Abstract
Rapid serogrouping of meningococci is essential for the effective public health management of cases of the disease and the contacts of infected patients. Here we describe an accurate nucleotide-sequencing method for the confirmation of serogroup Y and W135 meningococci by siaD gene analysis from cultures of Neisseria meningitidis.
Neisseria meningitidis is one of the major causes of bacterial meningitis, and serogroups A, B, C, Y, and W135 account for most disease worldwide (11). The rapid and accurate characterization of meningococci is crucial for effective public health management. Swift serogroup determination is essential, particularly during case clusters and outbreaks, while the changing epidemiology of this important pathogen makes serogrouping increasingly important. Meningococcal serogroup C conjugate vaccines have now been introduced in the United Kingdom and elsewhere, there have been two outbreaks associated with serogroup W135 disease and the Hajj pilgrimage (13) (M. K. Taha, M. Achtman, J. M. Alonso, B. Greenwood, M. Ramsay, A. Fox, S. Gray, and E. Kaczmarski, Letter, Lancet 356:2159, 2000), and serogroup Y now accounts for a significant percentage of disease in the United States (14).
Current methods for serogrouping meningococci rely on phenotypic methods, such as latex agglutination, or genotypic methods, such as PCR restriction fragment length polymorphism (RFLP) (2, 3). However, the former lacks sensitivity and specificity while the latter is based on the presence of RFLPs and, therefore, their longer-term accuracy is questionable with an organism that is both highly competent in the uptake of genetic material by transformation and undergoing epidemiological change worldwide (6, 9, 12). Nucleotide sequencing is being used increasingly in the modern microbiology laboratory and is accurate, digitally based, and, therefore, portable. We therefore developed a nucleotide-sequencing method which compares numerous nucleotide differences between serogroups Y and W135. The siaD gene is part of the operon in meningococci that is responsible for capsule synthesis and has been studied quite extensively (1, 3, 4, 8, 15). It is highly conserved between serogroup B and C meningococci but is less conserved among serogroup Y and W135 meningococci (3, 4, 8). In particular, there is a variable region towards the end of the first third of the gene in which numerous nucleotide differences occur between serogroup Y and W135. We therefore took advantage of this and designed oligonucleotide primers external to this variable region for PCR amplification and internal primers for nucleotide sequencing.
Ninety-six clinical isolates of N. meningitidis (isolated in Scotland between 1998 and 2002) were analyzed. Of the serogroups B, C, Y, and W135, 24 isolates of each were chosen. The serogroup Y isolates were from cerebrospinal fluid (n = 1), blood (n = 3), eye (n = 1), knee aspirate (n = 1), sputum (n = 8), throat (n = 8), urethra (n = 1), and unknown (n = 1). The cases of 6 patients were considered to be invasive serogroup Y infections, while the remaining 18 patients were carriers. The serogroup W135 isolates were from cerebrospinal fluid (n = 4), blood (n = 4), sputum (n = 4), throat (n = 6), vagina (n = 1), rectum (n = 1), and unknown (n = 4). The cases of 8 patients were considered to be invasive serogroup W135 infections, while the remaining 16 patients were carriers. Using standard methods as part of the service provided by the Scottish Meningococcus and Pneumococcus Reference Laboratory (3, 4, 7), all isolates had previously been serogrouped by latex agglutination and PCR-RFLP. PCR and nucleotide sequencing were performed as previously described (5) except that sequencing was performed on a MegaBACE 1000 automated DNA sequencer (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). Oligonucleotide primers WYxampF (5′-GCTGGAAGAAATGCATTCC-3′) and WYxampR (5′-CAGGATTGGTTAGCTCGA-3′) were used for PCR; the PCR cycle was 95°C for 5 min, followed by 35 cycles of 94°C for 25 s, 53°C for 40 s, and 72°C for 1 min, with a final extension step of 72°C for 5 min. Using oligonucleotide primers WyxseqF (5′-gAATCTTCCGAGCAGGAA-3′) and WyxseqR (5′-TGCGCGGAAGAATAGTG-3′), sequence labeling was performed as previously described; the sequence-labeling cycle was 30 cycles of 92°C for 20 s, 50°C for 15 s, and 60°C for 1 min. Nucleotide sequences from each strain were aligned using the AlignIR program (LICOR Biosciences, Cambridge, United Kingdom) and the ALIGN program at the Genestream Resource Center (http://www2.igh.cnrs.fr/bin/align-guess.cgi). These sequences were then compared against the consensus sequences gained from GenBank (accession numbers Y13969 and Y13970 for serogroups Y and W135, respectively). PCR was performed on all 96 strains, but only the 48 serogroup Y and W135 strains gave positive results. The PCR products were sequenced, and a total of 29 nucleotide differences were noted which could be used for determination of their serogroups by sequence alignment (Fig. 1). No polymorphism differences were noted between the test strains and the reference sequences in this study. However, as polymorphisms develop over time, an alignment database can be used to determine the serogroup (10). The nucleotide-sequencing results were compared with those gained from latex agglutination and PCR-RFLP. The results gained by genotyping were identical to those gained by phenotyping and PCR-RFLP, indicating that the test was 100% specific for serogroups Y and W135.
