Abstract
Recent events illustrate the imperative to rapidly and accurately detect and identify pathogens during disease outbreaks, whether they are natural or engineered. Particularly for our primary goal of detecting bioterrorist releases, detection techniques must be both species-wide (capable of detecting all known strains of a given species) and species specific. Due to classification restrictions on the publication of data for species that may pose a bioterror threat, we illustrate the challenges of finding such assays using five nonthreat organisms that are nevertheless of public health concern: human immunodeficiency virus (HIV) and four species of hepatitis viruses. Fluorogenic probe-based PCR assays (TaqMan; Perkin-Elmer Corp., Applied Biosystems, Foster City, Calif.) may be sensitive, fast methods for the identification of species in which the genome is conserved among strains, such as hepatitis A virus. For species such as HIV, however, the strains are highly divergent. We use computational methods to show that nine TaqMan primer and probe sequences, or signatures, are needed to ensure that all strains will be detected, but this is an unfeasible number, considering the cost of TaqMan probes. Strains of hepatitis B, C, and E viruses show intermediate divergence, so that two to three TaqMan signatures are required to detect all strains of each virus. We conclude that for species such as hepatitis A virus with high levels of sequence conservation among strains, signatures can be found computationally for detection by the TaqMan assay, which is a sensitive, rapid, and cost-effective method. However, for species such as HIV with substantial genetic divergence among strains, the TaqMan assay becomes unfeasible and alternative detection methods may be required. We compare the TaqMan assay with some of the alternative nucleic acid-based detection techniques of microarray, chip, and bead technologies in terms of sensitivity, speed, and cost.
One of the most critical first reactions to an outbreak of disease is to identify the pathogen responsible. Nucleic acid-based methods are rapid and sensitive, in contrast to current non-nucleic acid-based identification techniques. For example, culture growth requires days to weeks, and immune detection is not as sensitive and possibly not as specific as nucleic acid-based assays. Fast, sensitive, and accurate pathogen identification facilitates an appropriate response to determine the mode of transmission, the scope of quarantine, and the method of treatment. While we aim to detect both natural outbreaks and bioterrorist releases, our main charter is to rapidly and accurately discover bioterrorist releases. Because we cannot publish results of our analyses of threat organisms, we have illustrated our methods using species of public health concern for which sensitive and accurate assays are also valuable.
TaqMan fluorogenic assays have been shown to rapidly (within 2 h) and successfully identify four viruses that can infect humans, hepatitis viruses B and C (6, 9), Puumala hantavirus (1), and West Nile virus (2), as well as the fungal plant pathogens (12). Not only were these TaqMan assays more rapid than other methods, but they also had sensitivities equal to or greater than that of nested PCR (3).
TaqMan is a fluorogenic probe-based PCR assay in which, situated between two PCR primers, there is an internal oligonucleotide probe with a fluorescent label attached at the 5′ end and a quenching molecule that suppresses the fluorescent reporter at the 3′ end. During DNA replication in the PCR process, the internal oligonucleotide hybridizes to the template and is digested by the 5′-3′ endonuclease activity of the Thermus aquaticus (Taq) DNA polymerase as the PCR primer is extended. The internal oligonucleotide is digested only if DNA replication occurs, separating the fluorescent and quencher molecules. PCR products are detected within minutes by monitoring the increase in fluorescence that occurs exponentially with successive PCR amplification cycles. Thus, TaqMan assays require the design of three nucleic acid probes: two PCR primers and one internal oligonucleotide located between the two primers. One might think that empirical results argue forcefully for the development of TaqMan probes, or signatures, to distinguish any pathogens that pose threats to humans, livestock, or crops. Such signatures would require two things. First, they must be uniquely species specific to preclude false-positive results. Second, they must be species-wide (capable of detecting all known strains of a given species) to ensure positive results for all strains of that species, thus preventing false-negative results. Thus, we define a signature, for the purposes of this paper, as three probes suitable for a TaqMan assay that uniquely identify all strains of a given species. Some argue that species-wide signatures are unnecessary and that only common or currently circulating strains that cause human infection must be detected. However, because our primary focus is bioterrorism, we require species-wide signatures, as it is possible that a terrorist may release an old strain or engineer something into a strain that does not normally cause human infection. While we do have the ability to tailor our signatures to whatever level is required, our focus on defense against bioterrorism demands that we find species-wide signatures in any case.
While the empirical studies mentioned above developed TaqMan signatures for the strains and species tested, they do not demonstrate that the probes are species-wide or species specific compared to the sequences of a wide selection of other organisms. We note that it may also be desirable to develop strain- or serotype-specific signatures for exact identification purposes. Signatures specific to antibiotic resistance, virulence mechanisms, or other features would be useful for the detection of engineered organisms. These topics are not addressed in this paper.
Here, we present results of analyses to determine whether TaqMan signatures that are both species-wide and unique for human immunodeficiency virus (HIV) and hepatitis A, B, C, and E viruses could be developed. The results obtained with these species are presented to illustrate the complexity of the process of extracting high-quality signatures for many widely divergent viruses with high mutation rates (7). Our charter is to develop signatures for a number of viral and bacterial pathogens listed as threats by the Centers for Disease Control and Prevention and other government agencies, but we are prevented from discussing these organisms for obvious reasons of national security.
To briefly preview our results, we found strains of hepatitis A virus to be sufficiently similar that two signatures that were both species-wide and unique could be determined computationally. Thus, TaqMan assays are good candidates for detection of this species, as a single assay could detect all (sequenced) strains. Strains of hepatitis B, C, and E viruses and each of the subtypes of HIV, in contrast, differed sufficiently in that multiple TaqMan signatures would be required to span all clusters of strains in a given species. Considering the cost of TaqMan probes, we conclude that for divergent viruses like HIV, the requirement for multiple signatures to recognize all strains makes TaqMan assays economically unfeasible for large-scale use. However, TaqMan assays could still be feasible in a clinical setting, in which laboratory tests routinely cost $10 to $30.
