Abstract
The G and P genotypes of rotavirus stool isolates from 100 children were determined by reverse transcription-PCR and nucleotide sequencing. G1P[4] was the most prevalent genotype(41%), followed by G1P[8] (16%) and G4P[4] (14%). The G genoypes detected were G1 (73%), G4 (17.4%), G9 (6.3%), and G2 (2.8%). The P genotypes were P[4] (71%) and P[8] (29%). Coinfection with more than one G genotype occurred in 12 patients, and coinfection with more than one P genotype occurred in 11 patients.
Rotavirus (RV) is a major cause of gastroenteritis among young children worldwide and is associated with significant morbidity and mortality, particularly in developing countries (3). RV has an 11-segment double-stranded RNA genome and a triple-layer protein capsid. The outer layer of group A RV is composed of two proteins: VP4, which determines the P serotype; and VP7, which determines the G serotype (4). Reverse transcription-PCR (RT-PCR) and/or DNA sequencing has been used to define G and P types (5, 7). P specificity is more conservative than G specificity, with P[8] the most common type worldwide, followed by P[4] and P[6]. P[8] is most frequently associated with serotypes G1, G3, and G4, while P[4] is most frequently associated with G2 (6, 11).
Because antibodies to G protein constitute the major neutralizing antibodies, RV vaccines contained G1, G2, G3, and G4, the most common serotypes worldwide. The first oral RV vaccine (Rotashield) was clinically effective in five global trials (12, 15, 18, 20). However, it was withdrawn because of possible causative relationship to intussusception in infants (14). A new oral pentavalent human-bovine reassortant vaccine that contains G1 to G4 and a minor envelope serotype, P1, is presently being tested.
Recently, unusual serotypes, not included in RV vaccines, have been identified worldwide, such as G5, G8, and G9 (2, 10, 13, 17, 21). Recent studies worldwide, including the United States, suggest that G9 may be emerging as a new common serotype (16). In addition, reassortants of unusual VP7-VP4 gene combinations continue to emerge (9).
This study was undertaken to characterize the strains of RV that caused infection in children presenting to Children's Hospital of Michigan, Detroit, during the peak RV-associated gastroenteritis season of 2001(February and March) and to compare these strains to those used in the new pentavalent vaccine.
(This work was presented in part at the 39th Annual Meeting of the Infectious Diseases Society of America,, San Francisco, Calif., 25 to 28 October 2001 [abstr. 423]).
Fecal samples positive for RV by enzyme-linked immunosorbent assay (Rotazyme) were collected from 100 infants and children 2 days to 7 years old (mean age, 12.7 months; median age, 10 months). Fifty-six were boys. These children lived in 54 different zip codes within the tricounty metropolitan Detroit area and included African-American (56%), Caucasian (25%), Hispanic (4%), and other (15%) ethnicities. Rotavirus RNA was extracted from fecal samples according to the protocol for Trizol reagent (BRL). cDNA was synthesized with Ready-to-Go (Pharmacia Biotech) primed with random DNA hexamers (BRL).
Both P and G typing reactions were carried out as a two-step PCR as previously described (5, 7). A subset of DNA fragments was chosen to confirm the initial G and P typing. Presumptive PCR products were confirmed by direct nucleotide sequence analysis (ABI 3700 DNA analyzer). Phylogenetic analysis of the nucleotide sequences was confirmed by Basic Local Alignment Search Tool (BLAST) analysis at the National Center for Biotechnology Information.
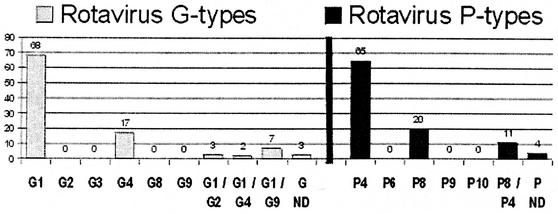
The most prevalent G-P combination was G1P[4] (41%), followed by G1P[8] (16%) and G4P[4] (14%). The G1P[4] genotype accounted for 30 to 50% of the isolates in each week of the study period. All but seven study samples could be assigned both a G genotype and a P genotype. Four could only be assigned a G genotype, and three could only be assigned a P genotype. Twelve patients had coinfections with more than one G genotype, including seven with G1/G9, three with G1/G2, and two with G1/G4. All mixed infections included the G1 genotype. The frequencies of G genotypes were as follows: G1, 73%; G4, 17.4%; G9, 6.3%; and G2, 2.8%. Of the seven G9 strains, three were associated with P[4], three were associated with P[8], and one was associated with both P[4] and P[8]. All three G2 genotypes were associated with the P[4] genotype. The G-P genotype distribution is shown in Table 1.
