Abstract
Pulsed-field gel electrophoresis (PFGE) was performed on 122 neonatal bloodstream isolates of group B streptococci (GBS) to further examine the relationship between macrolide resistance and serotype V GBS (GBS-V). Over one-third (35%) of macrolide-resistant GBS belonged to a single PFGE subtype of GBS-V, which was also the most common GBS-V subtype noted in previous Centers for Disease Control and Prevention surveillance studies. Erm methylase (ermA and ermB) was the most common resistance mechanism detected, present in 12 of 20 macrolide-resistant GBS.
Group B streptococci (GBS) cause significant morbidity and mortality in neonates and can also cause serious infections in adults (20, 21). Several serotypes of GBS exist (9), but only a few (serotypes Ia, Ib, Ic, II, and III) are common causes of human disease. Over the past decade, several investigators reported an increasing proportion of GBS disease to be due to serotype V (GBS-V) (1, 2, 8, 11, 21). During the same period, several investigators also reported an increase in macrolide (erythromycin) and clindamycin resistance among GBS isolates from U.S. hospitals (1, 13, 15, 17). In addition, several groups (including ours) have recently reported an association between macrolide resistance and GBS-V (1, 10, 12). It was found that, though GBS-V accounted for only ∼10% of neonatal GBS bloodstream isolates, 40% of erythromycin-resistant and 83% of clindamycin-resistant strains were GBS-V (1).
To further investigate the association between macrolide-lincosamide resistance and the emergence of GBS-V, we performed pulsed-field gel electrophoresis (PFGE) analysis of the SmaI chromosomal DNA digests of 122 neonatal bloodstream isolates of GBS for which antimicrobial susceptibilities and serotype distribution had previously been described (1). To determine if macrolide-resistant GBS-V represented a new subtype, we included a strain previously characterized by Elliott and colleagues (6) at the Centers for Disease Control and Prevention (CDC) and found to be the most common GBS-V PFGE pattern detected in the United States (representing 56% of isolates collected between 1986 and 1996).
The GBS bloodstream isolates in this study were collected as part of the SENTRY antimicrobial surveillance program (1). This report focuses on all neonatal bloodstream isolates of GBS obtained between January 1997 and December 1999 from 35 medical centers throughout the Western Hemisphere.
Upon receipt at the University of Iowa, GBS isolates were subcultured onto blood agar to ensure viability and purity. Confirmation of species identification was performed by conventional methods: all GBS in our study were confirmed by the Christie-Atkins-Munch-Petersen test. Antimicrobial susceptibility testing was performed by reference broth microdilution methods as described by the National Committee for Clinical Laboratory Standards (NCCLS) (14).
Serotyping was performed by an agglutination test with rabbit antisera to serotypes Ia, Ib, II, III, IV, and V, as described previously (1). For PFGE analysis, genomic DNA was prepared for restriction fragment analysis and then digested with SmaI by published techniques (16). Electrophoresis was performed on the CHEF-DR II apparatus (Bio-Rad Laboratories, Richmond, Calif.). Visual and computer-assisted (BioNumerics; Applied Maths, Kortrijk, Belgium) analysis of PFGE patterns was performed. Isolates were considered to be related (subtypes of the same PFGE type) if they varied by one to three bands and indistinguishable (same subtype) if they had all bands in common (19).
Erythromycin- or clindamycin-resistant isolates were also screened by PCR for the presence of ermA, ermB, ermC, and mefA resistance genes as previously described (18).
The antimicrobial susceptibility results and serotype distribution for these isolates have been described previously (1). In summary, all isolates were susceptible to penicillin, but 20 of the 122 (16%) were resistant to erythromycin, and seven of these (6%) were also nonsusceptible to clindamycin. Of the 71 isolates from the United States, 18 (25.4%) were resistant to erythromycin and 5 (7%) were resistant to clindamycin. Serotypes III and Ia were most frequent, accounting for over 75% of the isolates. Serotype V (9%) was the next most frequent. The rank order of serotype frequency was similar in all regions, although serotype V was more prevalent in the United States (11%) than in Latin America (5%). Three of the isolates were not typeable. There was no significant change in serotype frequency over the 3 years of the study (1).
The distribution of macrolide-lincosamide resistance genes according to erythromycin-clindamycin resistance phenotypes is reported in Table 1. Consistent with previous reports, methylase was the most common mechanism detected among macrolide-resistant isolates (3-5, 7). For all seven ermB-containing strains, the MIC of erythromycin was ≥8 μg/ml, and all were clindamycin intermediate or resistant. Strains containing either ermA or mefA possessed the same phenotype of clindamycin susceptibility with MICs of erythromycin being 1 to 4 μg/ml. One isolate could not be amplified with multiple erm and mef primers. The MIC of erythromycin was higher for this isolate, but the strain was clindamycin susceptible and may represent a ribosomal mutation, although the ribosomal DNA has not yet been examined.
TABLE 1.
