and 3 are the ones that bear the most conserved amino acids. Hence, this is compatible with the idea that the ATP would be facing the “top” of the structure depicted in Fig. 2. Moreover, amino acid residues located at the loops that are at the N terminus of the β-sheets in regions 2 and 3 are not conserved at all. The exception is in region 1; however, there is evidence by chemical modification and site-directed mutagenesis that this loop interacts with the activator fructose 1,6-bisphosphate in the E. coli ADP-Glc PPase (23).
The first pyrophosphorylase domain to be crystallized and solved was present in a bifunctional enzyme that is the product of the gene glmU (11). One domain of the GlmU protein is a UDP-N-acetylglucosamine pyrophosphorylase, and the other is an acetyltransferase. Later, other pyrophosphorylase domain structures were solved (8, 46, 65, 89). All these structures verify the predicted secondary-structure model of the ADP-Glc PPase (19). Regions 2, 3, and 4 are virtually identical. In region 4, the only difference is that two β-sheets were predicted rather than one because of the presence of a Gly (breaker). In the N-acetylglucosamine uridyltransferase, only one sheet is bent because of a Gly. Region 1 is very similar; there is a P-loop-like structure, but our model predicted an extra β-sheet. It is possible that the prediction is wrong or that different sugar nucleotide pyrophosphorylases vary in this region. When we predicted the secondary structure of GDP-mannose PPases, TDP-Glc PPases, CDP-Glc PPases, and UDP-Glc PPases, this was the region with the greatest variability. For this reason, the topology of the loop where Lys39 is present cannot be ascertained. To support the idea that the sugar-nucleotide pyrophophorylases have a similar catalytic domain, it was demonstrated that the homologous Glc 1-phosphate site is present in the GDP-mannose PPase from Pseudomonas aeruginosa (58).
The homology between ADP-Glc PPases and the N-acetylglucosamine uridyltransferase is extremely low. However, an alignment could be done using the predicted structure to match helices and sheets. Then, several residues were found conserved, all in loops that face the substrate in the N-acetylglucosamine uridyltransferase (Fig. 2). Lys195, the Glc 1-phosphate binding site in ADP-Glc PPases, is present in the N-acetylglucosamine uridyltransferase but shifted one position.
CATALYTIC RESIDUES
Despite the identification of several important residues on the structure of the ADP-Glc PPases, only recently was an amino acid identified as being mainly involved in catalysis (19). Comparison with the three-dimensional structures of known pyrophosphorylase domains and prediction of the structure led to the discovery of highly conserved residues throughout the superfamily of pyrophosphorylases despite the low homology. Asp142 in the E. coli ADP-Glc PPase was predicted to be close to the substrate site. Site-directed mutagenesis of this residue to Ala and Asn confirmed that the main role of Asp142 is catalytic (19). Kinetic analysis showed a decrease in specific activity of four orders of magnitude, whereas other kinetic parameters showed no significant changes.
In the pyrophosphorylase domain of the GlmU enzyme, it was proposed that Arg18 could be a catalytic residue (11). Even though this residue seems to be important in ADP-Glc PPases, it is not clear if it is directly involved in the catalytic reaction. Mutagenesis of the homologous Arg25 in the enzyme from Agrobacterium tumefaciens yielded an enzyme with an activity reduced by two orders of magnitude (29). Generally, it is expected that more dramatic effects would occur after mutation of catalytic residues.
DOMAIN CHARACTERIZATION
The central region of the protein has been identified as a substrate binding and catalytic domain by secondary-structure prediction, alignment with other sugar nucleotide pyrophosphorylase enzymes, and further site-directed mutagenesis. It has been proposed that the N and C termini are responsible for the distinctive regulatory properties of the different classes of ADP-Glc PPases (6). This is evident for the ADP-Glc PPases from oxygenic photosynthetic organisms because key residues for the regulation have been found on the C terminus of plant and cyanobacterial ADP-Glc PPases (4, 14, 18, 33, 85, 86). Also, several modifications on the C terminus caused modifications in the regulation of plant enzymes (26, 83). Residues that are critical for the binding of the activators have been found only on the N terminus of enzymes from heterotrophic bacteria (23). Two allosteric mutants (P295S and G336D) were found and characterized in the C terminus of the E. coli enzyme, but they had higher rather than decreased apparent affinities for the activator, which indicates that they are probably not involved in the binding of the regulators (59, 60). However, those mutants indicate the importance of the C terminus in regulation.