TABLE 1.
Frequency of RV G and P genotypes isolated from 100 stool samples
| Genotypea | % of Stool samples with genotype |
|---|---|
| G1/P4 | 41 |
| G1/P8 | 16 |
| G4/P4 | 14 |
| G1/P4/P8 | 7 |
| G9/G1/P8 | 3 |
| G9/G1/P4 | 3 |
| G1/G2/P4 | 3 |
| G4/P4/P8 | 2 |
| G1/G4/P4 | 1 |
| G1/G4/P4/P8 | 1 |
| G9/G1/P4/P8 | 1 |
| G4/P8 | 1 |
| G (ND)/P4 | 3 |
| G1/P (ND) | 4 |
| Total | 100 |
ND, not determined.
The VP4 genotypes detected were only P[4] (71%) and P[8] (29%). Dual P[4] and P[8] genotypes were detected in 11 patients. The distribution of G and P genotypes is shown in Fig. 1. In six samples, a signal for a common VP4 gene was detected, but no product was obtained in a second round of PCR to identify the specific P genotype. All of these samples were confirmed by direct DNA sequencing and BLAST analysis to have the P[8] genotype. In all of these P[8] isolates, similar nucleotide sequences were found in the nucleotide region 339 to 356 (P[8] primer binding site). However, mismatches were found between the P[8]-specific primer (1T-1) and its complementary region of the VP4 cDNA. These mismatches were found at four positions corresponding to nucleotides 342 (G:T), 345 (A:C), 347 (A:C), and 350 (T:C). This would explain the failure to detect the VP4 P[8] genotype by RT-PCR in this study.
FIG. 1.
Distribution of RV G and P genotypes isolated from 100 stool samples. ND, not determined.
Representative PCR products of G and P types including P[4] were verified by nucleotide sequencing. BLAST analysis of all of our genotypes showed 94 to 98% homology with reference strains (data not shown).
Of all RV isolates recovered from the 100 children, G1 was the predominant genotype, followed by G4, G9, and G2. The G3 genotype, one of the most common worldwide, was not detected in any stool sample. P[4] was the most prevalent P genotype. G1P[4], which is reported in the United States and other parts of the world at low frequencies (2, 8, 9, 16), was the most prevalent genotype in our patients (41%).
Recent Centers for Disease Control and Prevention (CDC) surveillance studies showed that the most frequent RV G types in the United States were G1 and G2, followed by G9 (8). G3 and G4 were detected less frequently and sporadically. Detroit was not included in these CDC studies. Our data show that G1 and G4 were the most frequent causative strains (73.4 and 17.4%, respectively). G9 accounted for 6.3%. This was higher than the most recent national average rate of 3%, but was not significantly different (P = 0.11) (8). The most common RV strains that have been globally associated with gastroenteritis are G1P[8] (53%), G3P[8] (5.4%), G4P[8] (14.3%), and G2P[4] (10.7%) (6, 11). These strains are significantly underrepresented in our area at rates of 16, 0, 1, and 3%, respectively (P < 0.05 for all). The G1P[4] genotype has never been reported as the most frequent circulating RV genotype in any other part of the world. The wide geographic area in which our patients lived and the detection of G1P[4] throughout the study period indicate that G1P[4] cases were not the result of horizontal transmission from the same index case. Considering the ethnic diversity in Detroit, it is also possible that G1P[4] reassortment may have emerged through the introduction of G2P[4] strains from other parts of the world.
In general, G9 strains have been associated with P[6], P[8], P[11], or P[19] (1, 8, 19, 21). In our patients, G9 serotypes were associated with P[4] or a combination of P[4] and P[8]. No G9-only serotypes were detected in our patients; all G9 serotypes were identified in combination with G1 serotypes. This indicates that G9 genotype may undergo constant reassortment in nature. Our results support the need for continuous surveillance of rotavirus G and P types. The increased prevalence of G9 in our community raises the possibility that G9 may represent an emerging strain that could escape vaccine-induced immunity and become more prevalent. The efficacy of the new pentavalent vaccine against reassorted virus variants involving G9 and one of the vaccine G types is still to be determined.
Acknowledgments
This work was supported by a grant from Children's Research Center of Michigan.
We thank Deborah Ryan and the nursing staff of 6 East/Children's Hospital of Michigan for their assistance in patient recruitment.