Distribution of macrolide-lincosamide resistance genotypes according to erythromycin and clindamycin resistance phenotypes
| Resistance gene | Erythromycin resistant-clindamycin susceptiblea | Erythromycin resistant clindamycin resistantb | Total n (%)c |
|---|---|---|---|
| ermA | 5 | 0 | 5 (25) |
| ermB | 0 | 7 | 7 (35) |
| ermC | 0 | 0 | 0 (0) |
| mef | 7 | 0 | 7 (35) |
| None | 1 | 0 | 1 (5) |
Susceptibility according to NCCLS breakpoints (14).
Includes one isolate that is clindamycin intermediate (MIC = 0.5 μg/ml).
Percentage of total of 20 macrolide-resistant isolates. No isolate possessed more than one resistance gene.
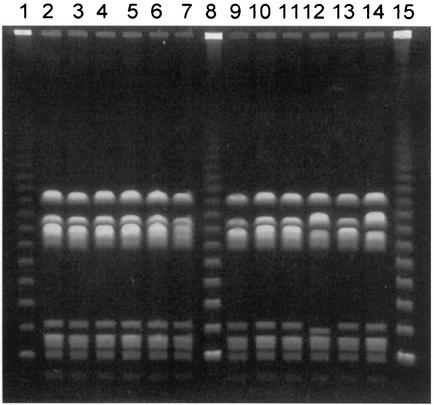
Over 50 PFGE patterns were identified among the 122 GBS isolates. Without exception, isolates within a serotype did not share PFGE patterns with isolates from other serotypes. Serotype Ia isolates demonstrated the most genetic heterogeneity, and serotype V strains demonstrated the least. Macrolide-resistant strains were found among serotypes Ia (two isolates), Ib (one), II (one), III (seven), and V (eight) and also included one nontypeable isolate. Overall, 10 different PFGE patterns were observed among the 20 macrolide-resistant GBS isolates. Figure 1 and Table 2 demonstrate the PFGE patterns of all GBS-V isolates, as well as macrolide susceptibility patterns and geographic locations of isolation. Seven of the eight macrolide-resistant GBS-V strains, isolated at seven centers from New Mexico to New York, were indistinguishable from the CDC GBS-V control strain (Fig. 1, lane 2) by PFGE. The remaining macrolide-resistant GBS-V strain, from a center in Canada, was of a related subtype.
FIG. 1.
PFGE patterns of neonatal bloodstream isolates of GBS-V. Lanes 1, 8, and 15, lambda ladder control; lanes 2 to 7 and 9 to 14, patterns of 12 different isolates (see Table 2 for descriptions of individual isolates). Lane 2 shows the pattern of a strain provided by the CDC and found to be the most common pattern of GBS-V in surveillance studies performed in the United States between 1986 and 1996 (6).
TABLE 2.
Neonatal bloodstream isolates of GBS-V for which PFGE patterns were determined
| Strain no.a | Yr isolated/location | Antibiotic susceptibility patternb | Genotype of isolate | PFGE type |
|---|---|---|---|---|
| 2 | 1995/Georgiac | S/S | A1 | |
| 3 | 1998/Illinois | R/R | ermB | A1 |
| 4 | 1997/Iowa | R/S | ermA | A1 |
| 5 | 1999/Kentucky | R/S | None | A1 |
| 6 | 1998/Indiana | R/R | ermB | A1 |
| 7 | 1998/New York | R/S | ermA | A1 |
| 9 | 1998/Brazil | S/S | A1 | |
| 10 | 1999/Texas | R/R | ermB | A1 |
| 11 | 1999/New Mexico | R/R | ermB | A1 |
| 12 | 1997/California | S/S | A2 | |
| 13 | 1999/Brazil | S/S | A1 | |
| 14 | 1997/Canada | R/R | ermB | A3 |
Strain numbers correspond to lane numbers of Fig. 1, where PFGE patterns for these isolates are shown.
Data are patterns of susceptibility to erythromycin/patterns of susceptibility to clindamycin. Abbreviations: S, susceptible; R, resistant.
Strain provided by the CDC, found to be the most common pattern of GBS-V in U.S. surveillance studies performed between 1986 and 1996 (6).
Our results indicate not only that much of the macrolide resistance among GBS was found among serotype V strains but that this resistance phenotype was geographically widespread and found in a GBS-V subtype that was the most commonly isolated in the United States and has been present in the population since 1975 (6). Although some diversity of PFGE patterns was found among GBS-V strains collected by the CDC from 1986 to 1996 (17 PFGE patterns among 45 isolates) (6), our findings are consistent with those of Blumberg and colleagues (2), who found little diversity within GBS-V strains in a 1992-1993 surveillance study in Atlanta, Ga. It would be of interest to examine the antimicrobial susceptibilities of GBS-V isolates collected over the past 2 to 3 decades to determine when macrolide resistance emerged in this GBS-V subtype. Given the fact that macrolides (especially erythromycin) are often used during pregnancy (because of their favorable safety record), we speculate that the acquisition of macrolide resistance by this common GBS-V subtype provided a selective advantage and may have contributed to the emergence of GBS-V as a more frequent human pathogen. Continued emergence of this subtype or other macrolide-resistant GBS would have obvious serious implications both for the formulation of treatment and prophylaxis recommendations and for the development of candidate GBS vaccines (9).