Recent experiments with chimeric enzymes between the ADP-Glc PPase from E. coli and A. tumefaciens suggest that the C terminus of the A. tumefaciens enzyme determines the high apparent affinity for the activator pyruvate, but the residues critical for the fructose 6-phosphate selectivity do not lie in this region (6). This agrees with site-directed mutagenesis experiments that suggested that the sites for the two activators are separate or overlap only partially (29). Experiments with the chimeric enzymes also supported the idea that the C terminus of the E. coli enzyme largely contributes to determining the selectivity for the activator fructose 1,6-bisphosphate. Since it has been found previously that Lys39 in the E. coli enzyme interacts with the allosteric activator (23), it is very possible that the regulation is determined by a combined arrangement between the N and C termini.
The N-terminal region of the ADP-Glc PPase, which is predicted to be a loop, may play a role as an “allosteric switch” to regulate enzyme activity. This loop possibly interferes with the transition between two different conformations of the enzyme (activated and nonactivated). A shorter N terminus may favor a conformation of the enzyme that facilitates activation. This was observed when the enzyme from E. coli and the small subunit from the potato tuber enzyme were truncated by 11 amino acids (5, 99, 100). It was necessary to remove at least 11 amino acids from the E. coli enzyme to observe this effect. When a truncation of 10 amino acids was generated in the small subunit of the potato tuber enzyme, the apparent affinity for the activator 3-PGA increased and the apparent affinity for the inhibitor Pi decreased (5). Similar results were observed when the large (modulatory) subunit was truncated by 17 amino acids in the N terminus (47).
EVOLUTION OF ADP-Glc PPases
ADP-Glc PPases seem to have a common pyrophosphorylase domain with other sugar nucleotide pyrophosphorylases, as suggested previously. ADP-Glc PPases are generally bigger because they have an extended C terminus (120 to 150 amino acids) and a slightly longer N terminus (10 to 40 amino acids). Differences in selectivity for the regulators of the ADP-Glc PPases play a key metabolic role in the organisms that use ADP-Glc for synthesis of polysaccharides as carbon and energy storage. It is possible that a common enzyme ancestor evolved to other forms having different regulatory properties accommodating different metabolic environments and developed into several classes of ADP-Glc PPase (Table 1).
It is very possible that a fragment of ≈150 amino acids at the C terminus was acquired to make it a regulated enzyme and/or to improve the rudimentary regulation that was already present. The other sugar nucleotide pyrophosphorylases are not considered allosteric enzymes, and this extended C terminus is either lacking or part of a completely different domain to form a bifunctional enzyme (11, 87). It is not known whether the regulatory sites are located in the same or distinct domains in the protein structure, but it seems that the C terminus plays an important role in all the ADP-Glc PPase classes. Some of these enzymes are relatively nonspecific in selectivity for allosteric regulators, which indicates a certain flexibility to undergo evolutionary changes for adaptation to a certain metabolism. This is evident because minimal changes in the protein sequence can alter the specificity for the regulators. A single mutation (K419Q) on the Anabaena ADP-Glc PPase changed the preference of the activator from 3-PGA to fructose 1,6-bisphosphate (14).
Experiments with directed molecular evolution demonstrated that a few mutations of the small subunit from the potato tuber enzyme could alter the selectivity for activators of the homotetrameric form (α4) (83). Construction of chimeric enzymes showed that a single “crossover” between two genes rendered two ADP-Glc PPases that would belong to different classes than their parents. From enzymes of class I (E. coli) and class IV (A. tumefaciens), two ADP-Glc PPases that could be included as class V (chimeric enzyme AE) and class VI (chimeric enzyme EA) were found (6). It has been observed that this plasticity is also present in the inhibitor site of the Anabaena enzyme. Mutants R294A, R294E, and R294Q changed the selectivity from Pi to NADPH (18). Unfortunately, the structure-function relationships of the regulatory site(s) in heterotrophic bacteria are far from clear. A more comprehensive characterization of the structure of the allosteric sites will be very important to understand the evolutionary mechanism.
Another important step in the evolution is the appearance of the β (large) subunit in eukaryotes, most probably by gene duplication. This allowed further divergence and specialization to obtain different polypeptides, a catalytic and a modulatory subunit. The catalytic subunit had more constraints to its evolution, and that could be why they show very high homology even among different plants. Moreover, cyanobacterial homotetrameric enzymes show higher homology with the small subunits from plants than the small and large subunits show between themselves. The small subunit kept the catalytic function but lost the ability to be activated efficiently in the absence of the large subunit. Replacement of several amino acids showed that this process could be reversed in vitro (83). The large subunits might have evolved to satisfy different requirements in the tissues (88). Later, the catalytic subunit on certain tissues might have acquired the ability to be regulated by thioredoxin (3).