MATERIALS AND METHODS
First approach: the MSA pipeline.
We used all available sequence data in GenBank for complete genomes of hepatitis A virus (7 complete genomes) and hepatitis E virus (17 complete genomes). An additional four complete genomes of hepatitis E virus listed as swine hepatitis E virus were not included in our analyses. Analyses were done with a collection of 30 HIV strains (HIV-30) that were selected to span a worldwide geographic range of strains. Similarly, a selection of 44 strains of hepatitis B virus and 27 strains of hepatitis C virus were selected. The computational limitations of multiple-sequence alignment (MSA) preclude alignments of significantly larger collections of sequence data. Analyses of HIV were also performed separately with sequence data for HIV subtype A (HIV-A; 10 strains), HIV-B (11 strains), and HIV-C (14 strains). The genomic identity numbers of all HIV and hepatitis virus strains that we examined are listed in Table 1.
TABLE 1.
Genbank sequence identification (genomic identity) numbers used to find species-wide probes
| GenBank genomic identity no. | |||||||
|---|---|---|---|---|---|---|---|
| HIV-30 | HIV-A | HIV-B | HIV-C | Hepatitis A virus | Hepatitis B virus | Hepatitis C virus | Hepatitis E virus |
| 11095910 | 14530236 | 11095910 | 13172878 | 9626732 | 21326584 | 11559442 | 21218079 |
| 1332335 | 14530245 | 1176374 | 1353860 | 19550900 | 22655598 | 22129792 | 21218075 |
| 13569297 | 14530254 | 13569277 | 13569227 | 603025 | 22651877 | 19568932 | 21218071 |
| 14041625 | 14530262 | 1772624 | 13569247 | 329606 | 22530871 | 15529110 | 21218067 |
| 14530236 | 2745742 | 3163929 | 13569257 | 329596 | 22415734 | 15487693 | 9626440 |
| 14530262 | 3808250 | 328658 | 13569277 | 329594 | 22135731 | 9930556 | 21929126 |
| 1772624 | 3808260 | 3694861 | 13569297 | 329582 | 22135726 | 7650265 | 21616617 |
| 2181211 | 4239648 | 4204997 | 13569307 | 22135721 | 464177 | 17974557 | |
| 2570232 | 5733950 | 4205033 | 13569327 | 22135716 | 221614 | 17974553 | |
| 2570307 | 7452908 | 665531 | 2194183 | 22135711 | 5918966 | 15320510 | |
| 3110554 | 8218025 | 3252927 | 22135706 | 5918960 | 12711849 | ||
| 3114562 | 3252966 | 22135701 | 5918930 | 7362938 | |||
| 325654 | 4324737 | 22135696 | 6010587 | 4033730 | |||
| 325762 | 6016887 | 22135691 | 6010581 | 1370097 | |||
| 4007991 | 22135686 | 6010579 | 1437484 | ||||
| 4262336 | 21624234 | 5748510 | 221701 | ||||
| 4324737 | 21624227 | 5532421 | 1209363 | ||||
| 4539033 | 18146697 | 5420376 | |||||
| 463057 | 18146691 | 4753720 | |||||
| 5733950 | 18146685 | 3550764 | |||||
| 6016887 | 21431678 | 3550758 | |||||
| 6090965 | 21388705 | 3098636 | |||||
| 6690750 | 21280301 | 2895898 | |||||
| 7452908 | 21280299 | 1030704 | |||||
| 747644 | 21280285 | 1183032 | |||||
| 8218025 | 21280281 | 329739 | |||||
| 8886632 | 21280275 | 329737 | |||||
| 9628880 | 21280271 | ||||||
| 9629357 | 15425696 | ||||||
| 995584 | 221497 | ||||||
| 12060438 | |||||||
| 21280267 | |||||||
| 21280261 | |||||||
| 21280255 | |||||||
| 21280251 | |||||||
| 21280243 | |||||||
| 19568072 | |||||||
| 19224211 | |||||||
| 18845081 | |||||||
| 18621118 | |||||||
| 18621103 | |||||||
| 18389985 | |||||||
| 11935071 | |||||||
| 16751309 | |||||||
We then generated an MSA for the species using Genomatix software (http://www.genomatix.de/), with hyphens used to distinguish deletions. We developed a program by taking as input an MSA that identifies conserved nucleotide bases at each position across strains, directing the output to a file that we call “gestalt.” We define conserved bases as those that are identical in at least y number of the strains but that may be deleted (indicated by a hyphen) in the remaining strains. Capital letters distinguish strings of 18 or more conserved bases. We based the cutoff of 18 on the minimum acceptable lengths of forward and reverse primers and internal probes for TaqMan assays. We set the parameter y equal to half of the number of strains for which we had sequence data, unless we indicate otherwise.
Sequences suitable for TaqMan assays must satisfy a number of additional specifications; for example, forward and reverse primers and the internal probe must be contained within a stretch of not more than 300 bp (5). Other requirements for the design of primers and probes are detailed at http://www.appliedbiosystems.com/support/techtools/pcropt/. We used the Primer3 program from the Massachusetts Institute of Technology (http://www-genome.wi.mit.edu/genome_software/other/primer3.html) to select our probes and internal oligonucleotides. Considerable effort went into tuning its many parameters to optimize performance for our needs.
We refer to a combination of forward and reverse primers and a probe that satisfy all TaqMan assay specifications as a “candidate signature.” On the basis of these guidelines, we developed code to select all acceptable TaqMan candidate signatures from a given gestalt file. We also searched the reverse complement of the gestalt file for probe sequences, since TaqMan assay restrictions on the G:C ratio might be satisfied by the reverse complement of a sequence but not the sequence itself. Some candidate signatures overlapped; for example, one candidate signature could share primers with another signature, but the probe of one would extend 2 bases longer than that of the other. Candidate signatures in which only the probe differed by virtue of being the forward versus the reverse complement of a sequence were also counted as different candidate signatures.
