Abstract
Corynebacterium freneyi is a recently described alpha-glucosidase-positive species of the genus Corynebacterium. To our knowledge, there is no description of human infection due to this species. We report on a case of bacteremia due to C. freneyi following vascular surgery.
CASE REPORT
Among the corynebacteria, Corynebacterium xerosis, Corynebacterium amycolatum, and Corynebacterium striatum are the species that are the most frequently isolated from clinical samples (8). Their natural habitat is human skin, and as a result, these species sometimes appear as sample contaminants. These species are frequently misidentified by biochemical identification (5, 6, 12, 14). Corynebacterium freneyi is closely related to these three species and was recently described by Renaud et al. (9). Those investigators have studied five strains isolated from clinical samples, but to our knowledge, there is no description of human infection due to this species. We report on a case of bacteremia due to C. freneyi after vascular surgery.
A-49-year-old man was hospitalized in April 2001 and June 2001 for acute ischemia of the right tibial artery and surgical recanalization. In August 2001, he suffered of acute pain in decubitus position. Upon examination, his feet were cold and had decreased sensitivity. On 18 August 2001, he received a graft of the cephalic left vein to create a femoral-pedal bridge. On 20 August, he suffered acute ischemia of his right leg and the leg was amputated at the metatarsal level. On the same day, he presented with a temperature of 38.5°C, and one pair of blood samples for culture (one sample for aerobic culture and one sample for anaerobic culture) were drawn and subcultured on sheep blood agar (bioMérieux, Marcy-l'Etoile, France) at 37°C. After blood culture and susceptibility testing, the patient was started on intravenous amoxicillin at 2 g/day. Apyrexia was obtained within 48 h. The patient was discharged on 27 August to a rest home.
In the case of our patient, after a culturing time of 48 h, 1-mm whitish colonies with irregular edges were observed from both the aerobic and the anaerobic blood culture bottles. Gram staining revealed gram-positive, non-spore-forming diphtheroids. Tests for catalase and alpha-glucosidase were positive. The API-CORYNE (bioMérieux) profile was 3110325, coding for C. striatum-C. amycolatum (API-CORYNE profiles book, 2nd ed., 1997).
Antibiotic susceptibility testing was performed on sheep blood Mueller-Hinton agar plates by the disk diffusion method according to the recommendations of the NCCLS (7). The MICs were 1 mg/liter for amoxicillin, 0.5 mg/liter for rifampin, 1 mg/liter for gentamicin, 2 mg/liter for vancomycin, 8 mg/liter for erythromycin, and 16 mg/liter for co-trimoxazole.
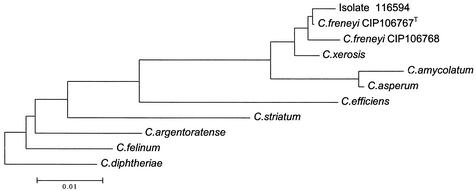
Further identification of the isolate was done by 16S RNA gene sequence analysis. The DNA of a single colony was extracted by using the Fast-prep DNA extraction kit and the Fast-prep DNA device as described by the supplier (Bio 101, Inc., La Jolla, Calif.) (3). The 16S rRNA gene was amplified by using primers FD1 (5′-AGAGTTTGATCCTGGCTGAG-3′) and RP2 (5′-ACGGCTACCTTGTTACGACTT-3′) (15). PCRs were performed with a Perkin-Elmer 9600 thermocycler under the following conditions: following a first denaturation step (95°C for 2 min), a three-step cycle of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min was repeated 35 times. Sequence determination was performed as described previously (2). The partial sequence (1,225 bp) of the 16S rRNA of this isolate was deposited in GenBank (accession number AY210513) and was aligned and compared with all eubacterial 16S rRNA gene sequences available in the GenBank and EMBL databases by multisequence analysis with the advanced BLAST software of the National Center for Biotechnology Information (1). The highest 16 rRNA gene sequence similarity value (99.7%) was obtained with the C. freneyi CIP106767T 16S rRNA gene sequence (EMBL AJ292762). The morphology and the biochemical characteristics of our isolate were similar to those of type strain CIP106767 studied by Renaud et al. (9). Following initial alignment of the sequences with the CUSTAL W program (version 1.8) (13), neighbor-joining analysis was performed by using PAUP software (version 4.0b1; Sinauer, Sunderland, Mass.). Figure 1 shows the dendrogram that we obtained. Our isolate, isolate 116594, is closely related to C. freneyi CIP106767T and C. freneyi CIP106768. The sequence of C. xerosis is the most similar to that of C. freneyi CIP106768 (similarity, 98.5%), followed by those of C. amycolatum (similarity, 98.0%), Corynebacterium asperum (similarity, 97.0%), Corynebacterium efficiens (similarity, 96.1%) (11), C. striatum (similarity, 94.0%), Corynebacterium argentoratense (similarity, 93.8%) (10), and Corynebacterium felinum (similarity, 92.0%) (4). These results are similar to those of Renaud et al. (9).