Acknowledgments
This work was supported in part by grants from the Department of Energy (DE-FG02-93ER20121) (J.P.), CONICET, Fundacion Antorchas, and ANPCyT (PICT'99 1-6074) (A.A.I.).
REFERENCES
- 1.Ball, K., and J. Preiss. 1994. Allosteric sites of the large subunit of the spinach leaf ADP-glucose pyrophosphorylase. J. Biol. Chem. 269:24706-24711. [PubMed] [Google Scholar]
- 2.Ball, S., T. Marianne, L. Dirick, M. Fresnoy, B. Delrue, and A. Decq. 1991. A Chlamydomonas reinhardtii low starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 185:17-26. [DOI] [PubMed] [Google Scholar]
- 3.Ballicora, M. A., J. B. Frueauf, Y. Fu, P. Schurmann, and J. Preiss. 2000. Activation of the potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J. Biol. Chem. 275:1315-1320. [DOI] [PubMed] [Google Scholar]
- 4.Ballicora, M. A., Y. Fu, N. M. Nesbitt, and J. Preiss. 1998. ADP-glucose pyrophosphorylase from potato tubers. Site-directed mutagenesis studies of the regulatory sites. Plant Physiol. 118:265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballicora, M. A., M. J. Laughlin, Y. Fu, T. W. Okita, G. F. Barry, and J. Preiss. 1995. Adenosine 5′-diphosphate-glucose pyrophosphorylase from potato tuber. Significance of the N terminus of the small subunit for catalytic properties and heat stability. Plant Physiol. 109:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballicora, M. A., J. I. Sesma, A. A. Iglesias, and J. Preiss. 2002. Characterization of chimeric ADP-glucose pyrophosphorylases of Escherichia coli and Agrobacterium tumefaciens. Importance of the C terminus on the selectivity for allosteric regulators. Biochemistry 41:9431-9437. [DOI] [PubMed] [Google Scholar]
- 7.Belanger, A. E., and G. F. Hatfull. 1999. Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J. Bacteriol. 181:6670-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blankenfeldt, W., M. Asuncion, J. S. Lam, and J. H. Naismith. 2000. The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA). EMBO J. 19:6652-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. Garcia-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 191:31-36. [DOI] [PubMed] [Google Scholar]
- 10.Branden, C. I. 1980. Relation between structure and function of alpha/beta-proteins. Q. Rev. Biophys. 13:317-338. [DOI] [PubMed] [Google Scholar]
- 11.Brown, K., F. Pompeo, S. Dixon, D. Mengin-Lecreulx, C. Cambillau, and Y. Bourne. 1999. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 18:4096-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charng, Y. Y., A. A. Iglesias, and J. Preiss. 1994. Structure-function relationships of cyanobacterial ADP-glucose pyrophosphorylase. Site-directed mutagenesis and chemical modification of the activator-binding sites of ADP-glucose pyrophosphorylase from Anabaena PCC 7120. J. Biol. Chem. 269:24107-24113. [PubMed] [Google Scholar]
- 13.Charng, Y. Y., G. Kakefuda, A. A. Iglesias, W. J. Buikema, and J. Preiss. 1992. Molecular cloning and expression of the gene encoding ADP-glucose pyrophosphorylase from the cyanobacterium Anabaena sp. strain PCC 7120. Plant Mol. Biol. 20:37-47. [DOI] [PubMed] [Google Scholar]
- 14.Charng, Y. Y., J. Sheng, and J. Preiss. 1995. Mutagenesis of an amino acid residue in the activator-binding site of cyanobacterial ADP-glucose pyrophosphorylase causes alteration in activator specificity. Arch. Biochem. Biophys. 318:476-480. [DOI] [PubMed] [Google Scholar]
- 15.Eidels, L., P. L. Edelmann, and J. Preiss. 1970. Biosynthesis of bacterial glycogen. VIII. Activation and inhibition of the adenosine diphosphoglucose pyrophosphorylases of Rhodopseudomonas capsulata and of Agrobacterium tumefaciens. Arch. Biochem. Biophys. 140:60-74. [DOI] [PubMed] [Google Scholar]
- 16.Espada, J. 1962. Enzymic synthesis of adenosine diphosphate glucose from glucose-1-phosphate and adenosine triphosphate. J. Biol. Chem. 237:3577-3581. [Google Scholar]
- 17.Frueauf, J. B., M. A. Ballicora, and J. Preiss. 2003. ADP-glucose pyrophosphorylase from potato tuber: site-directed mutagenesis of homologous aspartic acid residues in the small and large subunits. Plant J. 33:503-511. [DOI] [PubMed] [Google Scholar]