Once we identified suitable candidate signatures, i.e., those that were species-wide, we used the Vmatch software developed by S. Kurtz (S. Kurtz, personal communication [http://www.techfak.uni-bielefeld.de/∼kurtz/]) to find whether these potential signatures were unique compared to the sequence data available from GenBank. We compiled the GenBank sequence data into two database files, all_virus and all_microbes. The 137-Mb file all_virus contains 549 distinct species or strains, and the 632-Mb file all_microbes includes 194 species or strains of a wide range of species. To date, we have not included fungi in our analyses, although we hope to do so in the future.
The Vmatch software first builds an efficient computer data structure (suffix tree) that represents all possible substrings of the sequence contained in all_virus and all_microbes (the files that represent our present database of a representative wide variety of viral and microbial DNA sequences). Then we eliminated any of these substrings of 18 bases or more from our candidate signature sequences. The value for 18 bases was chosen to be appropriate for the TaqMan assay. For an all_virus and all_microbes database that completely represented all organisms found in nature, the probability of a false-positive result for a candidate signature would be reduced to 0. The advantage of using an analysis with Vmatch software over an analysis with BLAST software is that Vmatch scales well for large amounts of sequence data. The Vmatch software uses a custom virtual memory scheme to avoid the exhaustion of memory for jobs with large amounts of data. Moreover, the Vmatch software outputs data that are easier to parse than the output from the BLAST program to find all unique stretches of sequence. All signatures that our colleagues aim to use in field surveillance are validated empirically in rigorous screening experiments. Since the signatures that we present here will not be used to detect bioterrorist releases, we do not plan to test them empirically.
Modify the pipeline when multiple signatures are required for species-wide identification. (i) Second approach: PHYLIP clustering.
In some cases, the method described above yields no potential signatures. Therefore, we modified the process to determine if we could subdivide the strains of a species, each subset of which could be identified by at least one signature. We aimed to find a minimal set of candidate signatures that would represent all the strains of a target species. That is, all strains must be represented by at least one candidate signature. Some strains may be represented by more than one candidate signature.
Our first approach was to construct phylogenetic trees by using the PHYLIP software package (http://evolution.genetics.washington.edu/phylip.html), based on a number of measures of DNA distance (parsimony, maximum likelihood, fraction of shared bases or shared codons between each pair of strains, etc.). We used these trees to identify clustered subsets of strains. Then we created separate gestalt files of sequences that were conserved within each subset of strains. We hoped to identify a set of two to three signatures, the combination of which would identify all strains of the species under consideration. This phylogenetic clustering approach did not yield acceptable results, so we attempted an alternative solution.
(ii) Third approach: finding all shared candidate signatures, pruning the list, and specifying a minimal set.
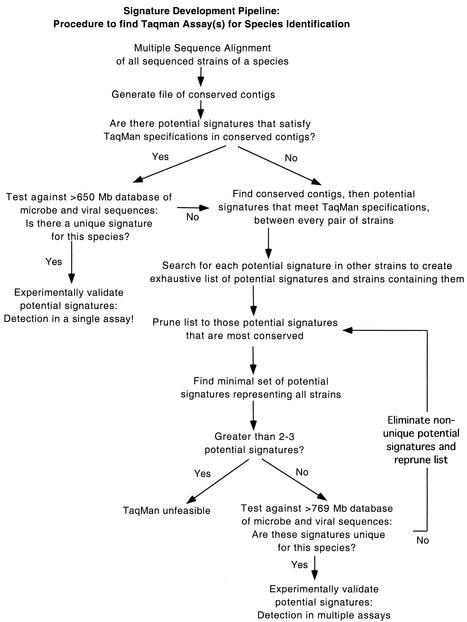
Our third approach is diagrammed in Fig. 1. For this approach, we again used the gestalt files generated for each pair of strains within a species to find candidate signatures shared by that pair. Then we searched for the primer and probe sequences of each candidate signature in all the other strains of that species. This generated an exhaustive list of candidate signatures and the strains containing each signature.
FIG. 1.
Flowchart outlining our computational approach for finding potential signatures from DNA sequence data.
We then created a “pruned list” by keeping the candidate signatures that were present in the largest number of strains (more conserved among strain clusters) and eliminating the candidate signatures that were less conserved in only a subset of strains than another, more conserved candidate signature. For example, if candidate signature 1 is shared by strains A, B, and C and candidate signature 2 is shared only by strains A and B, then we kept candidate signature 1 and removed candidate signature 2 from our list. If the same subset of strains shares two different candidate signatures (e.g., candidate signatures 1 and 2 are both shared only by strains A and B), then we retained only the first to appear in an ascii-sorted list, a selection criterion that does not affect a count of the minimal number of signatures necessary for species identification. Later, it is possible to select a different candidate signature shared by the same strains if the first does not pass the species-specificity test against all_virus and all_microbes by use of the Vmatch software. Since some strains did not share candidate signatures with any other strains, we also added to the pruned list entries representing each of those highly divergent strains. We believe that by using the most conserved candidate signatures of a cluster, we maximize the chances that newly sequenced strains of a rapidly evolving species will also contain these candidate signatures.