REFERENCES
- 1.Araújo, I. T., M. S. R. Ferreira, A. M. Fialho, R. M. Assis, C. M. Cruz, M. Rocha, and J. P. Leite. 2001. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 39:1999-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armah, G. E., C. T. Pager, R. H. Asmah, F. R. Anto, A. R. Oduro, F. Binka, and D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 3.Bern, C., J. Martines, I. de Zoysa, and R. I. Glass. 1992. The magnitude of the global problem of diarrhoeal disease: a ten-year update. Bull. W. H. O. 70:705-714. [PMC free article] [PubMed] [Google Scholar]
- 4.Estes, M. 1996. Rotaviruses and their replication, p. 1625-1655. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincot-Raven, Philadelphia, Pa.
- 5.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 7.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z.-Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin, D. D., C. D. Kirkwood, U. D. Parashar, P. A. Woods, J. S. Bresee, R. I. Glass, and J. R. Gentsch. 2000. Surveillance of rotavirus strains in the United States: identification of unusual strains. J. Clin. Microbiol. 38:2784-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iturriza-Gómara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iturriza-Gomara, M., D. Cubitt, D. Steele, J. Green, D. Brown, G. Kang, U. Desselberger, and J. Gray. 2000. Characterisation of rotavirus G9 strains isolated in the UK between 1995 and 1998. J. Med. Virol. 61:510-517. [DOI] [PubMed] [Google Scholar]
- 11.Koshimura, Y., T. Nakagomi, and O. Nakagomi. 2000. The relative frequencies of G serotypes of rotaviruses recovered from hospitalized children with diarrhea: a 10-year survey (1987-1996) in Japan with a review of globally collected data. Microbiol. Immunol. 44:499-510. [DOI] [PubMed] [Google Scholar]
- 12.Lanata, C. F., K. Midthun, R. E. Black, B. Butron, A. Huapaya, M. E. Penny, G. Ventura, A. Gil, M. Jett-Goheen, and B. L. Davidson. 1996. Safety, immunogenicity, and protective efficacy of one and three doses of the tetravalent rhesus rotavirus vaccine in infants in Lima, Peru. J. Infect. Dis. 174:268-275. [DOI] [PubMed] [Google Scholar]
- 13.Leite, J. P., A. A. Alfieri, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch. Virol. 141:2365-2374. [DOI] [PubMed] [Google Scholar]
- 14.Morbidity and Mortality Weekly Report. 1999. Withdrawal of rotavirus vaccine recommendation. Morb. Mortal. Wkly. Rep. 48:1007. [PubMed] [Google Scholar]
- 15.Perez-Schael, I., M. J. Guntinas, M. Perez, V. Pagone, A. M. Rojas, R. Gonzalez, W. Cunto, Y. Hoshino, and A. Z. Kapikian. 1997. Efficacy of the rhesus rotavirus-based quadrivalent vaccine in infants and young children in Venezuela. N. Engl. J. Med. 337:1181-1187. [DOI] [PubMed] [Google Scholar]
- 16.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. A. Woods, J. L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresee, and R. I. Glass. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran, M., C. D. Kirkwood, L. Unicomb, N. A. Cunliffe, R. L. Ward, M. K. Bhan, H. F. Clark, R. I. Glass, and J. R. Gentsch. 2000. Molecular characterization of serotype G9 rotavirus strains from a global collection. Virology 278:436-444. [DOI] [PubMed] [Google Scholar]
- 18.Rennels, M. B., R. I. Glass, P. H. Dennehy, D. I. Bernstein, M. E. Pichichero, E. T. Zito, M. E. Mack, B. L. Davidson, and A. Z. Kapikian. 1996. Safety and efficacy of high-dose rhesus-human reassortant rotavirus vaccines—report of the National Multicenter Trial. Pediatrics 97:7-13. [PubMed] [Google Scholar]
- 19.Santos, N., E. M. Volotão, C. C. Soares, M. C. M. Albuquerque, F. M. da Silva, T. R. B. de Carvalho, C. F. A. Pereira, V. Chizhikov, and Y. Hoshino. 2001. Rotavirus strains bearing genotype G9 or P[9] recovered from Brazilian children with diarrhea from 1997 to 1999. J. Clin. Microbiol. 39:1157-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santosham, M., L. H. Moulton, R. Reid, J. Croll, R. Weatherholt, R. Ward, J. Forro, E. Zito, M. Mack, G. Brenneman, and B. L. Davidson. 1997. Efficacy and safety of high-dose rhesus-human reassortant rotavirus vaccine in Native American populations. J. Pediatr. 131:632-638. [DOI] [PubMed] [Google Scholar]
- 21.Widdowson, M. A., G. J. van Doornum, W. H. van der Poel, A. S. de Boer, U. Mahdi, and V. Koopmans. 2000. Emerging group-A rotavirus and a nosocomial outbreak of diarrhoea. Lancet 356:1161-1162. [DOI] [PubMed] [Google Scholar]