- 18.Frueauf, J. B., M. A. Ballicora, and J. Preiss. 2002. Alteration of inhibitor selectivity by site-directed mutagenesis of Arg(294) in the ADP-glucose pyrophosphorylase from Anabaena PCC 7120. Arch. Biochem. Biophys. 400:208-214. [DOI] [PubMed] [Google Scholar]
- 19.Frueauf, J. B., M. A. Ballicora, and J. Preiss. 2001. Aspartate residue 142 is important for catalysis by ADP-glucose pyrophosphorylase from Escherichia coli. J. Biol. Chem. 276:46319-46325. [DOI] [PubMed] [Google Scholar]
- 20.Fu, Y., M. A. Ballicora, J. F. Leykam, and J. Preiss. 1998. Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J. Biol. Chem. 273:25045-25052. [DOI] [PubMed] [Google Scholar]
- 21.Fu, Y., M. A. Ballicora, and J. Preiss. 1998. Mutagenesis of the glucose-1-phosphate-binding site of potato tuber ADP-glucose pyrophosphorylase. Plant Physiol. 117:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furlong, C. E., and J. Preiss. 1969. Biosynthesis of bacterial glycogen synthesis. VII. Purification and properties of adenosine diphosphoglucose pyrophosphorylase of Rhodospirillum rubrum. J. Biol. Chem. 244:2539-2548. [PubMed] [Google Scholar]
- 23.Gardiol, A., and J. Preiss. 1990. Escherichia coli E-39 ADP-glucose synthetase has different activation kinetics from the wild-type allosteric enzyme. Arch. Biochem. Biophys. 280:175-180. [DOI] [PubMed] [Google Scholar]
- 24.Gentner, N., and J. Preiss. 1967. Activator-inhibitor interactions in the adenosine diphosphate glucose pyrophosphorylase of Escherichia coli B. Biochem. Biophys. Res. Commun. 27:417-423. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh, P., C. Meyer, E. Remy, D. Peterson, and J. Preiss. 1992. Cloning, expression, and nucleotide sequence of glgC gene from an allosteric mutant of Escherichia coli B. Arch. Biochem. Biophys. 296:122-128. [DOI] [PubMed] [Google Scholar]
- 26.Giroux, M. J., J. Shaw, G. Barry, B. J. Cobb, T. Greene, T. Okita, and L. C. Hannah. 1996. A single gene mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 93:5824-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Casati, D. F., M. A. Aon, and A. A. Iglesias. 2000. Kinetic and structural analysis of the ultrasensitive behaviour of cyanobacterial ADP-glucose pyrophosphorylase. Biochem. J. 350:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Casati, D. F., S. Cortassa, M. A. Aon, and A. A. Iglesias. 2003. Ultrasensitive behavior in the synthesis of storage polysaccharides in cyanobacteria. Planta 216:969-975. [DOI] [PubMed]
- 29.Gomez-Casati, D. F., R. Y. Igarashi, C. N. Berger, M. E. Brandt, A. A. Iglesias, and C. R. Meyer. 2001. Identification of functionally important amino-terminal arginines of Agrobacterium tumefaciens ADP-glucose pyrophosphorylase by alanine scanning mutagenesis. Biochemistry 40:10169-10178. [DOI] [PubMed] [Google Scholar]
- 30.Gomez-Casati, D. F., and A. A. Iglesias. 2002. ADP-glucose pyrophosphorylase from wheat endosperm. Purification and characterization of an enzyme with novel regulatory properties. Planta 214:428-434. [DOI] [PubMed] [Google Scholar]
- 31.Govons, S., N. Gentner, E. Greenberg, and J. Preiss. 1973. Biosynthesis of bacterial glycogen. XI. Kinetic characterization of an altered adenosine diphosphate-glucose synthase from a “glycogen-excess” mutant of Escherichia coli B. J. Biol. Chem. 248:1731-1740. [PubMed] [Google Scholar]
- 32.Greenberg, E., J. E. Preiss, V. M., and J. Preiss. 1983. Biosynthesis of bacterial glycogen: activator specificity of the ADP-glucose pyrophosphorylase of Rhodopseudomonads. Arch. Biochem. Biophys. 220:594-604. [DOI] [PubMed] [Google Scholar]
- 33.Greene, T. W., R. L. Woodbury, and T. W. Okita. 1996. Aspartic acid 413 is important for the normal allosteric functioning of ADP-glucose pyrophosphorylase. Plant Physiol. 112:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill, M. A., K. Kaufmann, J. Otero, and J. Preiss. 1991. Biosynthesis of bacterial glycogen. Mutagenesis of a catalytic site residue of ADP-glucose pyrophosphorylase from Escherichia coli. J. Biol. Chem. 266:12455-12460. [PubMed] [Google Scholar]