Next, we searched for a set of these candidate signatures of minimal size (minimal set) such that at least one candidate signature in this set would be present in every strain of the target species. First, we selected the two or three candidate signatures from the pruned list whose union maximized the strains represented. Then we added to this union another candidate signature that again maximized the strains represented and so on, until a minimal set was identified. Although this algorithm is not guaranteed to provide the smallest possible minimal set, it will in most cases identify something close to it. We verified that this technique did specify minimal sets for HIV-A, HIV-B, and HIV-C by examining all possible combinations of two, then three, and then four, etc., candidate signatures until we identified the smallest possible minimal set. For HIV-30, however, this algorithm indicated that a minimal set contains nine candidate signatures, which would have required an exhaustive search across the computationally unfeasible 669, which is equal to 5.6 × 1010, possible combinations to verify that it was indeed the true minimal set.
Finally, we used the Vmatch software to test whether all candidate signatures in the minimal set were unique to the species in question and yielded potential signatures comprising a minimal species-wide and species-specific set. Signatures for species of national security concern that will be used for detection in the field are rigorously tested by our colleagues in the laboratory (E. A. Vitalis, L. E. Danganan, J. R. Avila, A. Hubbell, L. Ott, T. A. Kuczmarski, L. Radnedge, N. K. Montgomery, C. L. Strout, T. R. Slezak, R. Meyers, and P. M. McCready, Int. Conf. Emerg. Infect. Dis., p. 163. 2002). Here, we present the results for the particular species evaluated as examples to illustrate our computational methods, but we will not use these assays in the field. Therefore, we are not planning to test these potential signatures in the laboratory.
RESULTS
First approach: the MSA pipeline.
The first process described, in which the MSA pipeline was used, yielded two potential signatures for the seven strains of hepatitis A virus but none for any of the strains of hepatitis B, C, or E virus, HIV, or any of the subtypes of HIV.
Second approach: PHYLIP clustering.
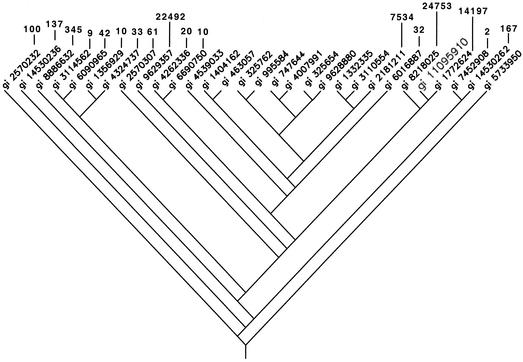
The second approach, in which we attempted to predict clusters based on phylogenetic subgroups (PHYLIP clustering), turned out to be unreliable. Some strains fell within a cluster but shared no candidate signatures satisfying all TaqMan requirements, while other strains were on a branch outside of a cluster yet still shared candidate signatures with members of that cluster (Fig. 2). Consequently, this “searching-by-hand” method was tedious and unfeasible as a means of finding a minimal set of signatures.
FIG. 2.
Phylogenetic tree describing the DNA sequence relationships among the HIV strains used in these analyses and constructed by using the dnapars and drawgram programs in the PHYLIP package. For those strains that share TaqMan assay results with the strain with genomic identity (gi) number 11095910 (enlarged text, fifth from right), the numbers of (potentially overlapping) assays that the pair shares are indicated. The strain with genomic identity number 11095910 shares TaqMan signatures with phylogenetically distant strains and does not necessarily share signatures with many of the more closely related strains, nor do the numbers of signatures shared follow any distinct pattern. Thus, phylogenetic proximity does not directly relate to whether strains share potential TaqMan signatures.
There is no definitive or consistent relationship between DNA distance and whether or not a pair of strains shares a candidate signature. That is, the DNA distance between two pairs of strains cannot be used as an indicator of whether those strains also share an amplicon. Logistic regression of the presence or absence of shared candidate signatures versus various measures of DNA distance verified that no single distance measure could reliably distinguish strain pairs with shared candidate signatures (results not presented). Using logistic regression and inverse prediction (JMP Statistical Package; SAS Institute), we determined thresholds of DNA distance below which shared candidate signatures between different strains were likely. These thresholds differed by an order of magnitude between different species and thus are not useful for the clustering of strains to find a minimum number of potential signatures.
Third approach: finding a minimal set of candidate signatures.
The third approach was successful at identifying a minimal set of signatures for each target species. Table 2 summarizes the total number of candidate signatures shared by at least two strains, the number of candidate signatures in the pruned list, and the number of signatures in the minimal set for detection of each species.
TABLE 2.
Number of candidate assays found and size of minimal set
| Species or group | Total no. of candidate assays | No. of candidate assays in pruned list | Minimal (no. of signatures) of candidate assays |
|---|---|---|---|
| HIV-30 | 2,923 | 66 | 9 |
| HIV-A | 286 | 21 | 3 |
| HIV-B | 1,107 | 7 | 2 |
| HIV-C | 473 | 55 | 2 |
| Hepatitis B virus | 36,671 | 205 | 2 |
| Hepatitis C virus | 14,595 | 26 | 2 |
| Hepatitis E virus | 7,182 | 3 | 3 |
Tables 3 and 4 provide the minimal set of TaqMan probes for the various groups of HIV and hepatitis viruses, with the proviso that they have not been screened empirically.
TABLE 3.