FIG. 1.
Dendrogram obtained by analysis of 16S rRNA gene sequences. Corynebacterium diphtheriae was used as the outgroup.
The isolate grew readily in pure culture, and no other C. freneyi strain was isolated in the same laboratory. The isolation of C. freneyi from a blood culture has never been reported. The isolate was recovered from blood when the patient presented with an acute onset of fever, with no other microorganism recovered from other specimens obtained at appropriate times. These facts suggest that this isolate was not a contaminant. This description could be relevant for infectious disease consulting.
Analysis of 16S rRNA gene sequences offers a reliable and straightforward tool for organism identification (12), and routine use of this method should increase our knowledge regarding the clinical spectrum of C. freneyi infections in humans.
Nucelotide sequence accession number.
The partial sequence (1,225 bp) of the 16S rRNA of patient isolate 116594 has been deposited in GenBank under accession number AY210513.
REFERENCES
- 1.Altschul, X., F. Stephen, L. Thomas, X. Madden, A. Alejandro, X. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beau, F., C. Bollet, T. Coton, E. Garnotel, and M. Drancourt. 1999. Molecular identification of a Nocardiopsis dassonvillei blood isolate J. Clin. Microbiol. 37:3366-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D., L. Hoyles, R. A. Hutson, G. Foster, and E. Falsen. 2001. Corynebacterium testudinoris sp. nov., from a tortoise, and Corynebacterium felinum sp. nov., from a Scottish wild cat. Int. J. Syst. E vol. Microbiol. 51:1349-1352. [DOI] [PubMed] [Google Scholar]
- 5.Funke, G., P. A. Lawson, K. A. Bernard, and M. D. Collins. 1996. Most Corynebacterium xerosis strains identified in the routine clinical laboratory correspond to Corynebacterium amycolatum. J. Clin. Microbiol. 34:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagrou, K., J. Verhaegen, M. Janssens, G. Wauters, and L. Verbist. 1998. Prospective study of catalase-positive coryneform organisms in clinical specimens: identification, clinical relevance, and antibiotic susceptibility. Diagn. Microbiol. Infect. Dis. 30:7-15. [DOI] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 1990. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 8.Renaud, F. N., M. Dutaur, S. Daoud, D. Aubel, P. Riegel, D. Monget, and J. Freney. 1998. Differentiation of Corynebacterium amycolatum, C. minutissimum, and C. striatum by carbon substrate assimilation tests. J. Clin. Microbiol. 36:3698-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renaud, F. N., D. Aubel, P. Riegel, H. Meugnier, and C. Bollet. 2001. Corynebacterium freneyi sp. nov., alpha-glucosidase-positive strains related to Corynebacterium xerosis. Int. J. Syst. E vol. Microbiol. 51:1723-1728. [DOI] [PubMed] [Google Scholar]
- 10.Riegel, P., R. Ruimy, D. De Briel, G. Prevost, F. Jehl, F. Bimet, R. Christen, and H. Monteil. 1995. Corynebacterium argentoratense sp. nov., from the human throat. Int. J. Syst. Bacteriol. 45:533-537. [DOI] [PubMed] [Google Scholar]
- 11.Seto, A., K. Yamada, E. Kimura, T. Nakamatsu, A. Hiraishi, and S. Yamanaka. 2002. Corynebacterium efficiens sp. nov., a glutamic-acid-producing species from soil and vegetables. Int. J. Syst. E vol. Microbiol. 52:1127-1131. [DOI] [PubMed] [Google Scholar]
- 12.Tang, Y. W., A. Von Graevenitz, M. G. Waddington, M. K. Hopkins, D. H. Smith, H. Li, C. P. Kolbert, S. O. Montgomery, and D. H. Persing. 2000. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis. J. Clin. Microbiol. 38:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wauters, G., B. Van Bosterhaut, M. Janssens, and J. Verhaegen. 1998. Identification of Corynebacterium amycolatum and other nonlipophilic fermentative corynebacteria of human origin. J. Clin. Microbiol. 36:1430-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]