- 35.Hylton, C., and A. M. Smith. 1992. The rb mutation of peas causes structural and regulatory changes in ADP-Glc pyrophosphorylase from developing embryos. Plant Physiol. 99:1626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igarashi, R. Y., and C. R. Meyer. 2000. Cloning and sequencing of glycogen metabolism genes from Rhodobacter sphaeroides 2.4.1. Expression and characterization of recombinant ADP-glucose pyrophosphorylase. Arch. Biochem. Biophys. 376:47-58. [DOI] [PubMed] [Google Scholar]
- 37.Iglesias, A. A., G. F. Barry, C. Meyer, L. Bloksberg, P. A. Nakata, T. Greene, M. J. Laughlin, T. W. Okita, G. M. Kishore, and J. Preiss. 1993. Expression of the potato tuber ADP-glucose pyrophosphorylase in Escherichia coli. J. Biol. Chem. 268:1081-1086. [PubMed] [Google Scholar]
- 38.Iglesias, A. A., Y. Y. Charng, S. Ball, and J. Preiss. 1994. Characterization of the kinetic, regulatory, and structural properties of ADP-glucose pyrophosphorylase from Chlamydomonas reinhardtii. Plant Physiol. 104:1287-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglesias, A. A., G. Kakefuda, and J. Preiss. 1992. Involvement of arginine residues in the allosteric activation and inhibition of Synechocystis PCC 6803 ADP-glucose pyrophosphorylase. J. Protein Chem. 11:119-128. [DOI] [PubMed] [Google Scholar]
- 40.Iglesias, A. A., G. Kakefuda, and J. Preiss. 1991. Regulatory and structural properties of the cyanobacterial ADP-glucose pyrophosphorylases. Plant Physiol. 97:1187-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iglesias, A. A., and F. E. Podestá. 1996. Photosynthate formation and partitioning in crop plants, p. 681-698. In M. Pessarakli (ed.), Handbook of photosynthesis. Marcel Dekker, Inc., New York, N.Y.
- 42.Iglesias, A. A., and J. Preiss. 1992. Bacterial glycogen and plant starch biosynthesis. Biochem. Educ. 20:196-203. [Google Scholar]
- 43.Kiel, J. A., J. M. Boels, G. Beldman, and G. Venema. 1994. Glycogen in Bacillus subtilis: molecular characterization of an operon encoding enzymes involved in glycogen biosynthesis and degradation. Mol. Microbiol. 11:203-218. [DOI] [PubMed] [Google Scholar]
- 44.Kleczkowski, L. A., P. Villand, E. Lüthi, O. A. Olsen, and J. Preiss. 1993. Insensitivity of barley endosperm ADP-Glc pyrophosphorylase to 3-phosphoglycerate and orthophosphate regulation. Plant Physiol. 101:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koshland, D. E. 1987. Switches, thresholds and ultrasensitivity. Trends Biochem. Sci. 12:225-229. [Google Scholar]
- 46.Kostrewa, D., A. D'Arcy, B. Takacs, and M. Kamber. 2001. Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg2+ at 1.96 A resolution. J. Mol. Biol. 305:279-289. [DOI] [PubMed] [Google Scholar]
- 47.Laughlin, M. J., S. E. Chantler, and T. W. Okita. 1998. N- and C terminus peptide sequences are essential for enzyme assembly, allosteric, and/or catalytic properties of ADP-glucose pyrophosphorylase. Plant J. 14:159-168. [DOI] [PubMed] [Google Scholar]
- 48.Laughlin, M. J., J. W. Payne, and T. W. Okita. 1998. Substrate binding mutants of the higher plant ADP-glucose pyrophosphorylase. Phytochemistry 47:621-629. [DOI] [PubMed] [Google Scholar]
- 49.Lee, Y. M., S. Mukherjee, and J. Preiss. 1986. Covalent modification of Escherichia coli ADP-glucose synthetase with 8-azido substrate analogs. Arch. Biochem. Biophys. 244:585-595. [DOI] [PubMed] [Google Scholar]
- 50.Lee, Y. M., and J. Preiss. 1986. Covalent modification of substrate-binding sites of Escherichia coli ADP-glucose synthetase. Isolation and structural characterization of 8-azido-ADP-glucose-incorporated peptides. J. Biol. Chem. 261:1058-1064. [PubMed] [Google Scholar]
- 51.Leloir, L. F. 1971. Two decades of research on the biosynthesis of saccharides. Science 172:1299-1303. [DOI] [PubMed] [Google Scholar]
- 52.Lemesle-Varloot, L., B. Henrissat, C. Gaboriaud, V. Bissery, A. Morgat, and J. P. Mornon. 1990. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D representation of protein sequences. Biochimie 72:555-574. [DOI] [PubMed] [Google Scholar]
- 53.Li, L., and J. Preiss. 1992. Characterization of ADP-glucose pyrophosphorylase from a starch-deficient mutant of Arabidopsis thaliana (L). Carbohydr. Res. 227:227-239. [Google Scholar]