Minimal sets of candidate TaqMan assays for different groups of HIVa
| HIV group | Forward primer | Internal oligonucleotide | Reverse primer | Strain containing signature sequences (genomic identity no.)b |
|---|---|---|---|---|
| HIV-30 | CACTTCCCCTTGGTTCTCTCAT | CCCATTCTGCAGCTTCCTCATTGATGG | GAAGGAGCCACCCCACAAG | 11095910 |
| 14530236 | ||||
| 14530262 | ||||
| 1772624 | ||||
| 2181211 | ||||
| 2570232 | ||||
| 8218025 | ||||
| 9629357 | ||||
| TGCCCCTTCACCTTTCCA | TCTATTATTCTTTCCCCTGCACTGTACCCCC | GACAGCAGTACAAATGGCAGTATTC | 13569297 | |
| 1772624 | ||||
| 2570307 | ||||
| 3114562 | ||||
| 5733950 | ||||
| 6016887 | ||||
| 6090965 | ||||
| 8886632 | ||||
| CCTGGGTGTTCCCTGCTAGA | CGGCTCCACGCTTGCTTGCTTAAA | GGCGGCGACTAGGAGAGAT | 325654 | |
| 325762 | ||||
| 4007991 | ||||
| 747644 | ||||
| 995584 | ||||
| TGTATTACTACTGCCCCTTCACCTTT | TGCTGTCCCTGTAATAAACCCGAAAATTTTGAATT | TGGCAGTATTCATTCACAATTTTAAAA | 14530236 | |
| 14530262 | ||||
| 4262336 | ||||
| 4539033 | ||||
| 6690750 | ||||
| 7452908 | ||||
| TTTGGAGGAATTGTATTTCTTGTTCTG:: | CCCTTCTTTTAAAATTCATGCAATGAACTGCCAT | TAGTAGAAGCAATGAATCACCACCTAA | 325654 | |
| 4007991 | ||||
| 9628880 | ||||
| CAATTATGTTGACAGGTGTAGGTCCTA | TTGATAAAACCTCCAATTCCCCCTATCATTTTTG | TAAATTTGCCAGGAAAATGGAAA | 4324737 | |
| 5733950 | ||||
| 6690750 | ||||
| GGGAGTGGCTAACCCTCAGA | TCTGAGCCCGGGAGCTCCCTG | AAGCAGCGGGTTCAGCTAGA | 14041625 | |
| 463057 | ||||
| TGGAAGGGATGTTTTACAGTGAGA | CAGAACTATACTCATGGGCCAGGAATAAGGTACCC | GAGTCTCAGTGTCCTCCCCTTCT | 3110554 | |
| ATAATACCAAAGGATTCTTCCCAGATT | CTTCAAACTGGTACCAATGGATCCATCAGAGGTA | TTGTTCTCTCCTTTATTGGCTTCCT | 1332335 | |
| HIV-A | GGAGCAGCAGGAAGCACTATG | CAATAACGCTGACGGTACAGGCCAGACAATTAT | TCTTGCCTGGAGCTGTTTAATG | 14530245 |
| 2745742 | ||||
| 3808250 | ||||
| 3808260 | ||||
| 4239648 | ||||
| 5733950 | ||||
| CTGACGGTACAGGCCAGACA | CAGCTCCAGGCAAGAGTCCTGGCT | GTGGTGCAGATGAGTTTTCCA | 14530254 | |
| 2745742 | ||||
| 3808250 | ||||
| 3808260 | ||||
| 7452908 | ||||
| GCAGCTATGCAAATGTTAAAAGATACC | CCACCAGGCCAGATGAGAGAACCAA | ATTTAATCCCAGGATTATCCATCTTTT | 14530236 | |
| 14530245 | ||||
| 14530262 | ||||
| 3808260 | ||||
| 4239648 | ||||
| HIV-B | CACCTAGAACTTTAAATGCATGGGTAA | CCACAAGATTTAAACACCATGCTAAACACAGTGG | GCTATGTCACTTCCCCTTGGTT | 11095910 |
| 13569277 | ||||
| 1772624 | ||||
| 3163929 | ||||
| 328658 | ||||
| 3694861 | ||||
| 4204997 | ||||
| 4205033 | ||||
| 665531 | ||||
| 8218025 | ||||
| CAGGGGCAAATGGTACATCAG | CATTATCAGAAGGAGCCACCCCACAAGATTT | CCCCACTGTGTTTAGCATGGT | 11095910 | |
| 1176374 | ||||
| 3163929 | ||||
| 328658 | ||||
| 3694861 | ||||
| 4204997 | ||||
| 4205033 | ||||
| 665531 | ||||
| HIV-C | CCATTTCTTTGGATGGGGTATG | CAGTACAGCCTATACAGCTGCCAGAAAAGGATAGC | GTAAATCTGACTTGCCCAGTTTAATTT | 1353860 |
| 13569247 | ||||
| 13569257 | ||||
| 2194183 | ||||
| 3252927 | ||||
| 4324737 | ||||
| GCAGGAAGATGGCCAGTCA | CTGAGCACCTTAAGACAGCAGTACAAATGGCA | CCCCCTTTTCTTTTAAAATTGTGAAT | 13172878 | |
| 13569227 | ||||
| 13569247 | ||||
| 13569277 | ||||
| 13569297 | ||||
| 13569307 | ||||
| 13569327 | ||||
| 3252927 | ||||
| 3252966 | ||||
| 6016887 |
Only one possible minimal set is given, although there are other (sometimes many) possible sets with the same total number of assays.
The genomic identity numbers refer to the GenBank sequence identification numbers.
TABLE 4.