Acknowledgments
We thank J. Elliott at the CDC for kindly providing the GBS type O strain for use in this study. We also thank Stacy Coffman and Rick Hollis for their laboratory expertise and Gary Doern for his helpful suggestions.
The SENTRY study was funded in part by an educational and research grant from Bristol-Myers Squibb Company.
REFERENCES
- 1.Andrews, J. I., D. J. Diekema, S. K. Hunter, P. R. Rhomberg, M. A. Pfaller, R. N. Jones, and G. V. Doern. 2000. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the Western Hemisphere. Am. J. Obstet. Gynecol. 183:859-862. [DOI] [PubMed] [Google Scholar]
- 2.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, et al. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 176:365-373. [DOI] [PubMed] [Google Scholar]
- 3.Culebras, E., I. Rodriguez-Avial, C. Betriu, M. Redondo, and J. Picazo. 2002. Macrolide and tetracycline resistance and molecular relationships of clinical strains of Streptococcus agalactiae. Antimicrob. Agents Chemother. 46:1574-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Azavedo, J., M. McGavin, C. Duncan, D. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Mouy, D., J. Cavallo, R. Leclercq, R. Fabre, and the AFORCOPI-BIO Network. 2001. Antimicrobial susceptibility and mechanisms of erythromycin resistance in clinical isolates of Streptococcus agalactiae: French multicenter study. Antimicrob. Agents Chemother. 45:2400-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitoussi, F., C. Loukil, I. Gros, O. Clermont, P. Mariani, S. Bonacorsi, I. Le Thomas, D. Deforche, and E. Bingen. 2001. Mechanisms of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob. Agents Chemother. 45:1889-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison, L. H., D. M. Dwyer, and J. A. Johnson. 1995. Emergence of serotype V group B streptococcal infection among infants and adults. J. Infect. Dis. 171:513. [DOI] [PubMed] [Google Scholar]
- 9.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, et al. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 10.Lin, F. C., P. H. Azami, L. E. Weisman, J. B. Philips, J. Regan, P. Clark, G. G. Rhoads, J. Clemens, J. Troendle, E. Pratt, R. A. Brenner, and V. Gill. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31:76-79. [DOI] [PubMed] [Google Scholar]
- 11.Lin, F. C., J. D. Clements, P. H. Azimi, J. A. Regan, L. E. Weisman, J. B. Philips, et al. 1998. Capsular polysaccharide types of group B streptococcal isolates from neonates with early-onset systemic infection. J. Infect. Dis. 177:790-792. [DOI] [PubMed] [Google Scholar]
- 12.Manning, S. D., B. Foxman, C. L. Pierson, P. Tallman, C. J. Baker, and M. D. Pearlman. 2003. Correlates of antibiotic resistant group B streptococcus isolated from pregnant women. Obstet. Gynecol. 101:74-79. [DOI] [PubMed] [Google Scholar]
- 13.Morales, W. J., S. S. Dickey, P. Bornick, and D. V. Lim. 1999. Change in antibiotic resistance of group B streptococcus: impact on intrapartum management. Am. J. Obstet. Gynecol. 181:310-314. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement, M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Pearlman, M. D., C. L. Pierson, and R. G. Faix. 1998. Frequent resistance of clinical group B streptococci isolates to clindamycin and erythromycin. Obstet. Gynecol. 92:258-261. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., R. J. Hollis, and H. S. Sader. 1994. PFGE analysis of chromosomal restriction fragments, p. 10.5.c.1-10.5.c.12. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 2, suppl. 1. American Society for Microbiology, Washington, D.C.
- 17.Rouse, D. J., W. W. Andrews, F. C. Lin, C. W. Mott, J. C. Ware, and J. B. Philips. 1998. Antibiotic susceptibility of group B streptococcus acquired vertically. Obstet. Gynecol. 92:931-934. [DOI] [PubMed] [Google Scholar]
- 18.Shortridge, V. D., R. K. Flamm, N. Ramer, J. Beyer, and S. K. Tanaka. 1996. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 26:73-78. [DOI] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wessels, M. R., and D. L. Kasper. 1993. The changing spectrum of group B streptococcal disease. N. Engl. J. Med. 328:1843-1844. [DOI] [PubMed] [Google Scholar]
- 21.Zaleznik, D. F., M. A. Rench, S. Hillier, M. A. Krohn, R. Platt, M. T. Lee, et al. 1999. Invasive disease due to group B streptococcus in pregnant women and neonates from diverse population groups. Clin. Infect. Dis. 30:276-281. [DOI] [PubMed] [Google Scholar]