- 54.Lin, T. P., T. Caspar, C. Somerville, and J. Preiss. 1988. Isolation and characterization of a starchless mutant of Arabidopsis thaliana L. Henyh lacking ADP-glucose pyrophosphorylase activity. Plant Physiol. 86:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin, T. P., T. Caspar, C. Somerville, and J. Preiss. 1988. A starch deficient mutant of Arabidopsis thaliana with low ADP-glucose pyrophosphorylase activity lacks one of the two subunits of the enzyme. Plant Physiol. 88:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin, M., D. Schneider, C. Bruton, K. Chater, and C. Hardisson. 1997. A glgC gene essential only for the first of two spatially distinct phases of glycogen synthesis in Streptomyces coelicolor A3(2). J. Bacteriol. 179:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuno, K., T. Blais, A. W. Serio, T. Conway, T. M. Henkin, and A. L. Sonenshein. 1999. Metabolic imbalance and sporulation in an isocitrate dehydrogenase mutant of Bacillus subtilis. J. Bacteriol. 181:3382-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.May, T. B., D. Shinabarger, A. Boyd, and A. M. Chakrabarty. 1994. Identification of amino acid residues involved in the activity of phosphomannose isomerase-guanosine 5′-diphospho-d-mannose pyrophosphorylase, a bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J. Biol. Chem. 269:4872-4877. [PubMed] [Google Scholar]
- 59.Meyer, C. R., J. A. Bork, S. Nadler, J. Yirsa, and J. Preiss. 1998. Site-directed mutagenesis of a regulatory site of Escherichia coli ADP-glucose pyrophosphorylase: the role of residue 336 in allosteric behavior. Arch. Biochem. Biophys. 353:152-159. [DOI] [PubMed] [Google Scholar]
- 60.Meyer, C. R., J. Yirsa, B. Gott, and J. Preiss. 1998. A kinetic study of site-directed mutants of Escherichia coli ADP-glucose pyrophosphorylase: the role of residue 295 in allosteric regulation. Arch. Biochem. Biophys. 352:247-254. [DOI] [PubMed] [Google Scholar]
- 61.Morell, M., M. Bloom, and J. Preiss. 1988. Affinity labeling of the allosteric activator site(s) of spinach leaf ADP-glucose pyrophosphorylase. J. Biol. Chem. 263:633-637. [PubMed] [Google Scholar]
- 62.Morell, M. K., M. Bloom, V. Knowles, and J. Preiss. 1987. Subunit structure of spinach leaf ADP-glucose pyrophosphorylase. Plant Physiol. 85:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakata, P. A., J. M. Anderson, and T. W. Okita. 1994. Structure and expression of the potato ADP-glucose pyrophosphorylase small subunit. J. Biol. Chem. 269:30798-30807. [PubMed] [Google Scholar]
- 64.Nakata, P. A., T. W. Greene, J. M. Anderson, B. J. Smith-White, T. W. Okita, and J. Preiss. 1991. Comparison of the primary sequences of two potato tuber ADP-glucose pyrophosphorylase subunits. Plant Mol. Biol. 17:1089-1093. [DOI] [PubMed] [Google Scholar]
- 65.Olsen, L. R., and S. L. Roderick. 2001. Structure of the Escherichia coli GlmU pyrophosphorylase and acetyltransferase active sites. Biochemistry 40:1913-1921. [DOI] [PubMed] [Google Scholar]
- 66.Parsons, T. F., and J. Preiss. 1978. Biosynthesis of bacterial glycogen. Incorporation of pyridoxal phosphate into the allosteric activator site and an ADP-glucose-protected pyridoxal phosphate binding site of Escherichia coli B ADP-glucose synthase. J. Biol. Chem. 253:6197-6202. [PubMed] [Google Scholar]
- 67.Parsons, T. F., and J. Preiss. 1978. Biosynthesis of bacterial glycogen. Isolation and characterization of the pyridoxal-phosphate allosteric activator site and the ADP-glucose-protected pyridoxal-phosphate binding site of Escherichia coli B ADP-glucose synthase. J. Biol. Chem. 253:7638-7645. [PubMed] [Google Scholar]
- 68.Plaskitt, K. A., and K. F. Charter. 1985. Influences of developmental genes on localized glycogen deposition colonies of mycelial prokaryote, Streptomyces coelicolor A3(2): a possible interface between metabolism and morphogenesis. Phil. Trans. R. Soc. Lond. B 347:105-121. [Google Scholar]
- 69.Plaxton, W. C., and J. Preiss. 1987. Purification and properties of non-proteolytically degraded ADP-glucose pyrophosphorylase from maize endosperm. Plant Physiol. 83:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Preiss, J. 1973. Adenosine diphosphoryl glucose pyrophosphorylase, p. 73-119. In P. D. Boyer (ed.), The enzymes, vol. 8. Academic Press, New York, N.Y.