Minimal sets of candidate TaqMan assays for four species of hepatitis viruses
| Hepatitis virus | Forward primer | Internal oligonucleotide | Reverse Primer | Strain containing signature sequences (genomic identity no.b) |
|---|---|---|---|---|
| Ac | GGTAGGCTACGGGTGAAACCT | AGACAAAAACCATTCAACGCCGGAGG | CTCAATGCATCCACTGGATGAG | 9626732 |
| 19550900 | ||||
| 603025 | ||||
| 329606 | ||||
| 329596 | ||||
| 329594 | ||||
| 329582 | ||||
| ATAGGGTAACAGCGGCGGATAT | AGACAAAAACCATTCAACGCCGGAGG | CTCAATGCATCCACTGGATGAG | 9626732 | |
| 19550900 | ||||
| 603025 | ||||
| 329606 | ||||
| 329596 | ||||
| 329594 | ||||
| 329582 | ||||
| B | GTCCAGAAGAACCAACAAGAAGATG | TGATAAAACGCCGCAGACACATCCA | TAGACTCGTGGTGGACTTCTCTCA | 11935071 |
| 12060438 | ||||
| 15425696 | ||||
| 16751309 | ||||
| 18146691 | ||||
| 18146697 | ||||
| 18621118 | ||||
| 18845081 | ||||
| 19224211 | ||||
| 19568072 | ||||
| 21280243 | ||||
| 21280251 | ||||
| 21280261 | ||||
| 21280267 | ||||
| 21280275 | ||||
| 21280281 | ||||
| 21280285 | ||||
| 21280299 | ||||
| 21326584 | ||||
| 21388705 | ||||
| 21431678 | ||||
| 21624227 | ||||
| 21624234 | ||||
| 22135686 | ||||
| 22135696 | ||||
| 22135701 | ||||
| 22135706 | ||||
| 22135711 | ||||
| 22135716 | ||||
| 22135721 | ||||
| 22135726 | ||||
| 22135731 | ||||
| 221497 | ||||
| 22415734 | ||||
| 22651877 | ||||
| CTCCCCGTCTGTGCCTTCT | TGTGCACTTCGCTTCACCTCTGCAC | GCCTCAAGGTCGGTCGTT | 12060438 | |
| 18146685 | ||||
| 18389985 | ||||
| 18621103 | ||||
| 18621118 | ||||
| 18845081 | ||||
| 21280243 | ||||
| 21280251 | ||||
| 21280255 | ||||
| 21280261 | ||||
| 21280267 | ||||
| 21280271 | ||||
| 21280275 | ||||
| 21280281 | ||||
| 21280285 | ||||
| 21280299 | ||||
| 21280301 | ||||
| 21326584 | ||||
| 21388705 | ||||
| 21431678 | ||||
| 21624227 | ||||
| 21624234 | ||||
| 22135686 | ||||
| 22135691 | ||||
| 221497 | ||||
| 22530871 | ||||
| 22651877 | ||||
| 22655598 | ||||
| C | TCTGCGGAACCGGTGAGT | ATTTGGGCGTGCCCCCGC | GCACTCGCAAGCACCCTAT | 1030704 |
| 11559442 | ||||
| 1183032 | ||||
| 15487693 | ||||
| 15529110 | ||||
| 19568932 | ||||
| 22129792 | ||||
| 221614 | ||||
| 2895898 | ||||
| 3098636 | ||||
| 329737 | ||||
| 329739 | ||||
| 3550758 | ||||
| 3550764 | ||||
| 464177 | ||||
| 4753720 | ||||
| 5420376 | ||||
| 5532421 | ||||
| 5918930 | ||||
| 5918960 | ||||
| 5918966 | ||||
| 6010579 | ||||
| 6010581 | ||||
| 6010587 | ||||
| 7650265 | ||||
| 9930556 | ||||
| AAAGAAAAACCAAACGTAACACCAA | CGCCCACAGGACGTCAAGTTCCC | ACCGCTCGGAAGTCTTCCT | 1030704 | |
| 15529110 | ||||
| 19568932 | ||||
| 22129792 | ||||
| 3098636 | ||||
| 329737 | ||||
| 329739 | ||||
| 4753720 | ||||
| 5420376 | ||||
| 5532421 | ||||
| 5748510 | ||||
| 5918960 | ||||
| 5918966 | ||||
| 7650265 | ||||
| 9930556 | ||||
| E | CAGGCCGGAAAACAAGCT | CGAACCACCACAGCATTCGCCA | CTCTAGCAGCGGCCAACTC | 21218067 |
| 21218071 | ||||
| 21929126 | ||||
| 7362938 | ||||
| GGAGGCCATGGTCGAGAA | CTCCGCCGTCCTTGAGCTCGA | TGCCCTGGCCCACTTTAC | 1209363 | |
| 12711849 | ||||
| 1370097 | ||||
| 1437484 | ||||
| 17974553 | ||||
| 17974557 | ||||
| 21616617 | ||||
| 221701 | ||||
| 4033730 | ||||
| 9626440 | ||||
| GCGCCCTAGGGCTGTTCT | TATGCTGCCCGCGCCACC | GGGCGAAGGGCTGAGAAT | 15320510 | |
| 21218075 | ||||
| 21218079 |
Only one possible minimal set is given, although there are other (sometimes many) possible sets with the same total number of assays.
The genomic identity numbers refer to GenBank sequence identification numbers.
For hepatitis A virus, each of the signatures is found in all complete hepatitis A virus genomes in GenBank.
DISCUSSION
The MSA pipeline.
TaqMan analyses are rapid and are among the most sensitive methods for determination of whether a given pathogen is present. Moreover, for organisms in which strains have diverged relatively little, a single, inexpensive assay can determine whether that organism is present. The computational search for potential signatures that we have described here minimizes the expensive and time-consuming in vitro work needed to identify signatures. Moreover, it provides greater assurance that these potential signatures are both more species specific and unique than signatures chosen on a purely empirical basis. Because we searched for signatures conserved among multiple existing strains, these signatures lie in regions of the genome likely to be conserved in newly evolving strains as well.
Without this care to select signatures that are conserved among strains, one runs the risk of false-negative results in a detection setting, with obvious, potentially calamitous consequences in a bioterror threat situation. The sequences of the forward primer, reverse primer, and probe used by Weinberger et al. (9) matched the sequences of only 17, 15, and 22 of the 44 strains, respectively, that we used in our analyses of hepatitis B virus. The signature used in the assay for hepatitis C virus published by Morris et al. (6) also failed to be conserved among all 27 strains that we examined: the sequences of only 24, 23, and 22 of the strains matched the forward primer, reverse primer, and probe sequences, respectively.