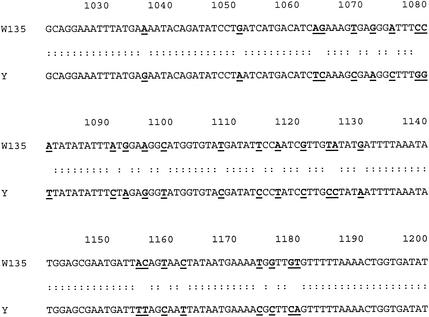
FIG. 1.
Partial sequence alignment of the siaD sequence from serogroups Y and W135. Sequences were aligned using the ALIGN program. Nucleotide numbers correspond to those in the published serogroup Y and W135 siaD sequences (accession numbers Y13969 and Y13970). Nucleotide identities are indicated by stacked dots. Nucleotide differences are shown in boldface and underlined.
Rapid tools are required for the serogroup determination of meningococci causing disease, and they must be accurate so that effective public health management can take place. Also, the handling of clusters and outbreaks, or even the description of new strains, relies on methods that currently depend on the analysis of RFLPs. Such analysis cannot be relied on as a robust, long-term method for serogrouping strains. In this study, a total of 50 nucleotide sequences were compared within the variable region of the siaD gene in serogroup Y and W135 meningococci. Of these sequences, 48 were from clinical meningococcal isolates which were serogrouped using phenotypic and genotypic methods. Nucleotide sequencing provides a more robust method, because it is based on the analysis of multiple nucleotide differences and takes into account nucleotide changes that can occur over time by mutation or recombination. It has been shown that this methodology can be applied to meningococcal cultures but could also possibly be applied to body fluids containing meningococcal DNA. Such a methodology is important for accurately serogrouping meningococcal strains that commonly cause disease in the United States and also those strains associated with the Hajj pilgrimage.
Acknowledgments
Funding for the liquid-handling robot and automated DNA sequencer was generously provided by the Meningitis Association (Scotland). This work was also supported, in part, by the Chief Scientist's Office of The Scottish Executive.
REFERENCES
- 1.Arreaza, L., B. Alcala, C. Salcedo, and J. A. Vazquez. 2001. Interruption of siaD in a meningococcal carrier isolate mediated by an insertion sequence. Clin. Diagn. Lab. Immunol. 8:465-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow, R., N. Andrews, D. Goldblatt, and E. Miller. 2001. Serological basis for use of meningococcal serogroup C conjugate vaccines in the United Kingdom: reevaluation of correlates of protection. Infect. Immun. 69:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow, R., H. Claus, U. Chaudhry, M. Guiver, E. B. Kaczmarski, M. Frosch, and A. J. Fox. 1998. siaD PCR ELISA for confirmation and identification of serogroup Y and W135 meningococcal infections. FEMS Microbiol. Lett. 159:209-214. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, R., H. Claus, M. Guiver, L. Smart, D. M. Jones, E. B. Kaczmarski, M. Frosch, and A. J. Fox. 1997. Non-culture diagnosis and serogroup determination of meningococcal B and C infection by a sialyltransferase (siaD) PCR ELISA. Epidemiol. Infect. 118:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, S. C., M. A. Diggle, and G. F. Edwards. 2001. Semiautomation of multilocus sequence typing for the characterization of clinical isolates of Neisseria meningitidis. J. Clin. Microbiol. 39:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, S. C., and G. F. Edwards. 2000. Implications for the serogroup incidence of meningococcal disease after the introduction of the MenC vaccine. Scott. Med. J. 45:67. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, S. C., J. Reid, L. Thom, B. C. Denham, and G. F. Edwards. 2002. Laboratory confirmation of meningococcal disease in Scotland, 1993-9. J. Clin. Pathol. 55:32-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claus, H., U. Vogel, M. Muhlenhoff, R. Gerardy-Schahn, and M. Frosch. 1997. Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol. Gen. Genet. 257:28-34. [DOI] [PubMed] [Google Scholar]
- 9.Diaz, P. S. 1999. The epidemiology and control of invasive meningococcal disease. Pediatr. Infect. Dis. J. 18:633-634. [DOI] [PubMed] [Google Scholar]
- 10.Diggle, M. A., and S. C. Clarke. 2002. Rapid assignment of nucleotide sequence data to allele types for multi-locus sequence analysis (MLSA) of bacteria using an adapted database and modified alignment program. J. Mol. Microbiol. Biotechnol. 4:515-517. [PubMed] [Google Scholar]
- 11.Hart, C. A., and T. R. Rogers. 1993. Meningococcal disease. J. Med. Microbiol. 39:3-25. [DOI] [PubMed] [Google Scholar]
- 12.Maiden, M. C., and B. G. Spratt. 1999. Meningococcal conjugate vaccines: new opportunities and new challenges. Lancet 354:615-616. [DOI] [PubMed] [Google Scholar]
- 13.Popovic, T., C. T. Sacchi, M. W. Reeves, A. M. Whitney, L. W. Mayer, C. A. Noble, G. W. Ajello, F. Mostashari, N. Bendana, J. Lingappa, R. Hajjeh, and N. E. Rosenstein. 2000. Neisseria meningitidis serogroup W135 isolates associated with the ET-37 complex. Emerg. Infect. Dis. 6:428-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 15.Vogel, U., H. Claus, G. Heinze, and M. Frosch. 1997. Functional characterization of an isogenic meningococcal α-2,3-sialyltransferase mutant: the role of lipooligosaccharide sialylation for serum resistance in serogroup B meningococci. Med. Microbiol. Immunol. (Berlin) 186:159-166. [DOI] [PubMed] [Google Scholar]