- 71.Preiss, J. 1996. ADP-glucose pyrophosphorylase: basic science and applications in biotechnology, p. 259-279. In M.R. El-Gewely (ed.), Biotechnology annual review. Elsevier Science, Amsterdam, The Netherlands. [DOI] [PubMed]
- 72.Preiss, J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38:419-458. [DOI] [PubMed] [Google Scholar]
- 73.Preiss, J. 1991. Biology and molecular biology of starch synthesis and its regulation, p. 59-114. In B. Mifflin (ed.), Oxford surveys of plant molecular and cell biology, vol. 7. Oxford University Press, Oxford, United Kingdom.
- 74.Preiss, J., C. Lammel, and E. Greenberg. 1976. Biosynthesis of bacterial glycogen. Kinetic studies of a glucose-1-phosphate adenylyltransferase (EC 2.7.7.27) from a glycogen-excess mutant of Escherichia coli B. Arch. Biochem. Biophys. 174:105-119. [DOI] [PubMed] [Google Scholar]
- 75.Preiss, J., and T. Romeo. 1994. Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 47:299-329. [DOI] [PubMed] [Google Scholar]
- 76.Preiss, J., L. Shen, E. Greenberg, and N. Gentner. 1966. Biosynthesis of bacterial glycogen. IV. Activation and inhibition of the adenosine diphosphate glucose pyrophosphorylase of Escherichia coli B. Biochemistry 5:1833-1845. [DOI] [PubMed] [Google Scholar]
- 77.Preiss, J., and M. N. Sivak. 1998. Biochemistry, molecular biology and regulation of starch synthesis. Genet. Eng. 20:177-223. [DOI] [PubMed] [Google Scholar]
- 78.Preiss, J., and M. N. Sivak. 1998. Starch and glycogen biosynthesis, p. 441-495. In B. M. Pinto (ed.), Comprehensive natural products chemistry, vol. 3. Pergamon Press, Oxford, United Kingdom.
- 79.Preiss, J., S. G. Yung, and P. A. Baecker. 1983. Regulation of bacterial glycogen synthesis. Mol. Cell. Biochem. 57:61-80. [DOI] [PubMed] [Google Scholar]
- 80.Rossman, M. G., D. Moras, and K. W. Olsen. 1974. Chemical and biological evolution of a nucleotide binding protein. Nature 250:194-199. [DOI] [PubMed] [Google Scholar]
- 81.Rost, B., and C. Sander. 1993. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. USA 90:7558-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rudi, H., D. N. P. Doan, and O. A. Olsen. 1997. A (His)6-tagged recombi-nant barley (Hordeum vulgare L.) endosperm ADP-glucose pyrophosphor-ylase expressed in the baculovirus-insect cell system is insensitive to regulation by 3-phosphoglycerate and inorganic phosphate. FEBS Lett. 419:124-130. [DOI] [PubMed] [Google Scholar]
- 83.Salamone, P. R., I. H. Kavakli, C. J. Slattery, and T. W. Okita. 2002. Directed molecular evolution of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. USA 99:1070-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saraste, M., P. R. Sibbald, and W. Wittinghofer. 1990. The P-loop -a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15:430-434. [DOI] [PubMed] [Google Scholar]
- 85.Sheng, J., Y. Y. Charng, and J. Preiss. 1996. Site-directed mutagenesis of lysine 382, the activator-binding site of ADP-glucose pyrophosphorylase from Anabaena PCC 7120. Biochemistry 35:3115-3121. [DOI] [PubMed] [Google Scholar]
- 86.Sheng, J., and J. Preiss. 1997. Arginine294 is essential for the inhibition of Anabaena PCC 7120 ADP-glucose pyrophosphorylase by phosphate. Biochemistry 36:13077-13084. [DOI] [PubMed] [Google Scholar]
- 87.Shinabarger, D., A. Berry, T. B. May, R. Rothmel, A. Fialho, and A. M. Chakrabarty. 1991. Purification and characterization of phosphomannose isomerase-guanosine diphospho-d-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J. Biol. Chem. 266:2080-2088. [PubMed] [Google Scholar]
- 88.Sivak, M. N., and J. Preiss. 1998. Starch: basic science to biotechnology, p. 1-199. In S. L. Taylor (ed.), Advances in food and nutrition research, vol. 41. Academic Press, San Diego, Calif.