Thus, our computations illustrate that strains of some species have diverged so much that multiple TaqMan signatures are required to preclude false-negative results in a detection assay. At a cost of approximately $2.30 per reaction mixture for a single-probe reaction or $2.50 per reaction mixture for two-probe reactions (12), a requirement for more than two to three TaqMan analyses with different signatures makes this technique unfeasible for large-scale use. Since a maximum of three assays may be carried out in a single reaction (with potential declines in the sensitivity of a triple-assay reaction compared to that of a single-assay reaction), our analyses suggest that a minimum of three reactions would be necessary to verify the presence of HIV, requiring nine signatures. The total cost for this would be over $7.50. In recent deployments with the signatures that we have developed, tens of thousands of assays were performed. Typically, samples are collected every 4 to 12 h from tens of detectors placed in numerous locations throughout a city. We anticipate widespread use of TaqMan assays in future deployments, perhaps spanning many cities nationally and internationally. Clearly, with hundreds of thousands of assays to be performed, it is essential that costs be kept down, and even with volume pricing, the cost for the detection of a single species is far too high. We are also investigating the use of primers containing degenerate bases, although the drawback of this approach is that degenerate bases decrease the sensitivity and selectivity of the reaction. Nevertheless, our laboratory colleagues have found assays containing degenerate bases to be successful.
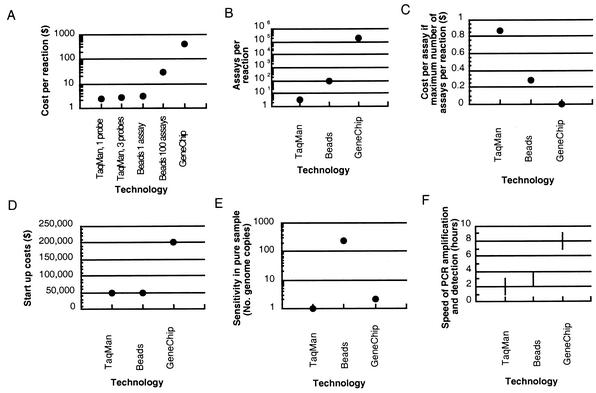
Instead of using a minimal set of multiple TaqMan assays or TaqMan assays with degenerate bases, alternative techniques should be considered (Fig. 3), such as custom DNA GeneChips (Affymetrix, Santa Clara, Calif.) or bead-based assays. GeneChips are a commercial product from Affymetrix that use the hybridization of unknown nucleic acids that have been amplified and labeled with fluorescent dyes to probes immobilized onto a solid surface. Hybrids are detected by the mapping of fluorescent signals on the surface to specific probes. GeneChips use 20- to 25-mers for highly specific hybridization to detect a sequence (4, 10). Although DNA microarrays that use longer probes of 500 to 5,000 bases may work to distinguish groups of closely related species, they may be less appropriate for strain or species identification, since some hybridization to nonidentical sequences occurs. This would be important, for example, if only one of a group of related species is virulent or drug resistant. Bead-based assays are another alternative (11). For example, the LabMAP system is commercially available from Luminex Corp. (Austin, Tex.). Bead-based technologies use uniquely color-coded microspheres onto the surface of which a capture probe with a specific antibody or complementary oligonucleotide is embedded. Binding of an analyte to a probe is detected via a fluorochrome-labeled reagent, which is measured by flow cytometry or microfluidics and lasers. Although other potential techniques for pathogen identification exist (enzyme-linked immunosorbent assays; other antibody tests with, for example, polystyrene optical fibers; and mass spectrometry, to name a few [www.spectrum.ieee.org/WEBONLY/publicfeature/oct01/bio2.html]), we limit our discussion to TaqMan, GeneChips, and bead-based assays.
FIG. 3.
Comparison of the TaqMan assay, bead-based assays, and GeneChips for pathogen detection in terms of costs per reaction (A), numbers of assays per reaction (B), the cost per assay if the maximum number of assays per reaction are performed (the value for GeneChips is $0.002) (C), start-up costs (D), sensitivity in a pure sample (number of genome copies) (E), and speed of detection after the DNA is prepared, that is, the amplification and detection steps (F).
The TaqMan assay is extremely sensitive, commonly detecting 5 to 25 genomes in an environmental sample (2). In a pure sample, one to five copies of DNA can be detected (9; http://www.cbi-biotech.com/hhv.htm). GeneChips are also sensitive, although they are not as sensitive as the TaqMan assay, in detecting down to 10,000 bacterial genomes in an environmental sample and down to 2 bacterial genomes (10 fg DNA) in a pure sample (W. J. Wilson, personal communication). The advantage of GeneChips and bead-based assays over the TaqMan assay, however, is that one may assay for many strains or species in a single reaction: one may perform 100 assays per reaction for beads or more than 200,000 assays per reaction for chips, whereas only 3 assays per reaction can be performed by the TaqMan assay. The ability to assay for multiple 20-mers by using GeneChips has the advantage that each probe is an independent detection assay for a particular species, which is represented by a number of probes on the chip. When the results of these independent assays are pooled and analyzed as a set, the test is extremely specific for the presence of that species. The gene to be detected is basically being sequenced by those multiple hybridization reactions, which are definitively testing for its presence. Consequently, the sensitivity of GeneChips has been shown to surpass that of standard PCR, which is 20 to 200 genomes in a pure sample (1 pg to 100 fg of DNA) (10; Wilson, personal communication).
The ability to perform many assays in a single reaction has the additional advantage that one may test for the presence of multiple species in a single reaction. Thus, GeneChips in particular may be especially appropriate for situations in which one does not have any prior knowledge of the pathogens that may be present, since they make an expansive search possible in a single reaction. Ideally, a single “pathogen chip” can be designed to discriminate among a number of different bacteria, viruses, and fungi. Using software similar to that which we have described, one could find 25-mers that discriminate at the level of the genus, species, or strain. This would enable clinicians to identify not only the species but also where that strain originated on the basis of strain-specific sequences. With chips that can hold up to 400,000 different 20-mers, this is a feasible aim. Such a chip might also contain spots to determine whether various antibiotic resistance genes or virulence genes are present.
At present, the first disadvantage of GeneChips is expense: the start-up costs of purchasing the equipment and making custom masks may surpass $200,000, depending on the institution, although with midi or mini arrays, this price could be reduced. Then, the cost per reaction is another $400 for chips with a large number of assays, again with some variation, depending on the institution and scale. With advances in technology, prices may drop. This compares with a start-up cost for TaqMan analyses of $47,000 for equipment and $2.30 per reaction for a single-probe assay, one-target reaction or $2.50 per reaction for a dual-probe assay, two-target reaction (Applied Biosystems) (12).
The second disadvantage of GeneChips compared to the TaqMan assay is the time required to get an answer. Rapid pathogen identification facilitates efforts to minimize exposure. The analysis time required for PCR amplification and detection is about 7 h for GeneChips but is only 1 to 3 h for the TaqMan assay (W. J. Wilson and K. S. Venkateswaran, personal communication). Bead-based assays can also be completed relatively rapidly (2 to 4 h).
Bead-based assays with DNA, for example, assays with Luminex microspheres, are both relatively rapid and relatively cheap, and in a multiplex reaction one can assay for up to 100 analytes, with only minor increments to the cost per reaction ($3.00 for a single analyte and an additional $0.25/analyte when multiplexing is used [K. S. Venkateswaran, personal communication]). However, this is 30 to 35% more expensive than a TaqMan reaction if one performs only one to three assays. In addition, one can test for protein as well as DNA by bead-based assays, unlike the TaqMan assay. Bead-based assays may be appropriate when one wants to detect whether any of a moderate number of species or toxins are present or when the species under consideration is too divergent for feasible detection by the TaqMan assay with only one or two probes. At present, the sensitivities of microsphere assays are limited by the extent of PCR amplification (Venkateswaran, personal communication). Such assays for HIV have been shown to have a lower sensitivity of 500 RNA molecules, or 250 virions (totaling approximately 10 fg of DNA) (8).
In conclusion, we described computational methods that are both species-wide and species specific for the identification of potential signatures for pathogen detection. These methods minimize the expensive and time-consuming in vitro work required to verify those signatures. Computational methods also distinguish species too divergent for feasible detection by the TaqMan assay, such as HIV and hepatitis E virus. We also discussed the relative merits of GeneChips and bead-based assays as alternative methods of pathogen detection.
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy by the Lawrence Livermore National Laboratory, University of California, under contract W-7405-Eng-48.
Information provided by T. Z. DeSantis, K. S. Venkateswaran, and W. J. Wilson was very helpful. We are grateful to S. Kurtz for generously providing us with software. Many thanks to W. J. Wilson and T. Z. DeSantis for comments on drafts of the manuscript.
REFERENCES
- 1.Garin, D., C. Peyrefitte, J. M. Crance, A. Le Faou, A. Jouan, and M. Bouloy. 2001. Highly sensitive TaqMan® PCR detection of Puumala hantavirus. Microbes Infect. 3:739-745. [DOI] [PubMed] [Google Scholar]
- 2.Hadfield, T. L., M. Turell, M. P. Dempsey, J. David, and E. J. Park. 2001. Detection of West Nile virus in mosquitoes by RT-PCR. Mol. Cell. Probes 15:147-150. [DOI] [PubMed] [Google Scholar]
- 3.Linssen, B., R. M. Kinney, P. Aguilar, K. L. Russell, D. M. Watts, O. R. Kaaden, and M. Pfeffer. 2000. Development of reverse transcription-PCR assays specific for detection of equine encephalitis viruses. J. Clin. Microbiol. 38:1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipshutz, R. J., S. P. A. Fodor, T. R. Gingeras, and D. J. Lockhart. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21:20-24. [DOI] [PubMed] [Google Scholar]
- 5.Livak, K., J. Marmaro, and S. Flood. 1995. Guidelines for designing TaqMan fluorogenic probes for 5′ nuclease assays. Perkin-Elmer, Boston, Mass.
- 6.Morris, T., B. Robertson, and M. Gallagher. 1996. Rapid reverse transcription-PCR detection of hepatitis C virus RNA in serum by using the TaqMan fluorogenic detection system J. Clin. Microbiol. 34:2933-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, D. B., J. McAllister, C. Casino, and P. Simmonds. 1997. Virus ‘quasi-species': making a mountain out of a molehill? J. Gen. Virol. 78:1511-1519. [DOI] [PubMed] [Google Scholar]
- 8.Wedemeyer, N., and T. Potter. 2001. Flow cytometry: an ′old' tool for novel applications in medical genetics. Clin. Genet. 60:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Weinberger, K. M., E. Wiedenmann, S. Bohm, and W. Jilg. 2000. Sensitive and accurate quantitation of hepatitis B virus DNA using a kinetic fluorescence detection system (TaqMan PCR). J. Virol. Methods 85:75-82. [DOI] [PubMed] [Google Scholar]
- 10.Wilson, W. J., C. L. Strout, T. Z. DeSantis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed]
- 11.Ye, F., M. Li, J. D. Taylor, Q. Nguyen, H. M. Colton, W. M. Casey, M. Wagner, M. P. Weiner, and J. Chen. 2001. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum. Mutat. 17:305-316. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, A. W., G. L. Hartman, B. Curio-Penny, W. L. Pedersen, and K. B. Becker. 1999. Molecular detection of Diaporthe phaseolorum and Phomopsis longicolla from soybean seeds. Phytopathology 89:796-804. [DOI] [PubMed] [Google Scholar]