- 89.Sivaraman, J., V. Sauve, A. Matte, and M. Cygler. 2002. Crystal structure of Escherichia coli glucose-1-phosphate thymidylyltransferase (RffH) complexed with dTTP and Mg2+. J. Biol. Chem. 277:44214-44219. [DOI] [PubMed] [Google Scholar]
- 90.Smidansky, E. D., M. Clancy, F. D. Meyer, S. P. Lanning, N. K. Blake, L. E. Talbert, and M. J. Giroux. 2002. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. USA 99:1724-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith-White, B. J., and J. Preiss. 1992. Comparison of proteins of ADP-glucose pyrophosphorylase from diverse sources. J. Mol. Evol. 34:449-464. [DOI] [PubMed] [Google Scholar]
- 92.Spatafora, G., K. Rohrer, D. Barnard, and S. Michalek. 1995. A Streptococcus mutans mutant that synthesizes elevated levels of intracellular polysaccharide is hypercariogenic in vivo. Infect. Immun. 63:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stark, D. M., K. P. Timmerman, G. F. Barry, J. Preiss, and G. M. Kishore. 1992. Role of ADP-glucose pyrophosphorylase in regulating starch levels in plant tissues. Science 258:287-292. [DOI] [PubMed] [Google Scholar]
- 94.Steiner, K. E., and J. Preiss. 1977. Biosynthesis of bacterial glycogen: genetic and allosteric regulation of glycogen biosynthesis in Salmonella typhimurium NT-2. J. Bacteriol. 129:246-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strange, R. E. 1968. Bacterial “glycogen” and survival. Nature 220:606-6077. [DOI] [PubMed] [Google Scholar]
- 96.Takata, H., T. Takaha, S. Okada, M. Takagi, and T. Imanaka. 1997. Characterization of a gene cluster for glycogen biosynthesis and a heterotetrameric ADP-glucose pyrophosphorylase from Bacillus stearothermophilus. J. Bacteriol. 179:4689-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uttaro, A. D., R. A. Ugalde, J. Preiss, and A. A. Iglesias. 1998. Cloning and expression of the glgC gene from Agrobacterium tumefaciens: purification and characterization of the ADP-glucose synthetase. Arch. Biochem. Biophys. 357:13-21. [DOI] [PubMed] [Google Scholar]
- 98.Weber, H., U. Heim, L. Borisjuk, and U. Wobus. 1995. Cell-type specific, coordinate expression of two ADP-glucose pyrophosphorylase genes in relation to starch biosynthesis during seed development in Vicia faba L. Planta 195:352-361. [DOI] [PubMed] [Google Scholar]
- 99.Wu, M. X., and J. Preiss. 1998. The N terminus region is important for the allosteric activation and inhibition of the Escherichia coli ADP-glucose pyrophosphorylase. Arch. Biochem. Biophys. 358:182-188. [DOI] [PubMed] [Google Scholar]
- 100.Wu, M. X., and J. Preiss. 2001. Truncated forms of the recombinant Escherichia coli ADP-glucose pyrophosphorylase: The importance of the N terminus region for allosteric activation and inhibition. Arch. Biochem. Biophys. 389:159-165. [DOI] [PubMed] [Google Scholar]
- 101.Yung, S. G., and J. Preiss. 1981. Biosynthesis of bacterial glycogen: purification and structural properties of Rhodospirillum tenue adenosine diphosphate glucose synthetase. J. Bacteriol. 147:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]